Abstract
Rhodothermus marinus is a halophilic extreme thermophile, with potential as a model organism for studies of the structural basis of antibiotic resistance. In order to facilitate genetic studies of this organism, we have surveyed the antibiotic sensitivity spectrum of R. marinus and identified spontaneous antibiotic-resistant mutants. R. marinus is naturally insensitive to aminoglycosides, aminocylitols and tuberactinomycins that target the 30S ribosomal subunit, but is sensitive to all 50S ribosomal subunit-targeting antibiotics examined, including macrolides, lincosamides, streptogramin B, chloramphenicol, and thiostrepton. It is also sensitive to kirromycin and fusidic acid, which target protein synthesis factors. It is sensitive to rifampicin (RNA polymerase inhibitor) and to the fluoroquinolones ofloxacin and ciprofloxacin (DNA gyrase inhibitors), but insensitive to nalidixic acid. Drug-resistant mutants were identified using rifampicin, thiostrepton, erythromycin, spiramycin, tylosin, lincomycin, and chloramphenicol. The majority of these were found to have mutations that are similar or identical to those previously found in other species, while several novel mutations were identified. This study provides potential selectable markers for genetic manipulations and demonstrates the feasibility of using R. marinus as a model system for studies of ribosome and RNA polymerase structure, function, and evolution.
1. Introduction
Extremophilic organisms are important model systems for investigating macromolecular structure, function and evolution. Macromolecular complexes such as the ribosome are important antibiotic targets and their structural studies have significantly advanced our understanding of antibiotic modes of action and mechanisms of antibiotic resistance [1,2,3]. Ribosomes from thermophiles have historically been attractive targets for structural studies due to their greater conformational homogeneity. Such investigations can potentially reveal the basis for adaptation to extreme environments, especially when coupled to genetic approaches. While the most thoroughly examined thermophilic organism is the bacterium Thermus thermophilus, studies of other, phylogenetically distant thermophiles could potentially facilitate a comparative approach. This is especially relevant to the extent that species-specific idiosyncrasies, such as differences in DNA repair patterns or codon usage bias, can influence the spectrum of mutants arising. Such idiosyncrasies provide a compelling motivation to explore novel model systems.
Rhodothermus marinus R-10T is a Gram-negative, non-motile, non-spore-forming, thermophilic and halophilic bacterium isolated from a submarine hot spring off the coast of Iceland [4]. It grows optimally at a temperature of 65 °C and a salinity of approximately 2%, making it both a model thermophile and a model halophile [5]. Based on 16S rRNA sequence comparisons, the genus Rhodothermus has been classified as a member of the Rhodothermaceae, branching deeply within the phylum Bacteroidetes, with its closest relative being the mesophilic, extremely halophilic genus Salinibacter [6,7]. Other genera of the Rhodothermaceae include Salisaeta [8], Rubricoccus [9], Rubrivirga [10], Longimonas [11], and Longibacter [12]. All members are halophilic and either aerobic or facultatively anaerobic; all but Rhodothermus are mesophilic, suggesting that adaptation of Rhodothermus to growth at high temperature is a derived rather than a primitive character. Its affinity with Salinibacter suggests that adaptation to hypersaline environments predates development of thermostability. This stands in contrast to members of the genus Thermus, which form part of a phylum that branches deeply in the universal phylogenetic tree, and for whom thermal adaptation is likely a primitive character. Interestingly, R. marinus was isolated from the same environmental sample as the halotolerant IB-21 strain of Thermus thermophilus [13], providing an opportunity to compare independently arising adaptations to the same thermal environment. This species is thus of great intrinsic interest from the standpoint of microbial evolution.
We have begun to develop R. marinus as a model system for genetic and structural studies of the ribosome and potentially other macromolecular complexes. Like other extremophiles, R. marinus has become an important subject of protein structural studies. Notable examples include a novel respiratory complex III [14] and the ribosomal protein uL16 arginyl hydroxylase [15]. Although structures of DNA gyrase, RNA polymerase, and the ribosome from R. marinus have yet to be solved, these would seem promising subjects for structural studies given their important roles as targets for major antibiotic classes. Here, we describe antibiotic-resistant mutants of R. marinus with alterations in cellular components responsible for gene expression.
R. marinus has a number of advantages as a potential model organism for the study of the protein synthesis. In contrast to most other bacteria, the R. marinus genome has a single rrn operon [16,17], facilitating the isolation of rRNA mutants with pure populations of mutant ribosomes. Early attempts to isolate E. coli rRNA mutants were hampered by the presence of seven rrn operons such that even dominant mutations arising in a single operon fail to express a selectable phenotype; isolation of such mutants required either expression of rRNA from multi-copy plasmids [18] or deletion of multiple rrn operons [19]. In general, ribosome structural studies can be impaired by the complication of mixed populations of mutant and wild-type ribosomes from species with multiple rrn operons. More recently, isolation of pure rRNA mutants of Mycobacterium spp. [20] or T. thermophilus [21] has been facilitated by deletion of one of only two rRNA operons. In the latter organism, isolation of antibiotic-resistant mutants could also arise by efficient homologous recombination between rRNA gene copies during antibiotic selection [22]. While R. marinus is not naturally competent for transformation, a method of DNA transfer by electroporation has been described [23] and targeted gene disruptions have been constructed [24]. There is thus significant potential for developing this species as a genetic system [25].
Here, we describe a collection of R. marinus mutants having base substitutions in rRNA, or amino acid substitutions or deletions in ribosomal proteins or RNA polymerase. In most instances, these mutations are similar or identical to those found in T. thermophilus or mesophilic bacteria. Some mutations, specifically those affecting ribosomal protein uL4, have not been previously observed.
2. Results
2.1. Spectrum of Antibiotic Sensitivity
Before selecting resistant mutants, we established the range of antibiotics inhibitory to R. marinus. Although the sensitivity of R. marinus to several antibiotic classes was reported in the initial description of the genus [4], we undertook a more expansive survey. This was done using a simple zone of inhibition disc assay (see Materials and Methods). Results from these assays are indicated in Table 1 and Table S2. We examined antibiotics targeting the ribosome and associated factors, as well as drugs targeting RNA polymerase or DNA gyrase. Assays showing no zone of inhibition were interpreted as indicating resistance.

Table 1.
