Dissection of Highly Prevalent qnrS1-Carrying IncX Plasmid Types in Commensal Escherichia coli from German Food and Livestock
Abstract
1. Introduction
2. Results and Discussion
2.1. qnrS1 Is Highly Prevalent on IncX Plasmids in Commensal E. coli
2.2. Three Prevalent IncX Plasmids, Carrying qnrS1 in German Livestock Were Detected
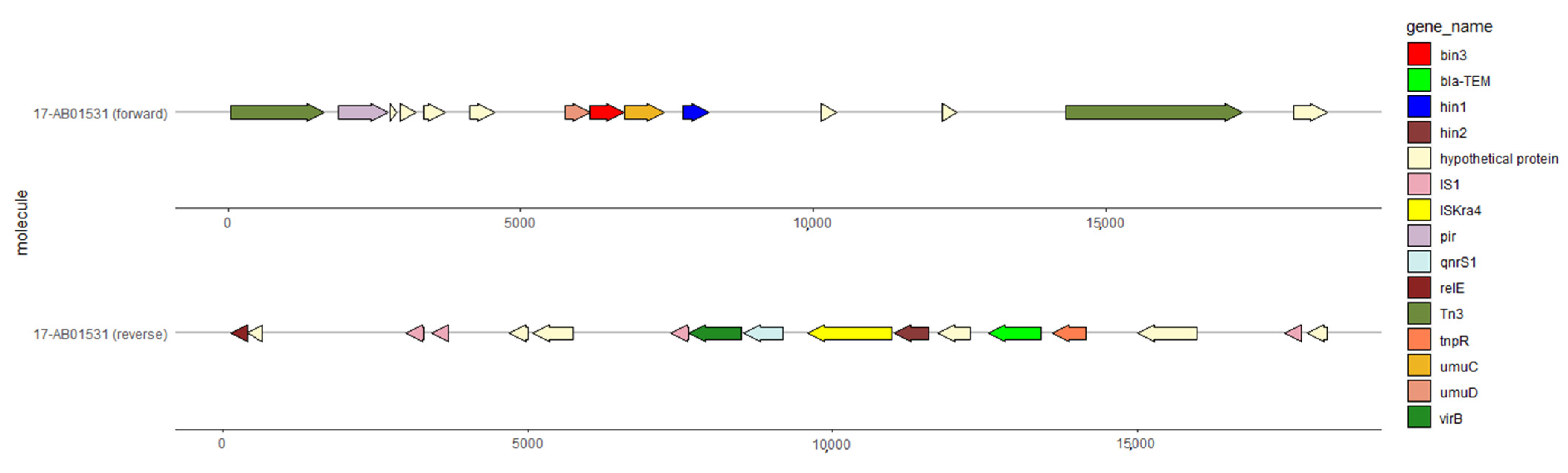
2.2.1. The Genomes of Prevalent qnrS1-Carrying IncX Plasmids
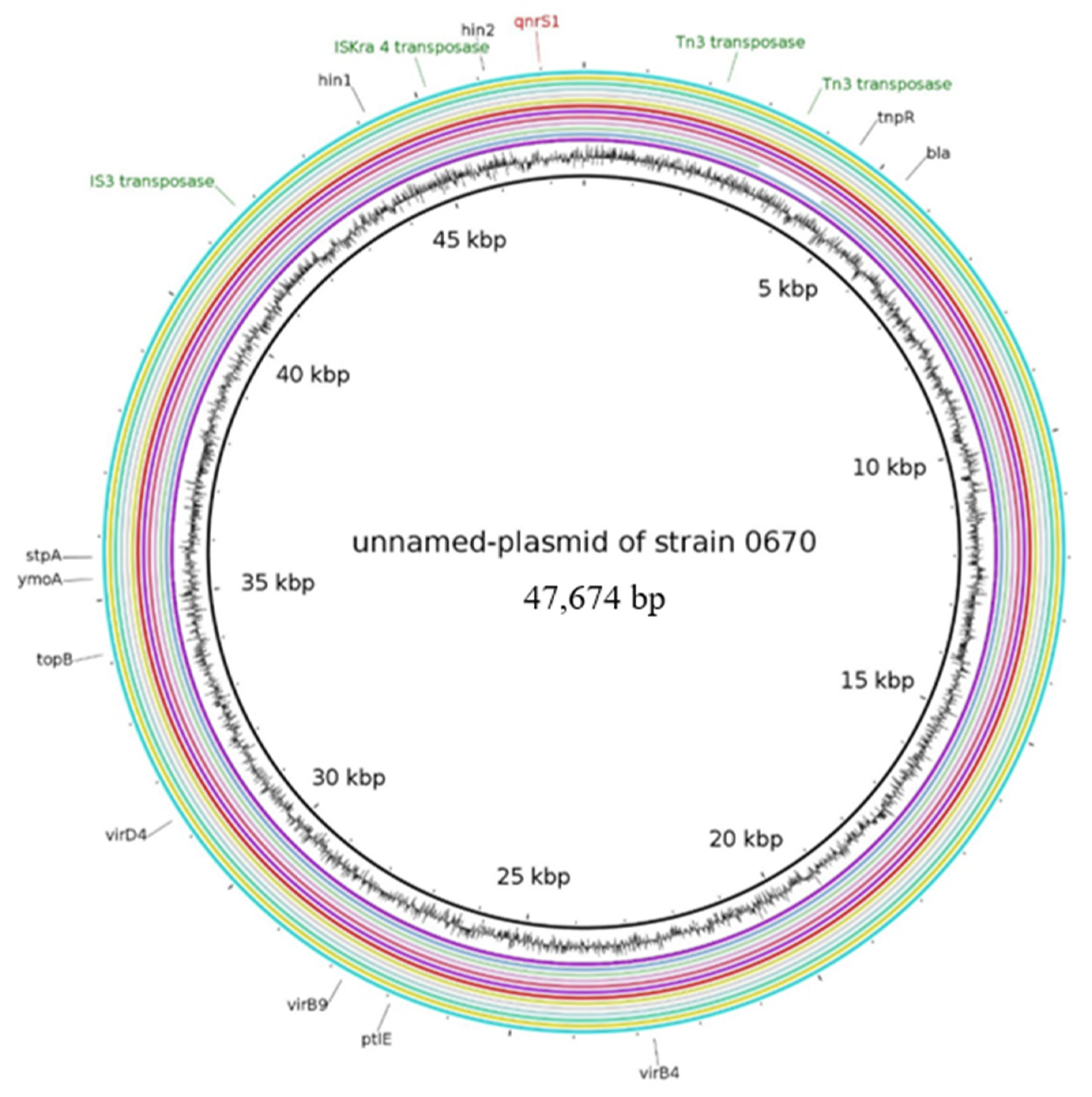
2.2.2. Characteristics of Plasmids Assigned to the Unnamed Reference Plasmid of the Strain 0670
2.2.3. Characteristics of Plasmids Assigned to the Reference Plasmid pKpvST101_6
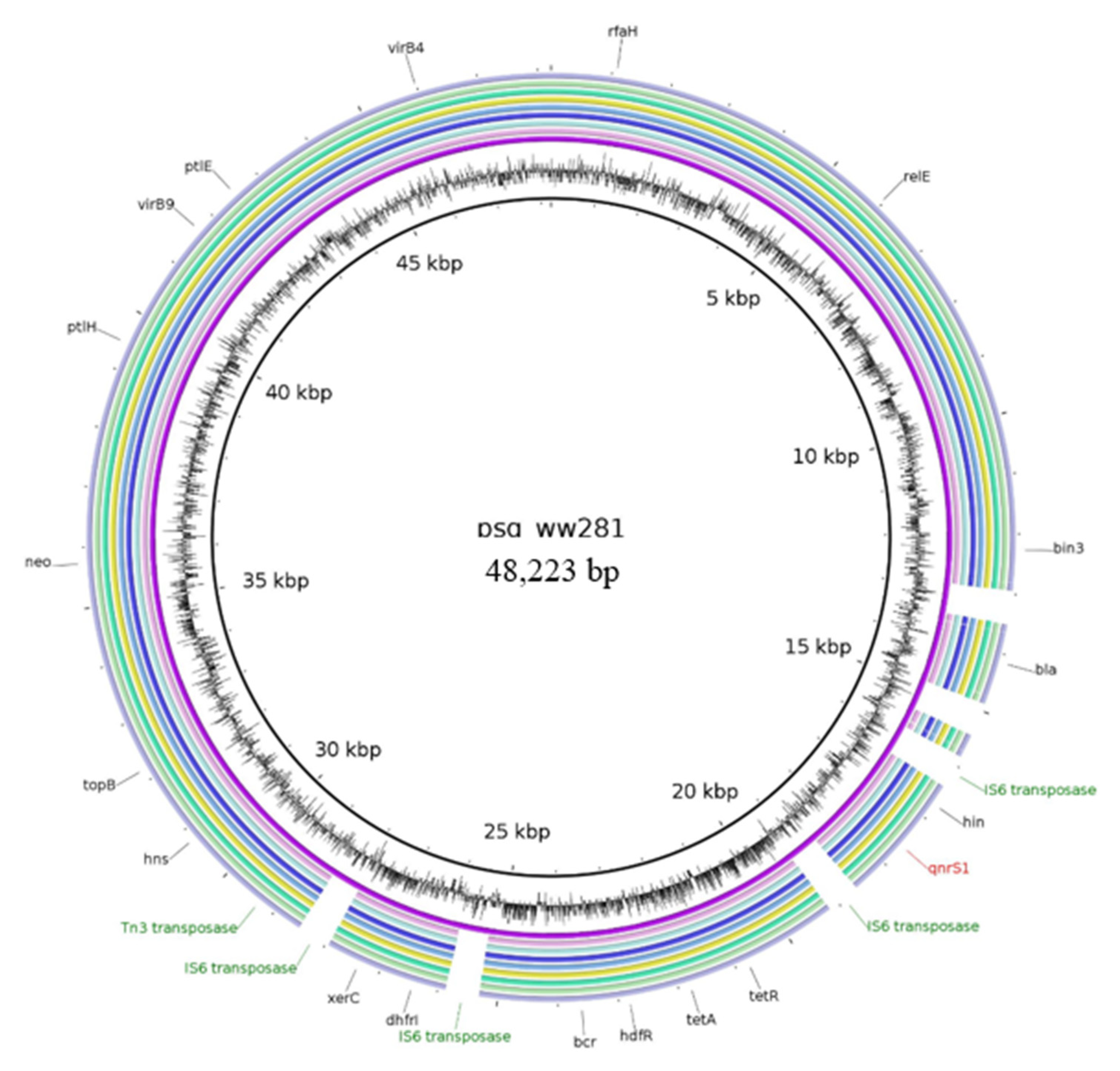
2.2.4. Characteristics of Plasmids Assigned to the Reference Plasmid psg_ww281
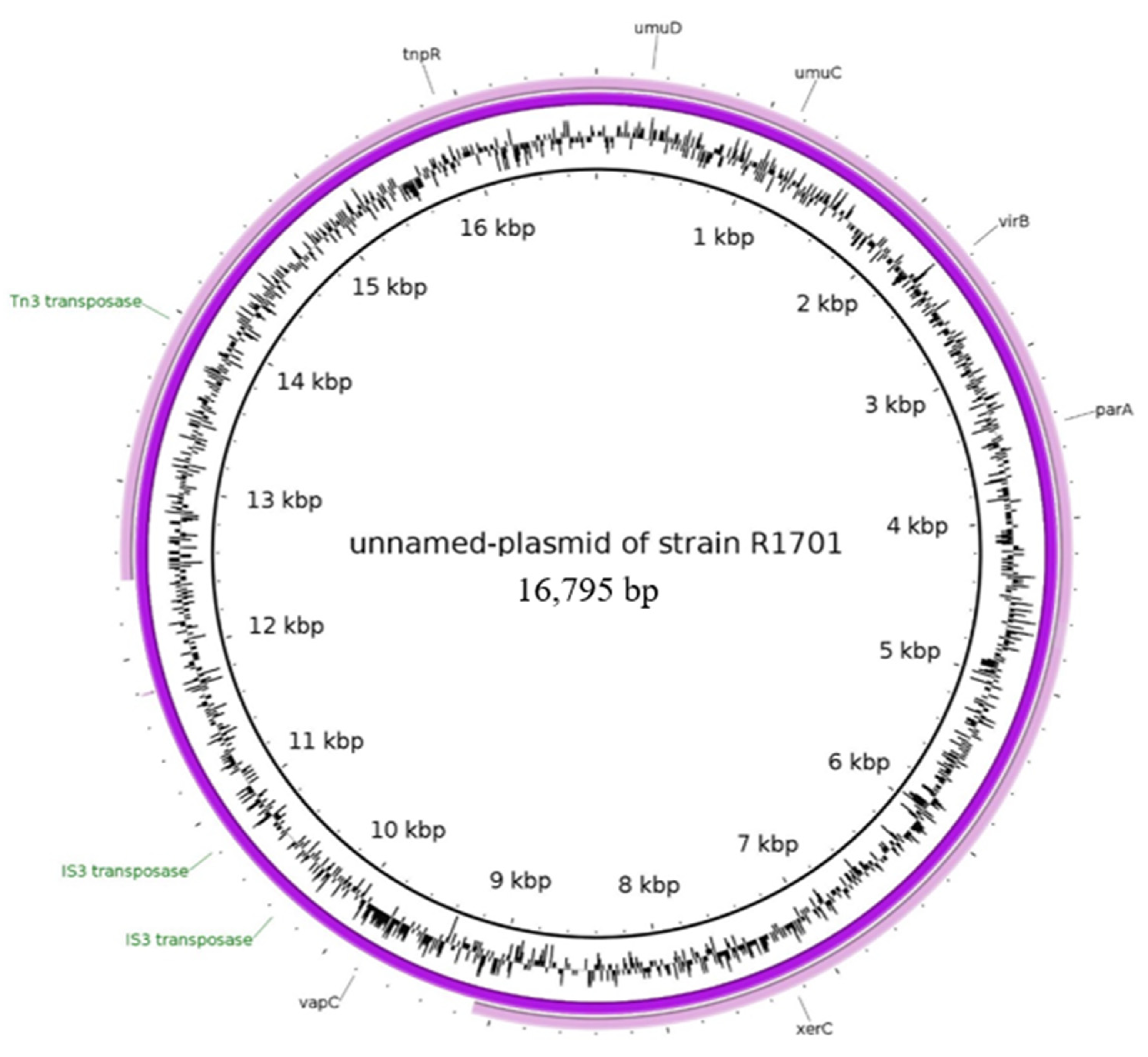
2.2.5. Characteristics of the Plasmid Assigned to the Unnamed Reference Plasmid of the Strain R1701
3. Materials and Methods
3.1. Isolate Characterization
3.2. DNA Extraction and Sequencing
3.3. Bioinformatics Analysis, Characterization and Visualization of the WGS Data
3.4. Conjugational Test
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Slettemeås, J.S.; Sunde, M.; Ulstad, C.R.; Norström, M.; Wester, A.L.; Urdahl, A.M. Occurrence and characterization of quinolone resistant Escherichia coli from Norwegian turkey meat and complete sequence of an IncX1 plasmid encoding qnrS1. PLoS ONE 2019, 14, e0212936. [Google Scholar] [CrossRef]
- WHO. Critically Important Antimicrobials for Human Medicine, 5th ed.; WHO: Geneva, Switzerland, 2017; p. 48. [Google Scholar]
- NORM/NORM-VET. Usage of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Norway; NORM-VET: Panama City Beach, FL, USA, 2016; pp. 1890–9965. [Google Scholar]
- Kaspersen, H.; Urdahl, A.M.; Simm, R.; Slettemeås, J.S.; Lagesen, K.; Norström, M. Occurrence of quinolone resistant E. coli originating from different animal species in Norway. Vet. Microbiol. 2018, 217, 25–31. [Google Scholar] [CrossRef]
- Yue, L.; Jiang, H.-X.; Liao, X.-P.; Liu, J.-H.; Li, S.-J.; Chen, X.-Y.; Chen, C.-X.; Lü, D.-H.; Liu, Y.-H. Prevalence of plasmid-mediated quinolone resistance qnr genes in poultry and swine clinical isolates of Escherichia coli. Vet. Microbiol. 2008, 132, 414–420. [Google Scholar] [CrossRef]
- Dolejska, M.; Duskova, E.; Rybarikova, J.; Janoszowska, D.; Roubalova, E.; Dibdakova, K.; Maceckova, G.; Kohoutova, L.; Literak, I.; Smola, J.; et al. Plasmids carrying blaCTX-M-1 and qnr genes in Escherichia coli isolates from an equine clinic and a horseback riding centre. J. Antimicrob. Chemother. 2011, 66, 757–764. [Google Scholar] [CrossRef]
