Integrating the Human and Animal Sides of Mycoplasmas Resistance to Antimicrobials
Abstract
:1. Introduction
1.1. Human Mycoplasmas
1.2. Animal Mycoplasmas
2. Assumed Active Antimicrobials and Usage
2.1. Intrinsic Resistance
2.2. Which Molecules Are Assumed to Be Active?
2.3. How Are They Used?
3. Mechanisms of Acquired Resistance
3.1. Mechanisms
3.2. Genetic Support
3.2.1. Chromosomal Mutations
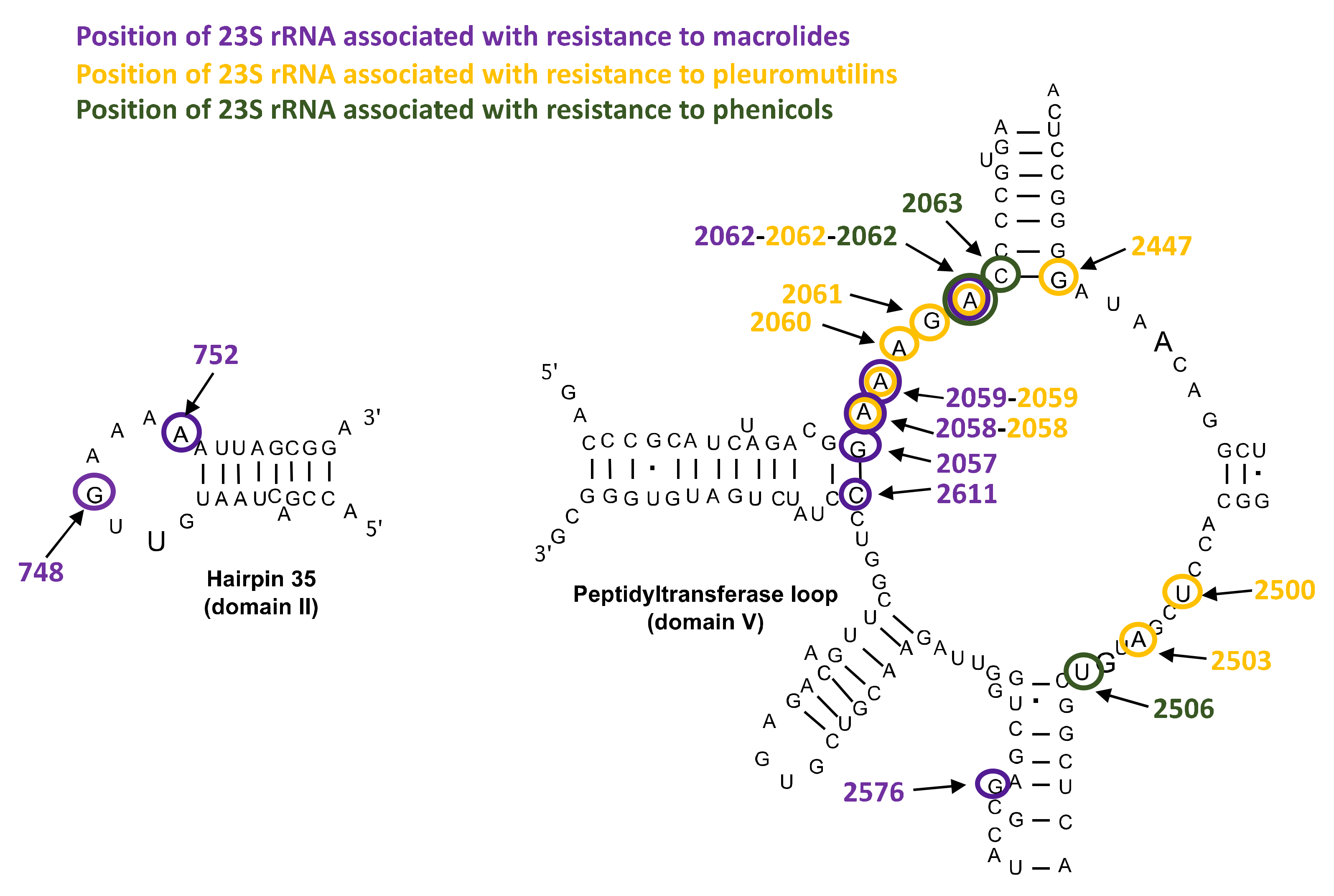
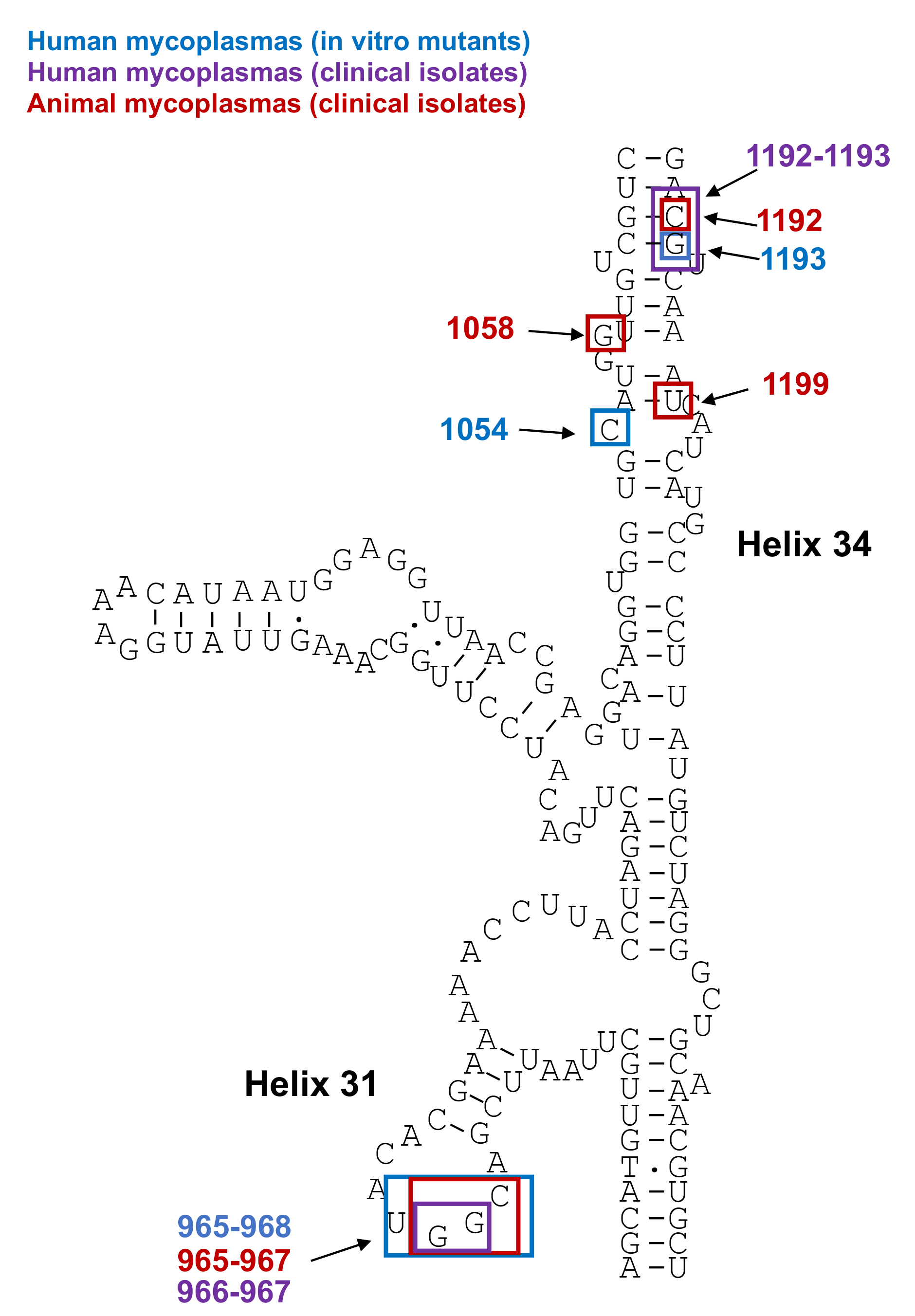
Mutations in 23S rRNA and in Ribosomal Proteins L4 and L22
Mutations in DNA Gyrase and Topoisomerase IV
Mutations in 16S rRNA
3.2.2. Acquisition of Mobile Genetic Elements
4. Susceptibility Testing and Epidemiology of Resistance
4.1. Methods for Determination of Antimicrobial Susceptibility
4.1.1. Phenotypic Methods
4.1.2. Genotypic Methods
4.2. Epidemiology of Resistance
4.2.1. Human Mycoplasmas
Prevalence of Resistance in M. pneumoniae
Prevalence of Resistance in Ureaplasma spp. and M. hominis
Prevalence of Resistance in M. genitalium
4.2.2. Animal Mycoplasmas
5. Critical Cases of Multiple Drug Resistance: Examples of Two Mycoplasma “Super” Bugs, M. bovis and M. genitalium
5.1. M. genitalium
5.2. M. bovis
6. Conclusions
- A lack of harmonized methodologies for AST of animal mycoplasmas, as well as the absence of clinical breakpoints, preventing data interpretation.
- The unavailability of European clinical breakpoints for human mycoplasmas that might be more adapted to European practices
- A lack of antibioresistance data on some specific mycoplasmas of cats, dogs and horses, as well as on non-cultivable haemotrophic mycoplasmas.
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Razin, S.; Yogev, D.; Naot, Y. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 1998, 62, 1094–1156. [Google Scholar] [CrossRef] [Green Version]
- Di Teodoro, G.; Marruchella, G.; Di Provvido, A.; D’Angelo, A.R.; Orsini, G.; Di Giuseppe, P.; Sacchini, F.; Scacchia, M. Contagious bovine pleuropneumonia: A comprehensive overview. Vet. Pathol. 2020, 57, 476–489. [Google Scholar] [CrossRef]
- Razin, S.; Hayflick, L. Highlights of mycoplasma research—An historical perspective. Biologicals 2010, 38, 183–190. [Google Scholar] [CrossRef]
- Brown, D.R. Phylum XVI. Tenericutes Murray 1984a, 356 VP. In Bergey’s Manual® of Systematic Bacteriology; Springer: New York, NY, USA, 2010. [Google Scholar]
- Matet, A.; Le Flèche-Matéos, A.; Doz, F.; Dureau, P.; Cassoux, N. Ocular Spiroplasma ixodetis in Newborns, France. Emerg. Infect. Dis. 2020, 26, 340–344. [Google Scholar] [CrossRef] [Green Version]
- Etienne, N.; Bret, L.; Le Brun, C.; Lecuyer, H.; Al, N.E.E.; Lanternier, F.; Hermine, O.; Ferroni, A.; Lecuit, M.; Pereyre, S.; et al. Disseminated Spiroplasma apis Infection in Patient with Agammaglobulinemia, France. Emerg. Infect. Dis. 2018, 24, 2382–2386. [Google Scholar] [CrossRef] [PubMed]
- Aquilino, A.; Masiá, M.; López, P.; Galiana, A.J.; Tovar, J.; Andrés, M.; Gutiérrez, F. First Human Systemic Infection Caused by Spiroplasma. J. Clin. Microbiol. 2014, 53, 719–721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, M.; Hitomi, S.; Goto, M.; Hasegawa, Y. Bloodstream Infection Due to Mycoplasma arginini in an Immunocompromised Patient. J. Clin. Microbiol. 2012, 50, 3133–3135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heller, M.; Schwarz, R.; Noe, G.; Jores, J.; Fischer, A.; Schubert, E.; Sachse, K. First human case of severe septicaemia associated with Mycoplasma capricolum subsp. capricolum infection. JMM Case Rep. 2015, 2. [Google Scholar] [CrossRef]
- White, C.P.; Jewer, D.D. Seal finger: A case report and review of the literature. Can. J. Plast. Surg. 2009, 17, 133–135. [Google Scholar] [CrossRef]
- Yuan, C.L.; Liang, A.B.; Yao, C.B.; Yang, Z.B.; Zhu, J.G.; Cui, L.; Yu, F.; Zhu, N.Y.; Yang, X.W.; Hua, X.G. Prevalence of Mycoplasma suis (Eperythrozoon suis) infection in swine and swine-farm workers in Shanghai, China. Am. J. Vet. Res. 2009, 70, 890–894. [Google Scholar] [CrossRef]
- Messick, J.B. Hemotrophic mycoplasmas (hemoplasmas): A review and new insights into pathogenic potential. Vet. Clin. Pathol. 2004, 33, 2–13. [Google Scholar] [CrossRef]
- Waites, K.; Talkington, D. New developments in human diseases due to mycoplasmas. In Mycoplasmas Molecular Biology Pathogenicity and Strategies for Control; Blanchard, A., Browning, G.F., Eds.; Horizon Bioscience: Norfolk, UK, 2005; pp. 289–354. [Google Scholar]
- Rosales, R.S.; Puleio, R.; Loria, G.R.; Catania, S.; Nicholas, R.A. Mycoplasmas: Brain invaders? Res. Vet. Sci. 2017, 113, 56–61. [Google Scholar] [CrossRef]
- Browning, G.F.; Noormohammadi, A.H.; Markham, P.F. Identification and characterization of virulence genes in mycoplasmas. In Mollicutes: Molecular Biology and Pathogenesis; Browning, G.F., Citti, C., Eds.; Caister Academic Press: Poole, UK, 2014; pp. 77–90. [Google Scholar]