Antibiotic sensitivity spectrum of R. marinus R-10T. Sensitivity was assessed using a disc assay to measure zones of inhibition.
We found R. marinus to be insensitive to all 30S ribosomal subunit antibiotics we tested. We confirmed the previous report of intrinsic resistance of R. marinus to the aminoglycosides streptomycin, kanamycin and gentamicin [4] and also found resistance to a number of other aminoglycosides (apramycin, tobramycin, neomycin, paromomycin, neamine, ribostamycin, and hygromycin B). R. marinus is also insensitive to the aminocylitols spectinomycin and kasugamycin. We observed resistance to capreomycin, a member of the tuberactinomycins, which binds at the 30S–50S subunit interface, consistent with the previously observed cross-resistance of aminoglycoside-resistant mutants to capreomycin [22,26], suggesting a common basis for insensitivity to both classes of drugs. All other antibiotics showed significant zones of inhibition. We confirmed sensitivity to lincomycin, erythromycin, chloramphenicol, and pristinamycin, all of which bind in or near the peptidyltransferase active site of the 50S subunit. Additional macrolides producing inhibition included the 14-atom macrolides oleandomycin, roxithromycin, and clarithromycin, the 15-atom macrolide azithromycin, and the 16-atom macrolides spiramycin, chalcomycin, tylosin, and carbomycin. The pleuromutilin tiamulin, another inhibitor of peptide bond formation, also inhibits growth of R. marinus. In summary, R. marinus is resistant to all 30S inhibitors tested and sensitive to all 50S subunit inhibitors tested. Sensitivity was also found to kirromycin and fusidic acid, which target protein synthesis factors EF-Tu and EF-G, respectively. Among non-ribosomal drugs, we found R. marinus to be sensitive to the RNA polymerase inhibitor rifampicin and to the DNA gyrase inhibitors ofloxacin and ciprofloxacin, but, as previously reported [4], insensitive to nalidixic acid.
2.2. Selection of Spontaneous Mutants
Selection of resistant mutants was attempted with a number of drugs, including the RNA polymerase inhibitor rifampicin, the protein synthesis inhibitors chloramphenicol, lincomycin, erythromycin, spiramycin, tylosin, oleandomycin, thiostrepton, and fusidic acid, and the gyrase inhibitors ofloxacin and ciprofloxacin. Resistant mutants arose on rifampicin, chloramphenicol, lincomyin, erythromycin, spiramycin, tylosin and thiostrepton. No mutants appeared on oleandomycin, fusidic acid, ofloxacin or ciprofloxacin, although a more exhaustive search on a wider range of drug concentrations could potentially reveal mutants resistant to these drugs. Individual isolates were purified and analyzed by sequencing the genes known from previous studies to be the likely sites of mutations. Based on our own studies with T. thermophilus [22], we had no reason to expect major differences in the general location of these mutations compared with mesophilic species.
2.3. Rifampicin-Resistance (RifR) Mutations in the RNA Polymerase β Subunit
Rifampicin inhibits bacterial transcription by binding to the β subunit of RNA polymerase, and mutations conferring resistance are generally found in rpoB, the gene encoding the β subunit. We identified three independent R. marinus RifR alleles of rpoB (locus tag RMAR_RS05525). These included rpoB1 (GTC to TTC) producing the amino acid substitution V146F; rpoB2 (GCC to GTC) producing A522V; and rpoB3 (GCA to TCA) producing H526Y (Figure 1A; E. coli amino acid residue numbering). Mutations at these positions have been found in other species (reviewed by [27]) and are located in or near the rifampicin binding site (Figure 1B; pdb entry 6ccv [28]).
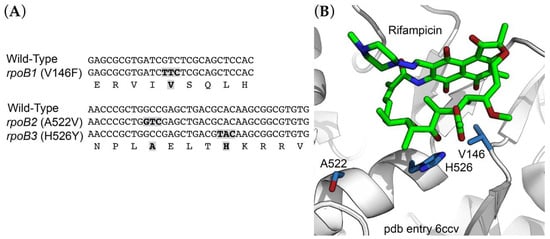
Figure 1.
Sites of RifR resistance mutations. (A) Alignment of a segment of RNA polymerase β-subunit sequence encompassing sites of RifR mutations. (B) Structure of the rifampicin binding site within Mycobacterium smegmatis RNA polymerase [28], showing positions corresponding to sites of RifR mutations in the R. marinus RNA polymerase β subunit.
2.4. Thiostrepton-Resistance Mutations in the 50S Ribosomal Subunit
Thiostrepton is a peptide antibiotic that acts as an inhibitor of EF-G-dependent translocation and binds to the 50S ribosomal subunit in a cleft formed by ribosomal protein uL11 and its binding site on 23S rRNA. The uL11 binding site itself is formed by two 23S rRNA loops (residues 1065–1073 and 1093–1098) brought together by a series of tertiary interactions (Figure 2A). Thiostrepton makes direct contact with A1067 and A1095 in these two loops and resistance can occur by base substitutions at either position, by enzymatic 2′-O-methylation of A1067, or by amino acid substitutions, deletions or insertions in uL11 (reviewed by [29]). We sequenced rrlA encoding 23S rRNA (locus tag RMAR_RS000900) and rplK encoding uL11 (locus tag RMAR05505) of the ThiR mutants. While we did not identify any changes in uL11, all mutants were found to have alterations at or near A1067 of 23S rRNA (Figure 2). These included an A1067C transversion, a deletion of A1069 (ΔA1069), a duplication of A1069 (A1069AA), or a duplication of G1071 (G1071GG). As seen in the Deinococcus radiodurans 50S ribosomal subunit-thiostrepton complex [29], A1067 is within a few Ångstroms of thiostrepton (Figure 2B), such that 2′-O-methylation could sterically block drug binding. The mechanism of resistance conferred by a base substitution at this position is not clear, but it presumably creates sufficient local distortion to decrease binding affinity. In contrast, changes at A1069 and G1071 are further away and must act more indirectly; residues 1067, 1068 and 1069 form a continuous base stack such that duplications or deletions of any of these residues could lead to repositioning of A1067. G1071 participates in a base triple with the G1091-C1100 base pair, which in turn is stacked on a base triple involving C1072, C1092 and G1099. Duplication of G1071 could destabilize the tertiary interaction between the two loops, thereby repositioning either A1067 or A1095, or both. This tertiary interaction is more directly stabilized by single hydrogen bonds between the N2 of G1068 and the O3′ of A1095, and between the O4′ of A1069 and the O2′ of A1096. Duplication or deletion of A1069 could easily disrupt these interactions and influence the positioning of A1067 and A1095.