- Monte, D.F.; Lincopan, N.; Berman, H.; Cerdeira, L.; Keelara, S.; Thakur, S.; Cray, P.; Landgraf, M. Genomic Features of High-Priority Salmonella enterica Serovars Circulating in the Food Production Chain, Brazil, 2000–2016. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Klemm, E.J.; Shakoor, S.; Page, A.J.; Qamar, F.N.; Judge, K.; Saeed, D.K.; Wong, V.K.; Dallman, T.J.; Nair, S.; Baker, S.; et al. Emergence of an Extensively Drug-Resistant Salmonella enterica Serovar Typhi Clone Harboring a Promiscuous Plasmid Encoding Resistance to Fluoroquinolones and Third-Generation Cephalosporins. mBio 2018, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kohler, V.; Vaishampayan, A.; Grohmann, E. Broad-host-range Inc18 plasmids: Occurrence, spread and transfer mechanisms. Plasmid 2018, 99, 11–21. [Google Scholar] [CrossRef]
- Hegstad, K.; Mikalsen, T.; Coque, T.M.; Werner, G.; Sundsfjord, A. Mobile genetic elements and their contribution to the emergence of antimicrobial resistant Enterococcus faecalis and Enterococcus faecium. Clin. Microbiol. Infect. 2010, 16, 541–554. [Google Scholar] [CrossRef] [PubMed]
- Iranzo, J.; Puigbò, P.; Lobkovsky, A.E.; Wolf, Y.I.; Koonin, E.V. Inevitability of Genetic Parasites. Genome Biol. Evol. 2016, 8, 2856–2869. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V. Horizontal gene transfer: Essentiality and evolvability in prokaryotes, and roles in evolutionary transitions. F1000Research 2016, 5, 1805. [Google Scholar] [CrossRef]
- Hall, J.P.J.; Brockhurst, M.A.; Harrison, E. Sampling the mobile gene pool: Innovation via horizontal gene transfer in bacteria. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160424. [Google Scholar] [CrossRef] [PubMed]
- Redondo-Salvo, S.; Fernández-López, R.; Ruiz, R.; Vielva, L.; de Toro, M.; Rocha, E.P.C.; Garcillán-Barcia, M.P.; De La Cruz, F. Pathways for horizontal gene transfer in bacteria revealed by a global map of their plasmids. Nat. Commun. 2020, 11, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Cerquetti, M.; García-Fernández, A.; Giufrè, M.; Fortini, D.; Accogli, M.; Graziani, C.; Luzzi, I.; Caprioli, A.; Carattoli, A. First Report of Plasmid-Mediated Quinolone Resistance Determinant qnrS1 in an Escherichia coli Strain of Animal Origin in Italy. Antimicrob. Agents Chemother. 2009, 53, 3112–3114. [Google Scholar] [CrossRef] [PubMed]