- Hoelzle, K.; Ade, J.; Hoelzle, L.E. Persistence in Livestock Mycoplasmas—A Key Role in Infection and Pathogenesis. Curr. Clin. Microbiol. Rep. 2020, 7, 81–89. [Google Scholar] [CrossRef]
- Waites, K.B.; Xiao, L.; Liu, Y.; Balish, M.F.; Atkinson, T.P. Mycoplasma pneumoniae from the Respiratory Tract and Beyond. Clin. Microbiol. Rev. 2017, 30, 747–809. [Google Scholar] [CrossRef] [Green Version]
- Pereyre, S.; Bébéar, C.; Bébéar, C. Mycoplasma hominis, M. genitalium, Ureaplasma spp. In Antimicrobial Therapy and Vaccines; Yu, V.L., Ed.; Esun Technologies: Pittsburgh, PA, USA, 2015; Volume I: Microbes. [Google Scholar]
- Horner, P.; Donders, G.; Cusini, M.; Gomberg, M.; Jensen, J.; Unemo, M. Should we be testing for urogenital Mycoplasma hominis, Ureaplasma parvum and Ureaplasma urealyticum in men and women?—A position statement from the European STI Guidelines Editorial Board. J. Eur. Acad. Dermatol. Venereol. 2018, 32, 1845–1851. [Google Scholar] [CrossRef] [Green Version]
- Waites, K.B.; Katz, B.; Schelonka, R.L. Mycoplasmas and Ureaplasmas as Neonatal Pathogens. Clin. Microbiol. Rev. 2005, 18, 687–702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jensen, J.; Cusini, M.; Gomberg, M.; Moi, H. 2016 European guideline on Mycoplasma genitalium infections. J. Eur. Acad. Dermatol. Venereol. 2016, 30, 1650–1656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srinivasan, S.; Chambers, L.C.; Tapia, K.A.; Hoffman, N.G.; Munch, M.M.; Morgan, J.L.; Domogala, D.; Lowens, M.S.; Proll, S.; Huang, M.-L.; et al. Urethral Microbiota in Men: Association of Haemophilus influenzae and Mycoplasma penetrans With Nongonococcal Urethritis. Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Neimark, H.; Johansson, K.E.; Rikihisa, Y.; Tully, J.G. Proposal to transfer some members of the genera Haemobartonella and Eperythrozoon to the genus Mycoplasma with descriptions of ‘Candidatus Mycoplasma haemofelis’, ‘Candidatus Mycoplasma haemomuris’, ‘Candidatus Mycoplasma haemosuis’ and ‘Candidatus Mycoplasma wenyonii’. Int. J. Syst. Evol. Microbiol. 2001, 51, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Ade, J.; Niethammer, F.; Schade, B.; Schilling, T.; Hoelzle, K.; Hoelzle, L.E. Quantitative analysis of Mycoplasma wenyonii and ‘Candidatus Mycoplasma haemobos” infections in cattle using novel gapN-based realtime PCR assays. Vet. Microbiol. 2018, 220, 1–6. [Google Scholar] [CrossRef]
- Neimark, H.; Hoff, B.; Ganter, M. Mycoplasma ovis comb. nov. (formerly Eperythrozoon ovis), an epierythrocytic agent of haemolytic anaemia in sheep and goats. Int. J. Syst. Evol. Microbiol. 2004, 54, 365–371. [Google Scholar] [CrossRef]
- Bébéar, C.M.; Kempf, I. Antimicrobial therapy and antimicrobial resistance. In Mycoplasmas Molecular Biology Pathogenicity and Strategies for Control; Blanchard, A., Browning, G.F., Eds.; Horizon Bioscience: Norfolk, UK, 2005; pp. 535–568. [Google Scholar]
- Gautier-Bouchardon, A.V. Antimicrobial resistance in Mycoplasma spp. Microbiol. Spectr. 2018, 6, 4. [Google Scholar] [CrossRef]
- Schultz, K.; Strait, E.; Erickson, B.; Levy, N. Optimization of an antibiotic sensitivity assay for Mycoplasma hyosynoviae and susceptibility profiles of field isolates from 1997 to 2011. Vet. Microbiol. 2012, 158, 104–108. [Google Scholar] [CrossRef] [PubMed]
- Kenny, G.E.; Hooton, T.M.; Roberts, M.C.; Cartwright, F.D.; Hoyt, J. Susceptibilities of genital mycoplasmas to the newer quinolones as determined by the agar dilution method. Antimicrob. Agents Chemother. 1989, 33, 103–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hannan, P.C.; O’Hanlon, P.J.; Rogers, N.H. In vitro evaluation of various quinolone antibacterial agents against veterinary mycoplasmas and porcine respiratory bacterial pathogens. Res. Vet. Sci. 1989, 46, 202–211. [Google Scholar] [CrossRef]
- Pereyre, S.; Gonzalez, P.; de Barbeyrac, B.; Darnige, A.; Renaudin, H.; Charron, A.; Raherison, S.; Bébéar, C. Mutations in 23S rRNA Account for Intrinsic Resistance to Macrolides in Mycoplasma hominis and Mycoplasma fermentans and for Acquired Resistance to Macrolides in M. hominis. Antimicrob. Agents Chemother. 2002, 46, 3142–3150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereyre, S.; Renaudin, H.; Charron, A.; Bébéar, C.M. Emergence of a 23S rRNA mutation in Mycoplasma hominis associated with a loss of the intrinsic resistance to erythromycin and azithromycin. J. Antimicrob. Chemother. 2006, 57, 753–756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organization. Critically Important Antimicrobials for Human Medicine; 6th Revision; World Health Organization: Geneva, Switzerland, 2019; Licence: CC BY-NC-SA 3.0 IGO. [Google Scholar]