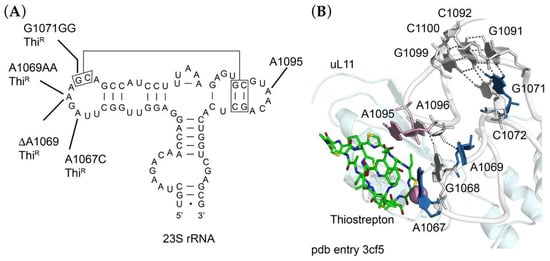
Figure 2.
Sites of mutations in 23S rRNA (A) Secondary structure of Rhodothermus marinus 23S rRNA in the vicinity of the thiostrepton binding site, indicating in blue the sites of single base substitution (A1067C), single base deletion (ΔA1069) and single nucleotide insertions (A1069AA and G1071GG), each conferring thiostrepton-resistance (ThiR). (B) Three-dimensional structure of the thiostrepton binding site in the crystal structure of the Deinococcus radiodurans 70S ribosome-thiostrepton complex (pdb entry 3cf5) [29]. Thiostrepton (green sticks); ribosomal protein uL11 (palecyan); 23S rRNA, white cartoon backbone with skyblue residues mutated to confer resistance in R. marinus. A1095 is colored pink. The 2′OH of A1067, site of methylation by resistance methyltransferases, is shown as a pink sphere.
2.5. Base Substitutions of 23S rRNA Residues in and around the Ribosomal Peptidyltransferase Active Site
The peptidyltransferase active site is situated deep within the 50S ribosomal subunit, and X-ray crystal structures of the bacterial ribosome in complex with transition state analogs indicate that all direct contacts with substrates in the active site involve residues of 23S rRNA [30]. Chloramphenicol, macrolides, lincosamides, streptogramin B, and various other antibiotics bind at or near this site, and in many cases mutually compete for binding (for an extensive review of antibiotic action, see [31]). While chloramphenicol, lincosamides, streptogramin B and some macrolides are bona fide peptidyltransferase inhibitors, other macrolides impair elongation of the nascent peptide by occluding the peptide exit channel; more recent studies have shown that macrolides inhibit the synthesis of a subset of proteins rather than inhibiting global translation (reviewed by [32]).
Chloramphenicol-resistant (ChlR) mutants of R. marinus were readily identified (Table 2) and occur at multiple sites within the central loop of 23S rRNA domain V, corresponding to the peptidyltransferase active site. These included A2059G, G2061C, A2062C, A2453C, U2500A, A2503C, A2503G, and U2504G. Each of these mutations is consistent with our previous studies with T. thermophilus. The most abundant mutation was A2058G, perhaps one of the most frequently described antibiotic-resistance mutation in the ribosome and was found independently in selections for erythromycin- (EryR), spiramycin- (SpiR), tylosin- (TylR), and lincomycin-resistant (LinR) mutants.

Table 2.
Mutations identified in this study. Abbreviations: Rep Strain, representative strain; Chl, chloramphenicol; Ery, erythromycin; Spi, spiramycin; Tyl, tylosin; Lnc, lincomycin; Thi, thiostrepton; Rif, rifampicin.
In addition, several double mutants were identified. These include A2453C/U2500A, selected on chloramphenicol; G2057A/A2062G, selected on erythromycin; and A2062G/A2503G, selected on spiramycin. The A2062G/A2503G double substitution is especially interesting in that A2062 and A2503 form a symmetrical base pair with one another via their Hoogsteen faces with two N6-N7 hydrogen bonds; either mutation alone could be isolated on chloramphenicol, while A2062G was isolated on either chloramphenicol or erythromycin (Figure 3). The appearance of multiple base substitutions is unusual and not easily explained but could be indicative of a high natural mutation frequency or stress-induced mutagenesis. Our protocols for selection of mutants are designed to minimize exposure to drug. Further studies will be needed to address this question. All of these residues are within close proximity to the corresponding drug binding sites as observed in T. thermophilus ribosome crystal structures (Figure 3B,C) [33].
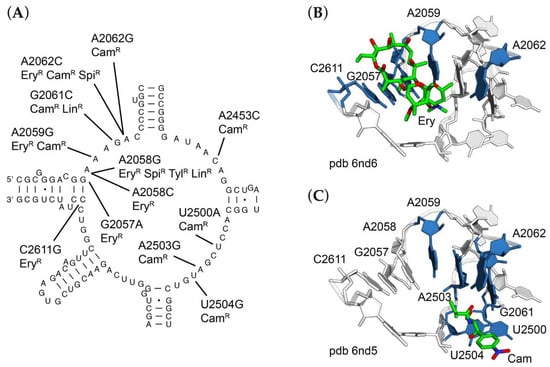
Figure 3.
Sites of mutations in or near the peptidyltransferase active site of 23S rRNA. (A) Secondary structure of Rhodothermus marinus central loop of domain V of 23S rRNA, with sites of mutations indicated, along with resistance phenotypes conferred. Three-dimensional structure of the peptidyltransferase center of Thermus thermophilus 70S ribosome in complex with (B) erythromycin (pdb entry 6nd6) and (C) chloramphenicol (pdb entry 6nd5). For panels (B,C), erythromycin or chloramphenicol are shown as green sticks; sites of mutations conferring resistance are colored blue.
2.6. Deletions in a Conserved Loop of Ribosomal Protein uL4 Conferring Erythromycin Resistance
While the peptidyltransferase active site is composed of rRNA, several proteins have globular domains situated on the subunit surface and extended structures that approach the active site, including uL2, uL3, uL4, and bL27 [30]. Proteins uL4 and uL22 form part of the polypeptide exit channel, near the binding site for macrolides. In a number of pathogenic organisms, resistance to erythromycin or other macrolides has been found to result from mutations in rplD or rplV, encoding 50S ribosomal proteins uL4 and uL22, respectively (reviewed by [34]). A large collection of mutations in both these proteins in E. coli reveal a wide range of mutations conferring macrolide resistance [35].