- Literak, I.; Literak, I.; Dolejska, M.; Janoszowska, D.; Hrusakova, J.; Meissner, W.; Rzyska, H.; Bzoma, S.; Cizek, A. Antibiotic-resistant Escherichia coli bacteria, including strains with genes encoding the extended-spectrum be-ta-lactamase and QnrS, in waterbirds on the Baltic Sea Coast of Poland. Appl. Environ. Microbiol. 2010, 76, 8126–8134. [Google Scholar] [CrossRef]
- Dobiasova, H.; Dolejska, M.; Jamborova, I.; Brhelova, E.; Blazkova, L.; Papousek, I.; Kozlova, M.; Klimes, J.; Cizek, A.; Literak, I. Extended spectrum beta-lactamase and fluoroquinolone resistance genes and plasmids among Escherichia coli isolates from zoo animals, Czech Republic. FEMS Microbiol. Ecol. 2013, 85, 604–611. [Google Scholar] [CrossRef]
- Botelho, L.A.B.; Kraychete, G.B.; Costa e Silva, J.L.; Regis, D.V.V.; Picão, R.C.; Moreira, B.M.; Bonelli, R.R. Widespread distribution of CTX-M and plasmid-mediated AmpC beta-lactamases in Escherichia coli from Brazilian chicken meat. Mem. Inst. Oswaldo Cruz. 2015, 110, 249–254. [Google Scholar] [CrossRef]
- Niero, G.; Bortolaia, V.; Vanni, M.; Intorre, L.; Guardabassi, L.; Piccirillo, A. High diversity of genes and plasmids encoding resistance to third-generation cephalosporins and quinolones in clinical Escherichia coli from commercial poultry flocks in Italy. Vet. Microbiol. 2018, 216, 93–98. [Google Scholar] [CrossRef]
- Fortini, D.; Fashae, K.; García-Fernández, A.; Villa, L.; Carattoli, A. Plasmid-mediated quinolone resistance and -lactamases in Escherichia coli from healthy animals from Nigeria. J. Antimicrob. Chemother. 2011, 66, 1269–1272. [Google Scholar] [CrossRef]
- Veldman, K.; Van Essen-Zandbergen, A.; Kant, A.; Mevius, D. Characterization of qnr-positive Escherichia coli isolates from food-producing animals in the Netherlands. J. Antimicrob. Chemother. 2011, 67, 239–240. [Google Scholar] [CrossRef]
- Mshana, S.E.; Falgenhauer, L.; Mirambo, M.M.; Mushi, M.F.; Moremi, N.; Julius, R.; Seni, J.; Imirzalioglu, C.; Matee, M.; Chakraborty, T. Predictors of blaCTX-M-15 in varieties of Escherichia coli genotypes from humans in community settings in Mwanza, Tanzania. BMC Infect. Dis. 2016, 16, 187. [Google Scholar] [CrossRef]
- Leclercq, R.; Derlot, E.; Duval, J.; Courvalin, P. Plasmid-Mediated Resistance to Vancomycin and Teicoplanin in Enterococcus Faecium. N. Engl. J. Med. 1988, 319, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Strahilevitz, J.; Jacoby, G.A.; Hooper, D.C.; Robicsek, A. Plasmid-Mediated Quinolone Resistance: A Multifaceted Threat. Clin. Microbiol. Rev. 2009, 22, 664–689. [Google Scholar] [CrossRef] [PubMed]