- Waites, K.B.; Crabb, D.M.; Duffy, L.B.; Jensen, J.S.; Liu, Y.; Paukner, S. In Vitro Activities of Lefamulin and Other Antimicrobial Agents against Macrolide-Susceptible and Macrolide-Resistant Mycoplasma pneumoniae from the United States, Europe, and China. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paukner, S.; Gruss, A.; Jensen, J.S. In Vitro Activity of Lefamulin against Sexually Transmitted Bacterial Pathogens. Antimicrob. Agents Chemother. 2018, 62, e02380-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waites, K.B.; Crabb, D.M.; Xiao, L.; Duffy, L.B.; Leal, S.M. In Vitro Activities of Eravacycline and Other Antimicrobial Agents against Human Mycoplasmas and Ureaplasmas. Antimicrob. Agents Chemother. 2020, 64. [Google Scholar] [CrossRef]
- Jensen, J.S.; Fernandes, P.; Unemo, M. In VitroActivity of the New Fluoroketolide Solithromycin (CEM-101) against Macrolide-Resistant and -Susceptible Mycoplasma genitalium Strains. Antimicrob. Agents Chemother. 2014, 58, 3151–3156. [Google Scholar] [CrossRef] [Green Version]
- Jensen, J.S.; Nørgaard, C.; Scangarella-Oman, N.; Unemo, M. In vitro activity of the first-in-class triazaacenaphthylene gepotidacin alone and in combination with doxycycline against drug-resistant and -susceptible Mycoplasma genitalium. Emerg. Microbes Infect. 2020, 9, 1388–1392. [Google Scholar] [CrossRef] [PubMed]
- Gouveia, A.C.D.; Unemo, M.; Jensen, J.S. In vitro activity of zoliflodacin (ETX0914) against macrolide-resistant, fluoroquinolone-resistant and antimicrobial-susceptible Mycoplasma genitalium strains. J. Antimicrob. Chemother. 2018, 73, 1291–1294. [Google Scholar] [CrossRef]
- Dallo, S.; Baseman, J. Intracellular DNA replication and long-term survival of pathogenic mycoplasmas. Microb. Pathog. 2000, 29, 301–309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bürki, S.; Frey, J.; Pilo, P. Virulence, persistence and dissemination of Mycoplasma bovis. Vet. Microbiol. 2015, 179, 15–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogl, G.; Plaickner, A.; Szathmary, S.; Stipkovits, L.; Rosengarten, R.; Szostak, M.P. Mycoplasma gallisepticum Invades Chicken Erythrocytes during Infection. Infect. Immun. 2008, 76, 71–77. [Google Scholar] [CrossRef] [Green Version]
- McEwen, S.A.; Collignon, P.J. Antimicrobial resistance: A one health perspective. Microbiol. Spectr. 2018, 6, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agency, E.M. European Surveillance of Veterinary Antimicrobial Consumption. ‘Sales of Veterinary Antimicrobial Agents in 31 European Countries in 2018’; European Medicines Agency: Amsterdam, The Netherlands, 2020; EMA/24309/2020. [Google Scholar]
- De Briyne, N.; Atkinson, J.; Borriello, S.P.; Pokludová, L. Antibiotics used most commonly to treat animals in Europe. Vet. Rec. 2014, 175, 325. [Google Scholar] [CrossRef] [Green Version]
- Blanco, P.; Hernando-Amado, S.; Reales-Calderon, J.A.; Corona, F.; Lira, F.; Alcalde-Rico, M.; Bernardini, A.; Sanchez, M.B.; Martinez, J.L. Bacterial Multidrug Efflux Pumps: Much More Than Antibiotic Resistance Determinants. Microorganisms 2016, 4, 14. [Google Scholar] [CrossRef] [Green Version]
- Raherison, S.; Gonzalez, P.; Renaudin, H.; Charron, A.; Bébeér, C. Evidence of Active Efflux in Resistance to Ciprofloxacin and to Ethidium Bromide by Mycoplasma hominis. Antimicrob. Agents Chemother. 2002, 46, 672–679. [Google Scholar] [CrossRef] [Green Version]
- Antunes, N.T.; Assunção, P.; Poveda, J.B.; Tavío, M.M. Mechanisms involved in quinolone resistance in Mycoplasma mycoides subsp. capri. Vet. J. 2015, 204, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Li, S.L.; Sun, H.M.; Zhu, B.L.; Liu, F.; Zhao, H.Q. Whole Genome Analysis Reveals New Insights into Macrolide Resistance in Mycoplasma pneumoniae. Biomed. Environ. Sci. 2017, 30, 343–350. [Google Scholar] [PubMed]
- Tatay-Dualde, J.; der Ham, M.P.-V.; Gaurivaud, P.; de la Fe, C.; Tardy, F. Efflux Might Participate in Decreased Susceptibility to Oxytetracycline in Contagious Agalactia-Causative Mycoplasma spp. Animals 2021, 11, 2449. [Google Scholar] [CrossRef] [PubMed]
- Raherison, S.; Gonzalez, P.; Renaudin, H.; Charron, A.; Bébéar, C. Increased Expression of Two Multidrug Transporter-Like Genes Is Associated with Ethidium Bromide and Ciprofloxacin Resistance in Mycoplasma hominis. Antimicrob. Agents Chemother. 2005, 49, 421–424. [Google Scholar] [CrossRef] [Green Version]
- Zhang, N.; Wu, Y.; Huang, Z.; Zhang, C.-Z.; Zhang, L.; Cai, Q.; Shen, X.; Jiang, H.; Ding, H. Relationship between danofloxacin PK/PD parameters and emergence and mechanism of resistance of Mycoplasma gallisepticum in In Vitro model. PLoS ONE 2018, 13, e0202070. [Google Scholar] [CrossRef] [PubMed]
- Lysnyansky, I.; Borovok, I. A GC-rich prophage-like genomic region of Mycoplasma bovirhinis HAZ141_2 carries a gene cluster encoding resistance to kanamycin and neomycin. Antimicrob. Agents Chemother. 2021, 65, e01010-20. [Google Scholar] [CrossRef]
- Daubenspeck, J.M.; Totten, A.H.; Needham, J.; Feng, M.; Balish, M.F.; Atkinson, T.P.; Dybvig, K. Mycoplasma genitalium Biofilms Contain Poly-GlcNAc and Contribute to Antibiotic Resistance. Front. Microbiol. 2020, 11, 585524. [Google Scholar] [CrossRef]
- Feng, M.; Schaff, A.C.; Balish, M.F. Mycoplasma pneumoniae biofilms grown in vitro: Traits associated with persistence and cytotoxicity. Microbiology 2020, 166, 629–640. [Google Scholar] [CrossRef]