We identified a number of R. marinus mutants selected for resistance to several macrolides, including erythromycin, spiramycin, and tylosin, and sequenced both rplD and rplV of each of these. No mutations in rplV were found in any mutant. Several EryR mutants were found to have deletions within rplD (locus tag RMAR_RS04205), initially noted by the diminished size of PCR products as viewed by agarose gel electrophoresis. Sequencing confirmed that the rplD genes of these mutants had deletions in the region corresponding to the loop of uL4 that extends toward the peptidyltransferase center and polypeptide exit channel where erythromycin binds.
Three distinct deletion mutations were identified in rplD (Figure 4). One of these, an out-of-frame deletion of the sequence 5′-GCTGTA-3′, results in both a K58N substitution and the deletion of L59 and Y60. A second allele, consisting of a 60-base pair, 20-amino acid deletion extending from A50 to R69, appears to be the result of homologous recombination between two short, direct repeats of the sequence 5′-CGGGCCG-3′. The third allele, a 54-bp, 18-amino acid deletion removing T65 to G82, appears to be the result of homologous recombination between direct repeats of the sequence 5′-GGTACGG-3′ (Figure S1). The original E. coli erythromycin-resistance mutation, eryA [36], resulted in a K63E substitution in the same region of uL4 [37]. Several site-directed mutagenesis studies of E. coli uL4 have demonstrated the malleability of this extended loop and its role in macrolide-ribosome interactions [38,39,40]. However, we are not aware of spontaneous deletions of uL4 of this magnitude having been previously reported.
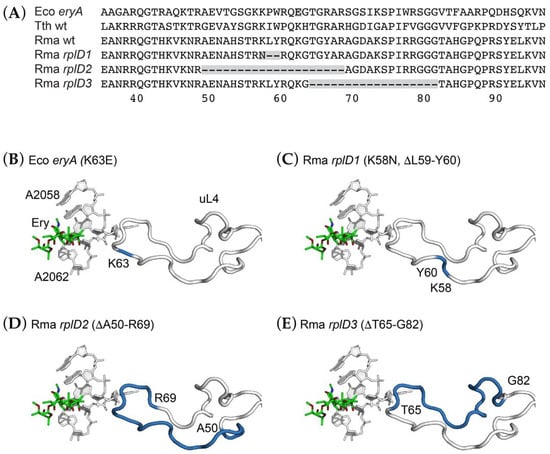
Figure 4.
Erythromycin-resistance mutations affecting ribosomal protein uL4. (A) Partial sequence alignment of ribosomal proteins uL4 from Escherichia coli (Eco), Thermus thermophilus (Tth), and Rhodothermus marinus (Rma) highlighting sights of erythromycin-resistance mutations. (B–E) Three-dimensional structure showing the interaction with the extended loop of ribosomal protein uL4 and the erythromycin binding site of 23S rRNA, illustrated using the Thermus thermophilus 70S ribosome-erythromycin complex (pdb entry 6nd6). For clarity, only several 23S rRNA nucleotides and the extended loops of uL4 are shown. Residues mutated are colored blue. (B) The eryA K63E mutation from E. coli. (C) The R. marinus rplD1 (K58N, ΔL59-Y60) mutation. (D) The R. marinus rplD2 (ΔA50-R69) mutation. (E) The R. marinus rplD3 (ΔT65-G82) mutation. Each of these mutations is expected to perturb RNA conformation in the macrolide binding site.
3. Discussion
R. marinus is both extremely thermophilic and halophilic, making it quite distinct physiologically from other organism that have been the subject of study of antibiotic action and mechanisms of resistance. Nevertheless, it is remarkable that many of the same mutations that confer antibiotic resistance in mesophiles also confer resistance in extremophiles such as T. thermophilus and R. marinus. Thus, the majority of the mutations we identified in this study are identical to those observed in a wide range of organisms, though our collection of R. marinus mutants is larger than most reported in individual species in a single study. Most of the base substitutions in the peptidyltransferase active site are at positions in close contact with various antibiotics, as observed in crystal structures of ribosome-antibiotic complexes (reviewed by [3]). The same can be said of the RNA polymerase mutants we found. This similarity is no doubt the result of the extreme conservation of rRNA sequence and structure in ribosome functional centers, and the catalytic center of RNA polymerase.
A number of high-resolution structures of the T. thermophilus RNA polymerase have been determined in various complexes [41,42,43], including with rifampicin [44]. More recently, the structure of the M. smegmatis RNA polymerase-rifampicin complex has been solved [28]. Of the R. marinus RNA polymerase residues mutated in RifR mutants, V146 and H526 are both quite conserved, whereas A522 is less so. Substitutions at H526 are among the more frequently observed RifR mutations in Mycobacterium tuberculosis [27]. As seen in the M. smegmatis RNA polymerase-rifampicin complex [28], all three residues are within the rifampicin binding site, although A522 is somewhat removed from rifampicin such that the mechanism of resistance caused by substitutions at this position is unclear (Figure 1B). It is perhaps worth noting that this position is a Ser in E. coli, M. tuberculosis, and T. aquaticus. While none of these residues make direct contact with the drug, their exchange with bulkier residues is likely to have a strong steric effect on drug binding.
Crystal structures of antibiotics bound to the peptidyltransferase center help to explain their mechanism of action [3]. As illustrated in Figure 3, base substitutions conferring drug resistance are components of the drug binding site. In the case of erythromycin-resistance, sites of mutations are located in close proximity to the drug, and the same is true for chloramphenicol-resistance. Consistent with this observation, mutations simultaneously conferring resistance to both drugs are located between the two drug binding sites. Further, crystal structures of ribosomes containing antibiotic-resistance mutations show small perturbations in local structure or indicate a loss of ribosome-drug contact [45]. In contrast to sites of rRNA mutations, ribosomal protein mutations are less similar across species. This is probably in part due to the lower conservation of ribosomal protein sequences, but more likely the lack of conservation at the DNA sequence level, which constrains the specific mutations that can occur. The deletion mutations in rplD observed in R. marinus are the result of recombination between short, fortuitously repeated DNA sequences. Synonymous codons at either of these repeats would presumably prevent these particular deletions from arising.