- Machuca, J.; Ortiz, M.; Recacha, E.; Díaz-De-Alba, P.; Docobo-Pérez, F.; Rodríguez-Martínez, J.-M.; Pascual, Á. Impact of AAC(6′)-Ib-cr in combination with chromosomal-mediated mechanisms on clinical quinolone resistance in Escherichia coli. J. Antimicrob. Chemother. 2016, 71, 3066–3071. [Google Scholar] [CrossRef] [PubMed]
- Machuca, J.; Briales, A.; Díaz-De-Alba, P.; Martínez-Martínez, L.; Pascual, Á.; Rodríguez-Martínez, J.-M. Effect of the efflux pump QepA2 combined with chromosomally mediated mechanisms on quinolone resistance and bacterial fitness in Escherichia coli: Table 1. J. Antimicrob. Chemother. 2015, 70, 2524–2527. [Google Scholar] [CrossRef]
- Poirel, L.; Pitout, J.D.; Calvo, L.; Rodriguez-Martinez, J.M.; Church, D.; Nordmann, P. In vivo selection of fluoroquinolone-resistant Escherichia coli isolates expressing plasmid-mediated quinolone resistance and expanded-spectrum beta-lactamase. Antimicrob. Agents Chemother. 2006, 50, 1525–1527. [Google Scholar] [CrossRef][Green Version]
- Tran, J.H.; Jacoby, G.A.; Hooper, D.C. Interaction of the Plasmid-Encoded Quinolone Resistance Protein Qnr with Escherichia coli DNA Gyrase. Antimicrob. Agents Chemother. 2005, 49, 118–125. [Google Scholar] [CrossRef]
- Hata, M.; Suzuki, M.; Matsumoto, M.; Takahashi, M.; Sato, K.; Ibe, S.; Sakae, K. Cloning of a Novel Gene for Quinolone Resistance from a Transferable Plasmid in Shigella flexneri 2b. Antimicrob. Agents Chemother. 2005, 49, 801–803. [Google Scholar] [CrossRef]
- Jacoby, G.A.; Walsh, K.E.; Mills, D.M.; Walker, V.J.; Oh, H.; Robicsek, A.; Hooper, D.C. qnrB, Another Plasmid-Mediated Gene for Quinolone Resistance. Antimicrob. Agents Chemother. 2006, 50, 1178–1182. [Google Scholar] [CrossRef]
- Novick, R.; Hoppensteadt, F. On plasmid incompatibility. Plasmid 1978, 1, 421–434. [Google Scholar] [CrossRef]
- Dolejska, M.; Villa, L.; Minoia, M.; Guardabassi, L.; Carattoli, A. Complete sequences of IncHI1 plasmids carrying blaCTX-M-1 and qnrS1 in equine Escherichia coli provide new insights into plasmid evolution. J. Antimicrob. Chemother. 2014, 69, 2388–2393. [Google Scholar] [CrossRef]
- Guo, S.; Tay, M.Y.F.; Thu, A.K.; Seow, K.L.G.; Zhong, Y.; Ng, L.C.; Schlundt, J. Conjugative IncX1 Plasmid Harboring Colistin Resistance Gene mcr-5.1 in Escherichia coli Isolated from Chicken Rice Retailed in Singapore. Antimicrob. Agents Chemother. 2019, 63, e01043-19. [Google Scholar] [CrossRef] [PubMed]