- Tassew, D.D.; Mechesso, A.F.; Park, N.-H.; Song, J.-B.; Shur, J.-W.; Park, S.-C. Biofilm formation and determination of minimum biofilm eradication concentration of antibiotics in Mycoplasma hyopneumoniae. J. Vet. Med. Sci. 2017, 79, 1716–1720. [Google Scholar] [CrossRef] [Green Version]
- Fürnkranz, U.; Walochnik, J.; Henrich, B. Mycoplasma hominis shows strain-dependent increase in resistance to selected antibiotics after symbiosis with Trichomonas vaginalis. J. Glob. Antimicrob. Resist. 2018, 14, 169–175. [Google Scholar] [CrossRef]
- Proctor, R.A.; Kriegeskorte, A.; Kahl, B.; Becker, K.; Lãffler, B.; Epeters, G. Staphylococcus aureus Small Colony Variants (SCVs): A road map for the metabolic pathways involved in persistent infections. Front. Cell. Infect. Microbiol. 2014, 4, 99. [Google Scholar] [CrossRef] [PubMed]
- Wagner, T.M.; Joshi, B.; Janice, J.; Askarian, F.; Skalko-Basnet, N.; Hagestad, O.C.; Mekhlif, A.; Wai, S.; Hegstad, K.; Johannessen, M. Enterococcus faecium produces membrane vesicles containing virulence factors and antimicrobial resistance related proteins. J. Proteom. 2018, 187, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Khalil, D.; Becker, C.A.; Tardy, F. Alterations in the Quinolone Resistance-Determining Regions and Fluoroquinolone Resistance in Clinical Isolates and Laboratory-Derived Mutants of Mycoplasma bovis: Not All Genotypes May Be Equal. Appl. Environ. Microbiol. 2016, 82, 1060–1068. [Google Scholar] [CrossRef] [Green Version]
- Der Ham, M.P.-V.; Tatay-Dualde, J.; Ambroset, C.; De la Fe, C.; Tardy, F. The moderate drift towards less tetracycline-susceptible isolates of contagious agalactia causative agents might result from different molecular mechanisms. Vet. Microbiol. 2018, 220, 39–46. [Google Scholar] [CrossRef]
- Rocha, E.P.C.; Blanchard, A. Genomic repeats, genome plasticity and the dynamics of Mycoplasma evolution. Nucleic Acids Res. 2002, 30, 2031–2042. [Google Scholar] [CrossRef]
- Chen, L.-L.; Chung, W.-C.; Lin, C.-P.; Kuo, C.-H. Comparative Analysis of Gene Content Evolution in Phytoplasmas and Mycoplasmas. PLoS ONE 2012, 7, e34407. [Google Scholar] [CrossRef] [Green Version]
- LeClerc, J.E.; Li, B.; Payne, W.L.; Cebula, T.A. High Mutation Frequencies Among Escherichia coli and Salmonella Pathogens. Science 1996, 274, 1208–1211. [Google Scholar] [CrossRef] [PubMed]
- Machalek, D.A.; Tao, Y.; Shilling, H.; Jensen, J.S.; Unemo, M.; Murray, G.; Chow, E.P.F.; Low, N.; Garland, S.M.; Vodstrcil, L.A.; et al. Prevalence of mutations associated with resistance to macrolides and fluoroquinolones in Mycoplasma genitalium: A systematic review and meta-analysis. Lancet Infect. Dis. 2020, 20, 1302–1314. [Google Scholar] [CrossRef]
- Pereyre, S.; Goret, J.; Bebear, C. Mycoplasma pneumoniae: Current Knowledge on Macrolide Resistance and Treatment. Front. Microbiol. 2016, 7, 974. [Google Scholar] [CrossRef] [Green Version]
- Pereyre, S.; Guyot, C.; Renaudin, H.; Charron, A.; Bébéar, C. In Vitro Selection and Characterization of Resistance to Macrolides and Related Antibiotics in Mycoplasma pneumoniae. Antimicrob. Agents Chemother. 2004, 48, 460–465. [Google Scholar] [CrossRef] [Green Version]
- Guschin, A.; Ryzhikh, P.; Rumyantseva, T.; Gomberg, M.; Unemo, M. Treatment efficacy, treatment failures and selection of macrolide resistance in patients with high load of Mycoplasma genitalium during treatment of male urethritis with josamycin. BMC Infect. Dis. 2015, 15, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tait-Kamradt, A.; Davies, T.; Cronan, M.; Jacobs, M.R.; Appelbaum, P.C.; Sutcliffe, J. Mutations in 23S rRNA and Ribosomal Protein L4 Account for Resistance in Pneumococcal Strains Selected In Vitro by Macrolide Passage. Antimicrob. Agents Chemother. 2000, 44, 2118–2125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prunier, A.-L.; Malbruny, B.; Tandé, D.; Picard, B.; Leclercq, R. Clinical Isolates of Staphylococcus aureus with Ribosomal Mutations Conferring Resistance to Macrolides. Antimicrob. Agents Chemother. 2002, 46, 3054–3056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereyre, S.; Métifiot, M.; Cazanave, C.; Renaudin, H.; Charron, A.; Bébéar, C. Characterisation of in vitro-selected mutants of Ureaplasma parvum resistant to macrolides and related antibiotics. Int. J. Antimicrob. Agents 2007, 29, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Waites, K.; Lysnyanski, I.; Bébéar, C. Emerging antimicrobial resistance in mycoplasmas of humans and animals. In Mollicutes: Molecular Biology and Pathogenesis; Browning, G.F., Citti, C., Eds.; Caister Academic Press: Norfolk, UK, 2014; pp. 289–322. [Google Scholar]
- Meygret, A.; Le Roy, C.; Renaudin, H.; Bébéar, C.; Pereyre, S. Tetracycline and fluoroquinolone resistance in clinical Ureaplasma spp. and Mycoplasma hominis isolates in France between 2010 and 2015. J. Antimicrob. Chemother. 2018, 73, 2696–2703. [Google Scholar] [CrossRef]