The finding of deletion mutations in uL4 is consistent with previously identified mutations in this protein. The original E. coli eryA allele of rplD was found to result in a single amino acid substitution, K63E [37] (Figure 4B). This mutation was subsequently found to cause an increase in the frequency of translational errors, including misreading errors and frameshifting [46]. While an assay for measuring translational accuracy does not yet exist for R. marinus, it should be possible to assess their effects by reconstructing the analogous deletions in E. coli rplD. While there is as yet no high-resolution structure of the R. marinus ribosome, structures of ribosomes from a variety of organisms indicate that the loop subjected to these deletion mutations is located in close proximity to 23S rRNA residues involved in erythromycin binding. A wide variety of amino acid substitutions as well as small deletions (1 to several residues) in this loop have been found in multiple species.
The lack of direct contact between uL4 and erythromycin demands consideration of an indirect mechanism of resistance, such as a local destabilization of rRNA conformation. The deletions could be mapped onto the T. thermophilus ribosome crystal structure and it is clear that they must abolish any direct contact with 23S rRNA in the erythromycin binding site (Figure 4C–E). This notion is consistent with a previous chemical probing study [47] and subsequent cryo-electron microscopic reconstruction [48] of the E. coli uL4 mutant, both of which showed significant structural distortion from a single amino acid substitution. We would expect the R. marinus deletions to make substantially greater distortions. A crystal structure of an archaeal 50S subunit bearing a 3 amino acid deletion in ribosomal protein uL22 shows the repositioning of several bases in the peptidyltransferase center [45]. Remarkably, the extended loops of both uL4 and uL22 of E. coli are dispensable for protein synthesis and growth but make important contributions to the kinetics and fidelity of 50S subunit assembly [40]. We expect that the deletions within uL4 will show extensive distortions, and future cryo-EM reconstructions of the R. marinus ribosome could be effective in testing this hypothesis.
The finding that R. marinus is resistant to aminoglycosides, capreomycin, kasugamycin, and spectinomycin, was unexpected. One possible explanation is an inability to import these drugs into the cell, and given the close structural relationship of the aminoglycosides, a common uptake mechanism seems plausible. Members of the Bacteroides genus are inherently resistant to aminoglycosides due to lack of an oxygen- or nitrate-dependent electron transport system [49]. This explanation is insufficient to explain resistance of R. marinus, which is obligately aerobic. Another possibility is natural variation in ribosome structure. Inspection of the 16S rRNA sequence revealed variation in the aminoglycoside binding site, a A1409-U1491 base pair, as opposed to the C1409-G1491 more frequently found in bacterial 16S rRNAs. Data retrieved from the Comparative RNA Web Site and Project database (http://www.rna.ccbb.utexas.edu; accessed on 9 August 2020) [50] indicate that among bacterial sequences, a C-G pair is found in 84.5% of bacterial 16S rRNA sequences, while an A-U base pair is found in 12.6% of sequences. These data also indicate that the adjacent base pair, 1410–1490, is C-G in R. marinus, a sequence found in only 0.1% of bacterial 16S rRNA sequences. The mesophilic extreme halophile Salinibacter ruber, whose closest relative is R. marinus, is also resistant to kanamycin and its 16S rRNA also has the A1409-U1491 base pair [6,7]. Based on secondary structure models retrieved from the RNAcentral database (http://rnacentral.org/; accessed on 9 August 2020) [51], this same A-U base pair is present in all members of the Rhodothermaceae. Whether or how either of these base pair identities might influence the aminoglycoside binding site is not obvious. Previous studies have found the aminoglycoside-resistance mutations C1409G of yeast mitochondrial 17S rRNA [52], or a G1491A of Tetrahymena thermophila 18S rRNA [53]. This would also explain the resistance to capreomycin, as these mutations confer cross-resistance to tuberactinomycins. In the absence of an in vitro protein synthesis system for R. marinus, it is not yet possible to distinguish between these two possible explanations for resistance to aminoglycosides and capreomycin.
4. Materials and Methods
4.1. Strains and Growth Conditions, Assessment of Antibiotic Sensitivity, and Isolation of Mutants
All mutants were derived from R. marinus R-10T ATCC 43812/DSM 4252 [4], which was a kind gift of JHD Cate, University of California, Berkeley. R. marinus was cultivated in liquid TEM medium (ATCC Medium 1598) containing 2% NaCl (referred to hereafter as TEMS medium) or on TMG medium containing 2% NaCl (referred to hereafter as TMGS medium). TMG medium consists of TEM lacking phosphate buffer and solidified with gelrite at a concentration of 1.1%. All cultures were grown at 65 °C under aerobic conditions with vigorous aeration at 200 rpm in a New Brunswick Innova 42 Shaker Incubator. Overnight cultures were typically cultivated in 20 mL of medium in 125 mL baffled culture flasks (Corning).
To assay antibiotic sensitivity, 100 μL of a saturated overnight culture grown in TMGS broth was spread-plated onto TMGS plates. A disc infused with 100 μg of antibiotic was placed onto the surface of the plate, which was then incubated at 65 °C overnight; zones of inhibition were subsequently measured. Spontaneous mutants were selected by spreading approximately 109 cells from a saturated overnight culture onto TMGS plates containing various antibiotic concentrations; chloramphenicol, 25, 50, or 100 μg/mL; erythromycin, 50, 100, or 200 μg/mL; tylosin, 100 μg/mL; spiramycin, 100 μg/mL; lincomycin, 100 μg/mL; thiostrepton, 25, 50, 100, or 200 μg/mL; rifampicin, 50, 100, or 200 μg/mL. Mutants were purified by restreaking onto TMGS medium containing antibiotic at the same concentration used in selection, then a second time on antibiotic-free TMGS. Mutants were never exposed to antibiotic after the initial single colony isolation. Single colonies were used to inoculate TEMS medium and shaken at 65 °C to saturation. Mutants were archived as 25% glycerol stocks at −80 °C.