- Rozwandowicz, M.; Brouwer, M.S.M.; Fischer, J.; Wagenaar, J.A.; Gonzalez-Zorn, B.; Guerra, B.; Mevius, D.J.; Hordijk, J. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 2018, 73, 1121–1137. [Google Scholar] [CrossRef] [PubMed]
- Dolejska, M.; Villa, L.; Hasman, H.; Hansen, L.H.; Carattoli, A. Characterization of IncN plasmids carrying blaCTX-M-1 and qnr genes in Escherichia coli and Salmonella from animals, the environment and humans. J. Antimicrob. Chemother. 2012, 68, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Su, J.; McElheny, C.L.; Stoesser, N.; Doi, Y.; Wang, M. IncX2 and IncX1-X2 Hybrid Plasmids Coexisting in a FosA6-Producing Escherichia coli Strain. Antimicrob. Agents Chemother. 2017, 61, e00536–e00617. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, J.; Wang, J.; Butaye, P.; Kelly, P.; Li, M.; Yang, F.; Gong, J.; Yassin, A.K.; Guo, W.; et al. Newly identified colistin resistance genes, mcr-4 and mcr-5, from upper and lower alimentary tract of pigs and poultry in China. PLoS ONE 2018, 13, e0193957. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Du, H.; Chen, L.; Chavda, K.D.; Pandey, R.; Zhang, H.; Xie, X.; Tang, Y.-W.; Kreiswirth, B.N. Genomic Characterization of Enterobacter cloacae Isolates from China That Coproduce KPC-3 and NDM-1 Car-bapenemases. Antimicrob. Agents Chemother. 2016, 60, 2519–2523. [Google Scholar] [CrossRef]
- Dobiasova, H.; Dolejska, M. Prevalence and diversity of IncX plasmids carrying fluoroquinolone and β-lactam resistance genes in Escherichia coli originating from diverse sources and geographical areas. J. Antimicrob. Chemother. 2016, 71, 2118–2124. [Google Scholar] [CrossRef]
- Liakopoulos, A.; van der Goot, J.; Bossers, A.; Betts, J.; Brouwer, M.S.M.; Kant, A.; Smith, H.; Ceccarelli, D.; Mevius, D. Genomic and functional characterisation of IncX3 plasmids encoding blaSHV-12 in Escherichia coli from human and animal origin. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.-H.; Liu, P.-P.; Wei, D.-D.; Liu, Y.; Wan, L.-G.; Xiang, T.-X.; Zhang, Y.-J. Clinical isolates of uropathogenic Escherichia coli ST131 producing NDM-7 metallo-β-lactamase in China. Int. J. Antimicrob. Agents 2016, 48, 41–45. [Google Scholar] [CrossRef]
- Ford, P.J.; Avison, M.B. Evolutionary mapping of the SHV beta-lactamase and evidence for two separate IS26-dependent blaSHV mobilization events from the Klebsiella pneumoniae chromosome. J. Antimicrob. Chemother. 2004, 54, 69–75. [Google Scholar] [CrossRef]