- Dégrange, S.; Renaudin, H.; Charron, A.; Pereyre, S.; Bébéar, C. Reduced susceptibility to tetracyclines is associated in vitro with the presence of 16S rRNA mutations in Mycoplasma hominis and Mycoplasma pneumoniae. J. Antimicrob. Chemother. 2008, 61, 1390–1392. [Google Scholar] [CrossRef]
- Le Roy, C.; Touati, A.; Balcon, C.; Garraud, J.; Molina, J.-M.; Berçot, B.; de Barbeyrac, B.; Pereyre, S.; Peuchant, O.; Bébéar, C. Identification of 16S rRNA mutations in Mycoplasma genitalium potentially associated with tetracycline resistance in vivo but not selected in vitro in M. genitalium and Chlamydia trachomatis. J. Antimicrob. Chemother. 2021, 76, 1150–1154. [Google Scholar] [CrossRef]
- Berçot, B.; Charreau, I.; Rousseau, C.; Delaugerre, C.; Chidiac, C.; Pialoux, G.; Capitant, C.; Bourgeois-Nicolaos, N.; Raffi, F.; Pereyre, S.; et al. High prevalence and high rate of antibiotic resistance of Mycoplasma genitalium infections in men who have sex with men: A substudy of the ANRS IPERGAY pre-exposure prophylaxis trial. Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Amram, E.; Mikula, I.; Schnee, C.; Ayling, R.D.; Nicholas, R.A.J.; Rosales, R.S.; Harrus, S.; Lysnyansky, I. 16S rRNA Gene Mutations Associated with Decreased Susceptibility to Tetracycline in Mycoplasma bovis. Antimicrob. Agents Chemother. 2014, 59, 796–802. [Google Scholar] [CrossRef] [Green Version]
- Khalil, D.; Becker, C.A.; Tardy, F. Monitoring the Decrease in Susceptibility to Ribosomal RNAs Targeting Antimicrobials and Its Molecular Basis in Clinical Mycoplasma bovis Isolates over Time. Microb. Drug Resist. 2017, 23, 799–811. [Google Scholar] [CrossRef]
- Chopra, I.; Roberts, M. Tetracycline antibiotics: Mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 2001, 65, 232–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donhofer, A.; Franckenberg, S.; Wickles, S.; Berninghausen, O.; Beckmann, R.; Wilson, D. Structural basis for TetM-mediated tetracycline resistance. Proc. Natl. Acad. Sci. USA 2012, 109, 16900–16905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kenny, G.E.; Cartwright, F.D. Susceptibilities of Mycoplasma hominis, M. pneumoniae, and Ureaplasma urealyticum to GAR-936, Dalfopristin, Dirithromycin, Evernimicin, Gatifloxacin, Linezolid, Moxifloxacin, Quinupristin-Dalfopristin, and Telithromycin Compared to Their Susceptibilities to Reference Macrolides, Tetracyclines, and Quinolones. Antimicrob. Agents Chemother. 2001, 45, 2604–2608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calcutt, M.J.; Foecking, M.F. An Excision-Competent and Exogenous Mosaic Transposon Harbors thetetMGene in Multiple Mycoplasma hominis Lineages. Antimicrob. Agents Chemother. 2015, 59, 6665–6666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chalker, V.J.; Sharratt, M.G.; Rees, C.L.; Bell, O.H.; Portal, E.; Sands, K.; Payne, M.S.; Jones, L.C.; Spiller, O.B. Tetracycline Resistance Mediated by tet (M) Has Variable Integrative Conjugative Element Composition in Mycoplasma hominis Strains Isolated in the United Kingdom from 2005 to 2015. Antimicrob. Agents Chemother. 2021, 65, e02513-20. [Google Scholar] [CrossRef]
- Henrich, B.; Hammerlage, S.; Scharf, S.; Haberhausen, D.; Fürnkranz, U.; Köhrer, K.; Peitzmann, L.; Fiori, P.L.; Spergser, J.; Pfeffer, K.; et al. Characterisation of mobile genetic elements in Mycoplasma hominis with the description of ICEHo-II, a variant mycoplasma integrative and conjugative element. Mob. DNA 2020, 11, 1–17. [Google Scholar] [CrossRef]
- Breton, M.; Tardy, F.; Dordet-Frisoni, E.; Sagne, E.; Mick, V.; Renaudin, J.; Sirand-Pugnet, P.; Citti, C.; Blanchard, A. Distribution and diversity of mycoplasma plasmids: Lessons from cryptic genetic elements. BMC Microbiol. 2012, 12, 257. [Google Scholar] [CrossRef] [Green Version]
- Citti, C.; Baranowski, E.; Dordet-Frisoni, E.; Faucher, M.; Nouvel, L.-X. Genomic Islands in Mycoplasmas. Genes 2020, 11, 836. [Google Scholar] [CrossRef]
- Meygret, A.; Peuchant, O.; Dordet-Frisoni, E.; Sirand-Pugnet, P.; Citti, C.; Bébéar, C.; Béven, L.; Pereyre, S. High Prevalence of Integrative and Conjugative Elements Encoding Transcription Activator-Like Effector Repeats in Mycoplasma hominis. Front. Microbiol. 2019, 10, 2385. [Google Scholar] [CrossRef]
- Faucher, M.; Nouvel, L.-X.; Dordet-Frisoni, E.; Sagné, E.; Baranowski, E.; Hygonenq, M.-C.; Marenda, M.; Tardy, F.; Citti, C. Mycoplasmas under experimental antimicrobial selection: The unpredicted contribution of horizontal chromosomal transfer. PLoS Genet. 2019, 15, e1007910. [Google Scholar] [CrossRef] [Green Version]