4.2. Identification of Mutations
Chromosomal DNA (gDNA) was prepared using Wizard Genomic DNA Kit (Promega, Madison, WI, USA). Oligonucleotide primers were synthesized by IDT and are described in Supplementary Table S1. All PCR reactions were performed using OneTaq DNA polymerase (New England Biolabs, Ipswich, MA, USA). Sanger sequencing of PCR products was performed by the Genomics and Sequencing Center at The University of Rhode Island. The rrnA operon encoding 16S rRNA (locus tag RMAR_RS00885), tRNAIle (locus tag RMAR_RS00890), tRNAAla (locus tag RMAR_RS00895), 23S rRNA (locus tag RMAR_RS00900), and 5S rRNA (locus tag RMAR_ RS00905), was amplified using primers Rma_rrnA_f3 and Rma_rrnA_r3, with a 58 °C annealing temperature and a 6 min extension time. The rplD gene encoding ribosomal protein uL4 (locus tag RMAR_RS04205) was amplified using primers Rma_rplD_f1 and Rma_rplD_r1, with a 52.5 °C annealing temperature and a 1 min extension time. The rplV gene encoding ribosomal protein uL22 (locus tag RMAR_RS04225) was amplified using primers Rma_rplV_f1 and Rma_rplV_r1, with a 49 °C annealing temperature and a 1 min extension time. The rplK gene encoding ribosomal protein uL11 (locus tag RMAR_RS05505) was amplified using primers Rma_rplK_f1 and Rma_rplK_r1, with a 60 °C annealing temperature and a 1 min extension time. The rpoB gene encoding the β-subunit of RNA polymerase (locus tag RMAR_RS05525) was amplified using primers Rma_rpoB_f1 and Rma_rpoB_r1, with a 49 °C annealing temperature and a 4 min extension time. Sequencing of the rrlA gene encoding 23S rRNA was performed using primers Rma_rrnA_f4, Rma_rrnA_f7, Rma_rrnA_f8, Rma_rrnA_r5, Rma_rrnA_r6, Rma_rrnA_r7.
5. Conclusions
Antibiotic sensitivity spectra and patterns of cross resistance can potentially be informative from both phylogenetic and ribosome structure-function perspectives. Extensive surveys of antibiotic-resistance mutations have been conducted for only a handful of species, making broad generalizations difficult. In this study, we have isolated and characterized a number of antibiotic-resistant mutants of a single species, potentially allowing direct comparisons of mutant phenotypes. Importantly, we find that mutations arising in a thermophilic-halophilic species closely resemble those found in mesophilic species, consistent with the extreme sequence conservation (and by implication, structural conservation) of antibiotic-binding sites in RNA polymerase and the ribosome. Surprising was the inherent resistance of this species to a range of structurally-unrelated 30S subunit inhibitors. The basis for this resistance remains to be determined. The ability to readily isolate rRNA mutations in this species makes it a candidate for future structural studies to address this question.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/antibiotics10111384/s1, Figure S1: Generation of rplD erythromycin-resistance mutations [54]; Table S1: Oligonucleotides used in this study [17]; Table S2: Antibiotic zones of inhibition.
Author Contributions
Conceptualization, S.T.G.; methodology and experimentation, S.S., S.A.D. and E.E.K.; validation, S.T.G., S.A.D. and E.E.K.; writing—original draft preparation, S.T.G.; writing—review and editing, S.T.G., E.E.K. and G.J.; funding acquisition, S.T.G. and G.J. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grant GM094157 (to G.J. and S.T.G.) from the U.S. National Institutes of Health, and by the USDA National Institute of Food and Agriculture, Hatch Formula project 1016013 (to S.T.G.). SAS was supported by the Rhode Island Institutional Development Award (IDeA) Network of Biomedical Research Excellence from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20GM103430. Sequencing was performed by a Rhode Island NSF EPSCoR research facility, the Genomics and Sequencing Center, which is supported in part by the National Science Foundation EPSCoR Cooperative Agreement #EPS-1004057.
Acknowledgments
The authors are extremely grateful to the Janet Atoyan for technical assistance and to Marianne Prior for logistical support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ma, C.; Yang, X.; Lewis, P.J. Bacterial transcription as a target for antibacterial drug development. Microbiol. Mol. Biol. Rev. 2016, 80, 139–160. [Google Scholar] [CrossRef] [PubMed]
- Murakami, K.S. Structural biology of bacterial RNA polymerase. Biomolecules 2015, 5, 848–864. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.N. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat. Rev. Microbiol. 2014, 12, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Alfredsson, G.A.; Kristjansson, J.K.; Hjörleifdottir, S.; Stetter, K.O. Rhodothermus marinus, gen. nov., sp. nov., a thermophilic, halophilic bacterium from submarine hot springs in Iceland. J. Gen. Microbiol. 1988, 134, 299–306. [Google Scholar] [CrossRef]
- Bjornsdottir, S.H.; Blondal, T.; Hreggvidsson, G.O.; Eggertsson, G.; Petursdottir, S.; Hjorleifsdottir, S.; Thorbjarnardottir, S.H.; Kristjansson, J.K. Rhodothermus marinus: Physiology and molecular biology. Extremophiles 2006, 10, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Antón, J.; Oren, A.; Benlloch, S.; Rodríguez-Valera, F.; Amann, R.; Rosselló-Mora, R. Salinibacter ruber gen. nov., sp. nov., a novel, extremely halophilic member of the Bacteria from saltern crystallizer ponds. Int. J. Syst. Evol. Microbiol. 2002, 52, 485–491. [Google Scholar] [CrossRef] [PubMed]