- Papagiannitsis, C.C.; Študentová, V.; Chudáčková, E.; Bergerová, T.; Hrabák, J.; Radej, J.; Novak, I. Identification of a New Delhi metallo-β-lactamase-4 (NDM-4)-producing Enterobacter cloacae from a Czech patient previously hospitalized in Sri Lanka. Folia Microbiol. 2013, 58, 547–549. [Google Scholar] [CrossRef] [PubMed]
- Espedido, B.A.; Dimitrijovski, B.; van Hal, S.; Jensen, S.O. The use of whole-genome sequencing for molecular epidemiology and antimicrobial surveillance: Identifying the role of IncX3 plasmids and the spread of blaNDM-4-like genes in the Enterobacteriaceae. J. Clin. Pathol. 2015, 68, 835–838. [Google Scholar] [CrossRef] [PubMed]
- Elena, A.; Quinteros, M.; Di Conza, J.; Gutkind, G.; Cejas, D.; Radice, M.A. Full characterization of an IncR plasmid harboring qnrS1 recovered from a VIM-11-producing Pseudomonas aeruginosa. Rev. Argent. Microbiol. 2020, 52, 298–304. [Google Scholar] [CrossRef]
- Jelocnik, M.; Bachmann, N.L.; Seth-Smith, H.; Thomson, N.R.; Timms, P.; Polkinghorne, A.M. Molecular characterisation of the Chlamydia pecorum plasmid from porcine, ovine, bovine, and koala strains indicates plasmid-strain co-evolution. Peer J. 2016, 4, e1661. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dortet, L.; Bonnin, R.A.; Pennisi, I.; Gauthier, L.; Jousset, A.B.; Dabos, L.; Furniss, R.C.D.; Mavridou, A.I.; Bogaerts, P.; Glupczynski, Y.; et al. Rapid detection and discrimination of chromosome- and MCR-plasmid-mediated resistance to polymyxins by MAL-DI-TOF MS in Escherichia coli: The MALDIxin test. J. Antimicrob. Chemother. 2018, 73, 3359–3367. [Google Scholar]
- Cattoir, V.; Poirel, L.; Rotimi, V.; Soussy, C.-J.; Nordmann, P. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J. Antimicrob. Chemother. 2007, 60, 394–397. [Google Scholar] [CrossRef] [PubMed]
- CDC. Standard Operating Procedure for PulseNet PFGE of Escherichia coli O157:H7, Escherichia coli non-O157 (STEC), Salmonella serotypes, Shigella sonnei and Shigella flexneri; PulseNet Methods & Protocols: Atlanta, GA, USA, 2017. [Google Scholar]
- Borowiak, M.; Fischer, J.; Hammerl, J.A.; Hendriksen, R.S.; Szabo, I.; Malorny, B. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J. Antimicrob. Chemother. 2017, 72, 3317–3324. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Deneke, C. BakCharak. 2018. Available online: https://gitlab.com/bfr_bioinformatics/bakcharak (accessed on 1 September 2020).