- Medvedeva, E.S.; Baranova, N.B.; Mouzykantov, A.A.; Grigorieva, T.Y.; Davydova, M.N.; Trushin, M.V.; Chernova, O.; Chernov, V.M. Adaptation of mycoplasmas to antimicrobial agents: Acholeplasma laidlawii extracellular vesicles mediate the export of ciprofloxacin and a mutant gene related to the antibiotic target. Sci. World J. 2014, 2014, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Rumbo, C.; Fernández-Moreira, E.; Merino, M.; Poza, M.; Mendez, J.A.; Soares, N.C.; Mosquera, A.; Chaves, F.; Bou, G. Horizontal Transfer of the OXA-24 Carbapenemase Gene via Outer Membrane Vesicles: A New Mechanism of Dissemination of Carbapenem Resistance Genes in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2011, 55, 3084–3090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulkarni, H.M.; Nagaraj, R.; Jagannadham, M.V. Protective role of E. coli outer membrane vesicles against antibiotics. Microbiol. Res. 2015, 181, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Waites, K.B.; Duffy, L.B.; Bébéar, C.M.; Matlow, A.; Talkington, D.F.; Kenny, G.E.; Totten, P.A.; Bade, D.J.; Zheng, X.; Davidson, M.K.; et al. Standardized methods and quality control limits for agar and broth microdilution susceptibility testing of Mycoplasma pneumoniae, Mycoplasma hominis, and Ureaplasma urealyticum. J. Clin. Microbiol. 2012, 50, 3542–3547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waites, K.B.; Bade, D.J.; Bébéar, C.; Brown, S.D.; Davidson, M.K.; Duffy, L.B.; Kenny, G.; Matlow, A.; Shortridge, D.; Talkington, D.; et al. Methods for Antimicrobial Susceptibility Testing for Human Mycoplasmas: Approved Guideline; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2011; Report No.: Document M43-A. [Google Scholar]
- Waites, K.; Bébéar, C. Mycoplasma. In Manual of Commercial Methods in Clinical Microbiology, 2nd ed.; Truant, A.L., Ed.; John Wiley & Sons Inc.: Hoboken, NJ, USA, 2016; pp. 195–213. [Google Scholar]
- Beeton, M.L.; Spiller, B. Antibiotic resistance among Ureaplasma spp. isolates: Cause for concern? J. Antimicrob. Chemother. 2016, 72, 330–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hannan, P.C. Guidelines and recommendations for antimicrobial minimum inhibitory concentration (MIC) testing against veterinary mycoplasma species. Vet. Res. 2000, 31, 373–395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Timofte, D.; Broens, E.M.; Guardabassi, L.; Pomba, C.; Allerton, F.; Ikonomopoulos, J.; Overesch, G.; Damborg, P. Driving laboratory standardization of bacterial culture and antimicrobial susceptibility testing in veterinary clinical microbiology in Europe and beyond. J. Clin. Microbiol. 2021, 59. [Google Scholar] [CrossRef] [PubMed]
- Jaÿ, M.; Poumarat, F.; Colin, A.; Tricot, A.; Tardy, F. Addressing the Antimicrobial Resistance of Ruminant Mycoplasmas Using a Clinical Surveillance Network. Front. Vet. Sci. 2021, 8. [Google Scholar] [CrossRef]
- Jelinski, M.; Kinnear, A.; Gesy, K.; Andrés-Lasheras, S.; Zaheer, R.; Weese, S.; McAllister, T.A. Antimicrobial Sensitivity Testing of Mycoplasma bovis Isolates Derived from Western Canadian Feedlot Cattle. Microorganisms 2020, 8, 124. [Google Scholar] [CrossRef] [Green Version]
- Waites, K.B.; Canupp, K.C.; Kenny, G.E. In Vitro Susceptibilities of Mycoplasma hominis to Six Fluoroquinolones as Determined by E Test. Antimicrob. Agents Chemother. 1999, 43, 2571–2573. [Google Scholar] [CrossRef] [Green Version]
- Francoz, D.; Fortin, M.; Fecteau, G.; Messier, S. Determination of Mycoplasma bovis susceptibilities against six antimicrobial agents using the E test method. Vet. Microbiol. 2005, 105, 57–64. [Google Scholar] [CrossRef]
- Filioussis, G.; Petridou, E.; Giadinis, N.D.; Kritas, S.K. In vitro susceptibilities of caprine Mycoplasma agalactiae field isolates to six antimicrobial agents using the E test methodology. Vet. J. 2014, 202, 654–656. [Google Scholar] [CrossRef]
- Touati, A.; Peuchant, O.; Jensen, J.S.; Bebear, C.; Pereyre, S. Direct detection of macrolide resistance in Mycoplasma genitalium isolates from clinical specimens from France by use of real-time PCR and melting curve analysis. J. Clin. Microbiol. 2014, 52, 1549–1555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salado-Rasmussen, K.; Jensen, J.S. Mycoplasma genitalium Testing Pattern and Macrolide Resistance: A Danish Nationwide Retrospective Survey. Clin. Infect. Dis. 2014, 59, 24–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kristiansen, G.Q.; Lisby, J.G.; Schønning, K. A 5′ nuclease genotyping assay for identification of macrolide-resistant Mycoplasma genitalium in clinical specimens. J. Clin. Microbiol. 2016, 54, 1593–1597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Roy, C.; Bébéar, C.; Pereyre, S. Clinical evaluation of three commercial PCR assays for the detection of macrolide resistance in Mycoplasma genitalium. J. Clin. Microbiol. 2020, 58, e01478-19. [Google Scholar] [CrossRef]
- Le Roy, C.; Bébéar, C.; Pereyre, S. Performance of three commercial molecular diagnostic assays for the simultaneous detection of Mycoplasma genitalium and macrolide resistance. J. Clin. Microbiol. 2021, 59, e00020-21. [Google Scholar] [CrossRef]