- Mongodin, E.F.; Nelson, K.E.; Daugherty, S.; Deboy, R.T.; Wister, J.; Khouri, H.; Weidman, J.; Walsh, D.A.; Papke, R.T.; Sanchez Perez, G.; et al. The genome of Salinibacter ruber: Convergence and gene exchange among hyperhalophilic bacteria and archaea. Proc. Natl. Acad. Sci. USA 2005, 102, 18147–18152. [Google Scholar] [CrossRef]
- Vaisman, N.; Oren, A. Salisaeta longa gen. nov., sp. nov., a red, halophilic member of the Bacteroidetes. Int. J. Syst. Evol. Microbiol. 2009, 59, 2571–2574. [Google Scholar] [CrossRef]
- Park, S.; Yoshizawa, S.; Kogure, K.; Yokota, A. Rubricoccus marinus gen. nov., sp. nov., of the family ‘Rhodothermaceae’, isolated from seawater. Int. J. Syst. Evol. Microbiol. 2011, 61, 2069–2072. [Google Scholar] [CrossRef]
- Park, S.; Song, J.; Yoshizawa, S.; Choi, A.; Cho, J.C.; Kogure, K. Rubrivirga marina gen. nov., sp. nov., a member of the family Rhodothermaceae isolated from deep seawater. Int. J. Syst. Evol. Microbiol. 2013, 63, 2229–2233. [Google Scholar] [CrossRef]
- Xia, J.; Zhou, Y.X.; Zhao, L.H.; Chen, G.J.; Du, Z.J. Longimonas halophila gen. nov., sp. nov., isolated from a marine solar saltern. Int. J. Syst. Evol. Microbiol. 2015, 65, 2272–2276. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Dunlap, C.A.; Flor-Weiler, L.; Rooney, A.P.; Chen, G.J.; Du, Z.J. Longibacter salinarum gen. nov., sp. nov., isolated from a marine solar saltern. Int. J. Syst. Evol. Microbiol. 2016, 66, 3287–3292. [Google Scholar] [CrossRef]
- Kristjansson, J.K.; Hreggvidsson, G.O.; Alfredsson, G.A. Isolation of halotolerant Thermus spp. from submarine hot springs in Iceland. Appl. Environ. Microbiol. 1986, 52, 1313–1316. [Google Scholar] [CrossRef] [PubMed]
- Sousa, J.S.; Calisto, F.; Langer, J.D.; Mills, D.J.; Refojo, P.N.; Teixeira, M.; Kühlbrandt, W.; Vonck, J.; Pereira, M.M. Structural basis for energy transduction by respiratory alternative complex III. Nat. Commun. 2018, 9, 1728. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, R.; Sekirnik, R.; Brissett, N.C.; Krojer, T.; Ho, C.H.; Ng, S.S.; Clifton, I.J.; Ge, W.; Kershaw, N.J.; Fox, G.C.; et al. Ribosomal oxygenases are structurally conserved from prokaryotes to humans. Nature 2014, 510, 422–426. [Google Scholar] [CrossRef] [PubMed]
- Andrésson, O.S.; Fridjónsson, O.H. The sequence of the single 16S rRNA gene of the thermophilic eubacterium Rhodothermus marinus reveals a distant relationship to the group containing Flexibacter, Bacteroides, and Cytophaga species. J. Bacteriol. 1994, 176, 6165–6169. [Google Scholar] [CrossRef]
- Nolan, M.; Tindall, B.J.; Pomrenke, H.; Lapidus, A.; Copeland, A.; Glavina Del Rio, T.; Lucas, S.; Chen, F.; Tice, H.; Cheng, J.F.; et al. Complete genome sequence of Rhodothermus marinus type strain (R-10). Stand. Genom. Sci. 2009, 1, 283–290. [Google Scholar] [CrossRef][Green Version]
- Sigmund, C.D.; Ettayebi, M.; Morgan, E.A. Antibiotic resistance mutations in 16S and 23S ribosomal RNA genes of Escherichia coli. Nucleic Acids Res. 1984, 12, 4653–4663. [Google Scholar] [CrossRef]
- Asai, T.; Zaporojets, D.; Squires, C.; Squires, C.L. An Escherichia coli strain with all chromosomal rRNA operons inactivated: Complete exchange of rRNA genes between bacteria. Proc. Natl. Acad. Sci. USA 1999, 96, 1971–1976. [Google Scholar] [CrossRef]
- Hobbie, S.N.; Bruell, C.; Kalapala, S.; Akshay, S.; Schmidt, S.; Pfister, P.; Böttger, E.C. A genetic model to investigate drug-target interactions at the ribosomal decoding site. Biochimie 2006, 88, 1033–1043. [Google Scholar] [CrossRef]
- Gregory, S.T.; Dahlberg, A.E. Genetic and structural analysis of base substitutions in the central pseudoknot of Thermus thermophilus 16S ribosomal RNA. RNA 2009, 15, 215–223. [Google Scholar] [CrossRef][Green Version]
- Gregory, S.T.; Carr, J.F.; Rodriguez-Correa, D.; Dahlberg, A.E. Mutational analysis of 16S and 23S rRNA genes of Thermus thermophilus. J. Bacteriol. 2005, 187, 4804–4812. [Google Scholar] [CrossRef] [PubMed]
- Bjornsdottir, S.H.; Thorbjarnardottir, S.H.; Eggertsson, G. Establishment of a gene transfer system for Rhodothermus marinus. Appl. Microbiol. Biotechnol. 2005, 66, 675–682. [Google Scholar] [CrossRef] [PubMed]
- Bjornsdottir, S.H.; Fridjonsson, O.H.; Hreggvidsson, G.O.; Eggertsson, G. Generation of targeted deletions in the genome of Rhodothermus marinus. Appl. Environ. Microbiol. 2011, 77, 5505–5512. [Google Scholar] [CrossRef][Green Version]
- Bjornsdottir, S.H.; Fridjonsson, O.H.; Kristjansson, J.K.; Eggertsson, G. Cloning and expression of heterologous genes in Rhodothermus marinus. Extremophiles 2007, 11, 283–293. [Google Scholar] [CrossRef] [PubMed]
- Maus, C.E.; Plikaytis, B.B.; Shinnick, T.M. Molecular analysis of cross-resistance to capreomycin, kanamycin, amikacin, and viomycin in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2005, 49, 3192–3197. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, B.P. Resistance to rifampicin: A review. J. Antibiot. 2014, 67, 625–630. [Google Scholar] [CrossRef] [PubMed]
- Peek, J.; Lilic, M.; Montiel, D.; Milshteyn, A.; Woodworth, I.; Biggins, J.B.; Ternei, M.A.; Calle, P.Y.; Danziger, M.; Warrier, T.; et al. Rifamycin congeners kanglemycins are active against rifampicin-resistant bacteria via a distinct mechanism. Nat. Commun. 2018, 9, 4147. [Google Scholar] [CrossRef]
- Harms, J.M.; Wilson, D.N.; Schluenzen, F.; Connell, S.R.; Stachelhaus, T.; Zaborowska, Z.; Spahn, C.M.; Fucini, P. Translational regulation via L11: Molecular switches on the ribosome turned on and off by thiostrepton and micrococcin. Mol. Cell 2008, 30, 26–38. [Google Scholar] [CrossRef]