- Alikhan, N.-F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef]
- NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018, 46, D8–D13. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed]
- GeneiousPrime, version 2020.2.2. Software: Geneious Prime. The Biomatters Develoment Team: Auckland, New Zealand, 2020.
- Robertson, J.B.K. Mob-Suite. 2020. Available online: https://github.com/phac-nml/mob-suite (accessed on 16 August 2021).
- Irrgang, A.; Pauly, N.; Tenhagen, B.-A.; Grobbel, M.; Kaesbohrer, A.; Hammerl, A.J.A. Spill-Over from Public Health? First Detection of an OXA-48-Producing Escherichia coli in a German Pig Farm. Microorganisms 2020, 8, 855. [Google Scholar] [CrossRef] [PubMed]
- Hammerl, J.A.; Klein, I.; Lanka, E.; Appel, B.; Hertwig, S. Genetic and Functional Properties of the Self-Transmissible Yersinia enterocolitica Plasmid pYE854, Which Mobilizes the Virulence Plasmid pYV. J. Bacteriol. 2008, 190, 991–1010. [Google Scholar] [CrossRef] [PubMed]

| Isolate | ST | Resistance Genes * | Source | Phenotypic Resistance Profile |
|---|---|---|---|---|
| 17-AB00542 | 1288 | aph(3′′)-Ib, aph(6)-Id, blaEC, blaTEM-1, qnrS1, tet(B) | calf, faeces | TET |
| 17-AB00544 | 155 | aph(3′)-Ia, blaEC-18, blaTEM-176, dfrA14, floR+, qnrS1, tet(A) | calf, faeces | AMP, CHL, CIP, TET, TMP |
| 17-AB00639 | 10 | aac(3)-IVa, aph(3″)-Ib, aph(4)-Ia, aph(6)-Id, blaCTX-M-1, blaEC, blaTEM-1, dfrA5, mph(A), qnrS1, sul2 | pig, faeces | AMP, CIP, FOT, GEN, SMX, TAZ, TMP |
| 17-AB00742 | 10 | aadA1, blaEC-15, blaTEM-1, qnrS1 | pig, faeces | AMP, CIP |
| 17-AB00995 | 392 | aph(3″)-Ib, aph(6)-Id, blaEC-18, blaTEM-1, qnrS1, sul2, tet(B) | calf, faeces | AMP, CIP, SMX, TET |
| 17-AB01005 | 1244 | aadA1, aph(3″)-Ib, aph(6)-Id, blaEC, blaSHV-12, qnrS1, tet(B) | calf, faeces | FEP, FOT, TAZ |
| 17-AB01006 | 10 | blaEC-15, blaSHV-12, qnrS1 | calf, faeces | FEP, FOT, TAZ |
| 17-AB01018 | 88 | aph(3″)-Ib, aph(3′)-Ia, aph(6)-Id, blaEC-13, blaSHV-12, blaTEM-1, dfrA5, floR, qnrS1, sul2, tet(A) | pig, faeces | AMP, CHL, CIP, FOT, SMX, TAZ, TET, TMP |
| 17-AB01105 | 58 | aadA5, blaCTX-M-1, blaEC-18, blaTEM-1, dfrA17, dfrA5, qnrS1, sul2, tet(A) | pig, faeces | FEP, FOT, TAZ |
| 17-AB01352 | 88 | blaEC-13, blaTEM-1, qnrS1, tet(A) | calf, meat | AMP, CIP, TET |
| 17-AB01531 | 34 | blaEC, blaTEM-1, qnrS1, sul2, | pig, faeces | AMP, CIP, SMX |
| 17-AB01539 | 10 | aadA1, blaEC, blaTEM-1, qnrS1 | pig, faeces | AMP, CIP |
| 17-AB01619 | 10 | aac(3)-IIa, aadA5, aph(3′)-Ia, blaCTX-M-15, blaEC, blaTEM-176, dfrA14, dfrA17, floR+, mph(A), qacE∆1, qnrS1, sul1, sul2, tet(A), tet(B) | calf, faeces | AMP, AZI, CHL, CIP, FOT, GEN, NAL, SMX, TAZ, TET, TMP |
| 17-AB01686 | 1288 | aph(3″)-Ib, aph(3′)-Ia, aph(6)-Id, blaCTX-M-15, blaEC, blaTEM-176, dfrA14, floR+, qnrS1, tet(A), tet(B) | calf, faeces | AMP, CHL, CIP, FOT, SMX, TAZ, TET, TMP |
| 17-AB01707 | 10 | aph(3′)-Ia, blaEC, blaTEM-176, dfrA14, floR+, qnrS1, tet(A) | calf, faeces | AMP, CHL, CIP, TET, TMP |
| 17-AB01752 | 641 | aph(3″)-Ib, aph(6)-Id, blaEC-13, blaTEM-1, qnrS1, tet(B) | pig, faeces | AMP, CIP, TET |
| 17-AB01792 | 101 | aadA5, blaCTX-M-1, blaEC-18, blaTEM-1, dfrA17, qnrS1, sul2 | pig, faeces | FEP, FOT, TAZ |
| 17-AB01795 | 10 | blaEC-15, blaTEM-1, qnrS1 | pig, faeces | AMP, CIP |