- Spiller, O.B.; Rees, C.L.; Morris, D.J.; Davies, R.L.; Jones, L. Mycoplasma genitalium prevalence in Welsh sexual health patients: Low antimicrobial resistance markers and no association of symptoms to bacterial load. Microb. Pathog. 2019, 139, 103872. [Google Scholar] [CrossRef]
- Huerta, M.F.; Bodiyabadu, K.; Esperalba, J.; Bradshaw, C.S.; Serra-Pladevall, J.; Garland, S.M.; Fernández-Naval, C.; Jensen, J.S.; Pumarola, T.; Ebeyan, S.; et al. Multicenter clinical evaluation of a novel multiplex real-time PCR (qPCR) assay for detection of fluoroquinolone resistance in Mycoplasma genitalium. J. Clin. Microbiol. 2019, 57, e00886-19. [Google Scholar] [CrossRef] [Green Version]
- Nijhuis, R.H.T.; Duinsbergen, R.G.; Pol, A.; Godschalk, P.C.R. Prevalence of Chlamydia trachomatis, Neisseria gonorrhoeae, Mycoplasma genitalium and Trichomonas vaginalis including relevant resistance-associated mutations in a single center in the Netherlands. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 40, 591–595. [Google Scholar] [CrossRef] [PubMed]
- Murray, G.L.; Doyle, M.; Bodiyabadu, K.; Vodstrcil, L.A.; Garland, S.M.; Danielewski, J.; Machalek, D.A.; McGuinness, C.; Plummer, E.L.; De Petra, V.; et al. Evaluation of ResistancePlus MG FleXible, a ‘near-patient’ test for the detection of Mycoplasma genitalium and macrolide resistance mutations, using freshly collected clinical samples. J. Med. Microbiol. 2021, 70, jmm001271. [Google Scholar] [CrossRef] [PubMed]
- Conway, R.; Cook, S.; Malone, C.; Bone, S.; Hassan-Ibrahim, M.O.; Soni, S. Clearance of Mycoplasma genitalium infection with moxifloxacin in the presence of quinolone resistance–associated mutations. Sex. Transm. Dis. 2019, 47, 197–198. [Google Scholar] [CrossRef] [PubMed]
- Sulyok, K.M.; Bekő, K.; Kreizinger, Z.; Wehmann, E.; Jerzsele, A.; Rónai, Z.; Turcsányi, I.; Makrai, L.; Szeredi, L.; Jánosi, S.; et al. Development of molecular methods for the rapid detection of antibiotic susceptibility of Mycoplasma bovis. Vet. Microbiol. 2018, 213, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Bekő, K.; Kreizinger, Z.; Yvon, C.; Saller, O.; Catania, S.; Feberwee, A.; Gyuranecz, M. Development of molecular assays for the rapid and cost-effective determination of fluoroquinolone, macrolide and lincosamide susceptibility of Mycoplasma synoviae isolates. PLoS ONE 2020, 15, e0241647. [Google Scholar] [CrossRef] [PubMed]
- Ben Shabat, M.; Mikula, I.; Gerchman, I.; Lysnyansky, I. Development and evaluation of a novel single-nucleotide-polymorphism real-time PCR assay for rapid detection of fluoroquinolone-resistant Mycoplasma bovis. J. Clin. Microbiol. 2010, 48, 2909–2915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morozumi, M.; Tajima, T.; Sakuma, M.; Shouji, M.; Meguro, H.; Saito, K.; Iwata, S.; Ubukata, K. Sequence Type Changes Associated with Decreasing Macrolide-Resistant Mycoplasma pneumoniae, Japan. Emerg. Infect. Dis. 2020, 26, 2210–2213. [Google Scholar] [CrossRef] [PubMed]
- Kenri, T.; Suzuki, M.; Sekizuka, T.; Ohya, H.; Oda, Y.; Yamazaki, T.; Fujii, H.; Hashimoto, T.; Nakajima, H.; Katsukawa, C.; et al. Periodic Genotype Shifts in Clinically Prevalent Mycoplasma pneumoniae Strains in Japan. Front. Cell. Infect. Microbiol. 2020, 10, 385. [Google Scholar] [CrossRef]
- Zhou, Z.; Li, X.; Chen, X.; Luo, F.; Pan, C.; Zheng, X.; Tan, F. Macrolide-Resistant Mycoplasma pneumoniae in Adults in Zhejiang, China. Antimicrob. Agents Chemother. 2014, 59, 1048–1051. [Google Scholar] [CrossRef] [Green Version]
- Xiao, L.; Ratliff, A.E.; Crabb, D.M.; Mixon, E.; Qin, X.; Selvarangan, R.; Tang, Y.-W.; Zheng, X.; Bard, J.D.; Hong, T.; et al. Molecular characterization of Mycoplasma pneumoniae isolates in the United States from 2012 to 2018. J. Clin. Microbiol. 2020, 58, e00710-20. [Google Scholar] [CrossRef]
- Waites, K.B.; Ratliff, A.; Crabb, D.M.; Xiao, L.; Qin, X.; Selvarangan, R.; Tang, Y.-W.; Zheng, X.; Bard, J.D.; Hong, T.; et al. Macrolide-resistant Mycoplasma pneumoniae in the United States as determined from a national surveillance program. J. Clin. Microbiol. 2019, 57, e00968-19. [Google Scholar] [CrossRef] [Green Version]
- Dumke, R.; Ziegler, T. Long-term low rate of macrolide-resistant Mycoplasma pneumoniae strains in Germany. Antimicrob. Agents Chemother. 2019, 63, e00455-19. [Google Scholar] [CrossRef] [Green Version]
- Kawai, Y.; Miyashita, N.; Kubo, M.; Akaike, H.; Kato, A.; Nishizawa, Y.; Saito, A.; Kondo, E.; Teranishi, H.; Ogita, S.; et al. Therapeutic efficacy of macrolides, minocycline, and tosufloxacin against macrolide-resistant Mycoplasma pneumoniae pneumonia in pediatric patients. Antimicrob. Agents Chemother. 2013, 57, 2252–2258. [Google Scholar] [CrossRef] [Green Version]
- Beeton, M.L.; Chalker, V.J.; Jones, L.C.; Maxwell, N.C.; Spiller, O.B. Antibiotic Resistance among Clinical Ureaplasma Isolates Recovered from Neonates in England and Wales between 2007 and 2013. Antimicrob. Agents Chemother. 2016, 60, 52–56. [Google Scholar] [CrossRef] [Green Version]