- Ban, N.; Nissen, P.; Hansen, J.; Moore, P.B.; Steitz, T.A. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 2000, 289, 905–920. [Google Scholar] [CrossRef]
- Gale, E.F.; Cundliffe, P.E.; Reynolds, M.H.; Richmond, M.J. Waring. In The Molecular Basis of Antibiotic Action; John Wiley & Sons: London, UK, 1981. [Google Scholar]
- Vázquez-Laslop, N.; Mankin, A.S. How macrolide antibiotics work. Trends Biochem. Sci. 2018, 43, 668–684. [Google Scholar] [CrossRef] [PubMed]
- Svetlov, M.S.; Plessa, E.; Chen, C.W.; Bougas, A.; Krokidis, M.G.; Dinos, G.P.; Polikanov, Y.S. High-resolution crystal structures of ribosome-bound chloramphenicol and erythromycin provide the ultimate basis for their competition. RNA 2019, 25, 600–606. [Google Scholar] [CrossRef] [PubMed]
- Fyfe, C.; Grossman, T.H.; Kerstein, K.; Sutcliffe, J. Resistance to macrolide antibiotics in public health pathogens. Cold Spring Harb. Perspect. Med. 2016, 6, a025395. [Google Scholar] [CrossRef]
- Diner, E.J.; Hayes, C.S. Recombineering reveals a diverse collection of ribosomal proteins L4 and L22 that confer resistance to macrolide antibiotics. J. Mol. Biol. 2009, 386, 300–315. [Google Scholar] [CrossRef]
- Wittmann, H.G.; Stoffler, G.; Apirion, D.; Rosen, L.; Tanaka, K.; Tamaki, M.; Takata, R.; Dekio, S.; Otaka, E.; Osawa, S. Biochemical and genetic studies of two different types of erythromycin resistant mutants of Escherichia coli with altered ribosomal proteins. Mol. Gen. Genet. 1973, 127, 175–189. [Google Scholar] [CrossRef] [PubMed]
- Chittum, H.S.; Champney, W.S. Ribosomal protein gene sequence changes in erythromycin-resistant mutants of Escherichia coli. J. Bacteriol. 1994, 176, 6192–6198. [Google Scholar] [CrossRef]
- Zengel, J.M.; Jerauld, A.; Walker, A.; Wahl, M.C.; Lindahl, L. The extended loops of ribosomal proteins L4 and L22 are not required for ribosome assembly or L4-mediated autogenous control. RNA 2003, 9, 1188–1197. [Google Scholar] [CrossRef] [PubMed]
- Zaman, S.; Fitzpatrick, M.; Lindahl, L.; Zengel, J. Novel mutations in ribosomal proteins L4 and L22 that confer erythromycin resistance in Escherichia coli. Mol. Microbiol. 2007, 66, 1039–1050. [Google Scholar] [CrossRef]
- Lawrence, M.G.; Shamsuzzaman, M.; Kondopaka, M.; Pascual, C.; Zengel, J.M.; Lindahl, L. The extended loops of ribosomal proteins uL4 and uL22 of Escherichia coli contribute to ribosome assembly and protein translation. Nucleic Acids Res. 2016, 44, 5798–5810. [Google Scholar] [CrossRef]
- Murakami, K.S.; Masuda, S.; Darst, S.A. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 Å resolution. Science 2002, 296, 1280–1284. [Google Scholar] [CrossRef]
- Vassylyev, D.G.; Sekine, S.; Laptenko, O.; Lee, J.; Vassylyeva, M.N.; Borukhov, S.; Yokoyama, S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature 2002, 417, 712–719. [Google Scholar] [CrossRef]
- Murakami, K.S.; Darst, S.A. Bacterial RNA polymerases: The wholo story. Curr. Opin. Struct. Biol. 2003, 13, 31–39. [Google Scholar] [CrossRef]
- Campbell, E.A.; Korzheva, N.; Mustaev, A.; Murakami, K.; Nair, S.; Goldfarb, A.; Darst, S.A. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 2001, 104, 901–912. [Google Scholar] [CrossRef]
- Tu, D.; Blaha, G.; Moore, P.B.; Steitz, T.A. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell 2005, 121, 257–270. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, M.; Gregory, S.T.; Dahlberg, A.E. Multiple defects in translation associated with altered ribosomal protein L4. Nucleic Acids Res. 2004, 32, 5750–5756. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gregory, S.T.; Dahlberg, A.E. Erythromycin resistance mutations in ribosomal protein L22 and L4 perturb the higher order structure of 23S ribosomal RNA. J. Mol. Biol. 1999, 289, 827–834. [Google Scholar] [CrossRef]
- Gabashvili, I.S.; Gregory, S.T.; Valle, M.; Grassucci, R.; Worbs, M.; Wahl, M.C.; Dahlberg, A.E.; Frank, J. The polypeptide tunnel system in the ribosome and its gating in erythromycin resistance mutants of L4 and L22. Mol. Cell 2001, 8, 181–188. [Google Scholar] [CrossRef]
- Bryan, L.E.; Kowand, S.K.; Van Den Elzen, H.M. Mechanism of aminoglycoside antibiotic resistance in anaerobic bacteria: Clostridium perfringens and Bacteroides fragilis. Antimicrob. Agents Chemother. 1979, 15, 7–13. [Google Scholar] [CrossRef]
- Cannone, J.J.; Subramanian, S.; Schnare, M.N.; Collett, J.R.; D’Souza, L.M.; Du, Y.; Feng, B.; Lin, N.; Madabusi, L.V.; Müller, K.M.; et al. The comparative RNA web (CRW) site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinform. 2002, 3, 2. [Google Scholar]
- The RNAcentral Consortium; Petrov, A.I.; Kay, S.J.E.; Kalvari, I.; Howe, K.L.; Gray, K.A.; Bruford, E.A.; Kersey, P.J.; Cochrane, G.; Finn, R.D.; et al. RNAcentral: A comprehensive database of non-coding RNA sequences. Nucleic Acids Res. 2017, 45, D128–D134. [Google Scholar] [PubMed]
- Li, M.; Tzagoloff, A.; Underbrink-Lyon, K.; Martin, N.C. Identification of the paromomycin-resistance mutation in the 15 S rRNA gene of yeast mitochondria. J. Biol. Chem. 1982, 257, 5921–5928. [Google Scholar] [CrossRef]
- Spangler, E.A.; Blackburn, E.H. The nucleotide sequence of the 17S ribosomal RNA gene of Tetrahymena thermophila and the identification of point mutations resulting in resistance to the antibiotics paromomycin and hygromycin. J. Biol. Chem. 1985, 260, 6334–6340. [Google Scholar] [CrossRef]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).