| 17-AB01798 | 641 | aadA1, aadA2, aph(3″)-Ib, aph(6)-Id, blaEC-13, blaSHV-12, blaTEM-1, cmlA1, dfrA32, ere(A), mef(B), qacE∆1, qacL, qnrS1, sul1, sul3, tet(A) | pig, faeces | FEP, FOT, TAZ |
| 17-AB01875 | 10 | aph(3′)-Ia, blaCTX-M-1, blaEC, blaTEM-176, dfrA14, floR+, mph(A), qnrS1, tet(A) | calf, faeces | AMP, CHL, CIP, FOT, TAZ, TET, TMP |
| 17-AB01969 | 711 | blaEC-18, blaTEM-1, mph(A), mph(E), msr(E), qnrS1 | calf, faeces | AMP, AZI, CIP |
| 17-AB02071 | 58 | blaEC-18, blaSHV-12, qnrS1 | calf, faeces | AMP, CIP, FOT, TAZ |
| 17-AB02090 | 48 | blaCTX-M-1, blaEC-15, blaTEM-1, qnrS1 | calf, faeces | AMP, CIP, FOT, TAZ |
| 17-AB02355 | 2230 | aadA1, aadA2, aph(6)-Id, blaEC-13, blaSHV-12, blaTEM-1, cmlA1, dfrA14, qacL, qnrS1, sul2, sul3, tet(A) | pig, faeces | AMP, CIP, FOT, SMX, TAZ, TET, TMP |
| 17-AB02707 | 58 | aac(3)-IVa, aph(3″)-Ib, aph(6)-Id, blaCTX-M-1, blaEC-18, blaTEM-1, qnrS1 | calf, faeces | AMP, CIP, FOT, TAZ |
| 17-AB02711 | 7469 | aadA2, aph(3′)-Ia, blaCTX-M-1, blaEC-8, blaTEM-176, dfrA14, floR+, lnu(F), mph(A), qnrS1, tet(A) | calf, faeces | AMP, CIP, FOT, SMX, TAZ, TET, TMP |
| 17-AB02721 | 7469 | aadA2, aph(3′)-Ia, blaCTX-M-1, blaEC-8, blaTEM-176, dfrA14, floR+, lnu(F), mph(A), qnrS1, tet(A) | calf, faeces | FEP, FOT, TAZ |
| 17-AB02726 | 7469 | aadA2, aph(3′)-Ia, blaCTX-M-1, blaEC-8, blaTEM-147, dfrA14, floR+, lnu(F), mph(A), qnrS1, tet(A) | calf, faeces | FEP, FOT, TAZ |
| 17-AB02951 | 2496 | aadA1, aadA5, aph(3″)-Ib, aph(6)-Id, blaCTX-M-1, blaEC-15, blaTEM-1, dfrA1, dfrA17, mef(C), mph(B), mph(G), qac∆1, qnrS1, sul1, sul2 | pig, faeces | FEP, FOT, TAZ |
| Plasmid Name | Reference Plasmid | AMR Genes | Inc | bp | |
|---|---|---|---|---|---|
 | unnamed plasmid of strain R17071 | NZ_CP039972.1 | blaTEM * | IncR, IncX1 | 16,795 |
| pKpvST101_6 | NZ_CP031373.1 | blaSHV, qnrS1 | IncX3 | 43,670 | |
| unnamed plasmid of strain 0670 | NZ_CP020088.1 | blaTEM-1, qnrS1 | IncX1, IncX3 | 47,674 | |
| psg_ww281 plasmid | NZ_CP037995.1 | aph(3′)-Ia, blaTEM-176, dfrA14, floR, qnrS1, tet(A) | IncX1 | 48,223 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Juraschek, K.; Käsbohrer, A.; Malorny, B.; Schwarz, S.; Meemken, D.; Hammerl, J.A. Dissection of Highly Prevalent qnrS1-Carrying IncX Plasmid Types in Commensal Escherichia coli from German Food and Livestock. Antibiotics 2021, 10, 1236. https://doi.org/10.3390/antibiotics10101236
Juraschek K, Käsbohrer A, Malorny B, Schwarz S, Meemken D, Hammerl JA. Dissection of Highly Prevalent qnrS1-Carrying IncX Plasmid Types in Commensal Escherichia coli from German Food and Livestock. Antibiotics. 2021; 10(10):1236. https://doi.org/10.3390/antibiotics10101236
Chicago/Turabian StyleJuraschek, Katharina, Annemarie Käsbohrer, Burkhard Malorny, Stefan Schwarz, Diana Meemken, and Jens André Hammerl. 2021. "Dissection of Highly Prevalent qnrS1-Carrying IncX Plasmid Types in Commensal Escherichia coli from German Food and Livestock" Antibiotics 10, no. 10: 1236. https://doi.org/10.3390/antibiotics10101236
APA StyleJuraschek, K., Käsbohrer, A., Malorny, B., Schwarz, S., Meemken, D., & Hammerl, J. A. (2021). Dissection of Highly Prevalent qnrS1-Carrying IncX Plasmid Types in Commensal Escherichia coli from German Food and Livestock. Antibiotics, 10(10), 1236. https://doi.org/10.3390/antibiotics10101236