- Morris, D.J.; Jones, L.; Davies, R.L.; Sands, K.; Portal, E.; Spiller, O.B. MYCO WELL D-ONE detection of Ureaplasma spp. and Mycoplasma hominis in sexual health patients in Wales. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 2427–2440. [Google Scholar] [CrossRef]
- Valentine-King, M.A.; Brown, M.B. Antibacterial Resistance in Ureaplasma Species and Mycoplasma hominis Isolates from Urine Cultures in College-Aged Females. Antimicrob. Agents Chemother. 2017, 61, e01104-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernández, J.; Karau, M.J.; Cunningham, S.A.; Greenwood-Quaintance, K.E.; Patel, R. Antimicrobial susceptibility and clonality of clinical ureaplasma isolates in the United States. Antimicrob. Agents Chemother. 2016, 60, 4793–4798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawai, Y.; Nakura, Y.; Wakimoto, T.; Nomiyama, M.; Tokuda, T.; Takayanagi, T.; Shiraishi, J.; Wasada, K.; Kitajima, H.; Fujita, T.; et al. In Vitro Activity of five quinolones and analysis of the quinolone resistance-determining regions of gyrA, gyrB, parC, and parE in Ureaplasma parvum and Ureaplasma urealyticum clinical isolates from perinatal patients in Japan. Antimicrob. Agents Chemother. 2015, 59, 2358–2364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, L.; Liu, A.; Li, R.; Zhao, S. Antimicrobial resistance, genetic characterization, and molecular epidemiology of Ureaplasma species in males with infertility. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 2177–2183. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Pan, L.; Wu, N.; Wang, L.; Liu, Z.; Kong, Y.; Ruan, Z.; Xie, X.; Zhang, J. Antimicrobial resistance in clinical Ureaplasma spp. and Mycoplasma hominis and structural mechanisms underlying quinolone resistance. Antimicrob. Agents Chemother. 2020, 64, e02560-19. [Google Scholar] [CrossRef]
- Ahmadi, M.H. Resistance to tetracyclines among clinical isolates of Mycoplasma hominis and Ureaplasma species: A systematic review and meta-analysis. J. Antimicrob. Chemother. 2020, 76, 865–875. [Google Scholar] [CrossRef]
- Sweeney, M.T.; Lubbers, B.V.; Schwarz, S.; Watts, J.L. Applying definitions for multidrug resistance, extensive drug resistance and pandrug resistance to clinically significant livestock and companion animal bacterial pathogens. J. Antimicrob. Chemother. 2018, 73, 1460–1463. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.; Carmeli, Y.; Falagas, M.; Giske, C.; Harbarth, S.; Hindler, J.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaÿ, M.; Ambroset, C.; Tricot, A.; Colin, A.; Tardy, F. Population structure and antimicrobial susceptibility of Mycoplasma ovipneumoniae isolates in France. Vet. Microbiol. 2020, 248, 108828. [Google Scholar] [CrossRef] [PubMed]
- Ayling, R.D.; Bisgaard-Frantzen, S.; March, J.B.; Godinho, K.; Nicholas, R.A.J. Assessing the In Vitro Effectiveness of Antimicrobials against Mycoplasma mycoides subsp. mycoides Small-Colony Type To Reduce Contagious Bovine Pleuropneumonia Infection. Antimicrob. Agents Chemother. 2005, 49, 5162–5165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Provost, A.; Perreau, P.; Breard, A.; Le Goff, C.; Martel, J.L.; Cottew, G.S. Contagious bovine pleuropneumonia. Rev. Sci. Tech. Off. Int. Epizoot. 1987, 6, 625–679. [Google Scholar]
- Gerchman, I.; Levisohn, S.; Mikula, I.; Manso-Silván, L.; Lysnyansky, I. Characterization of in vivo-acquired resistance to macrolides of Mycoplasma gallisepticum strains isolated from poultry. Vet. Res. 2011, 42, 90. [Google Scholar] [CrossRef] [Green Version]
- Lysnyansky, I.; Gerchman, I.; Levisohn, S.; Mikula, I.; Feberwee, A.; Ferguson, N.; Noormohammadi, A.; Spergser, J.; Windsor, H. Discrepancy between minimal inhibitory concentration to enrofloxacin and mutations present in the quinolone-resistance determining regions of Mycoplasma gallisepticum field strains. Vet. Microbiol. 2012, 160, 222–226. [Google Scholar] [CrossRef]
- Tully, J.; Cole, R.; Taylor-Robinson, D.; Rose, D. A Newly discovered mycoplasma in the human urogenital tract. Lancet 1981, 317, 1288–1291. [Google Scholar] [CrossRef]
- Manhart, L.E.; Broad, J.M.; Golden, M.R. Mycoplasma genitalium: Should we treat and how? Clin. Infect. Dis. 2011, 53, S129–S142. [Google Scholar] [CrossRef] [Green Version]
- Lis, R.; Rowhani-Rahbar, A.; Manhart, L.E. Mycoplasma genitalium Infection and Female Reproductive Tract Disease: A Meta-analysis. Clin. Infect. Dis. 2015, 61, 418–426. [Google Scholar] [CrossRef] [Green Version]
- Pereyre, S.; Nadalié, C.L.; Bébéar, C.; Arfeuille, C.; Beby-Defaux, A.; Bercot, B.; Boisset, S.; Bourgeois, N.; Carles, M.-J.; Decré, D.; et al. Mycoplasma genitalium and Trichomonas vaginalis in France: A point prevalence study in people screened for sexually transmitted diseases. Clin. Microbiol. Infect. 2017, 23, 122.e1–122.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manhart, L.E.; Holmes, K.K.; Hughes, J.P.; Houston, L.S.; Totten, P.A. Mycoplasma genitalium Among Young Adults in the United States: An Emerging Sexually Transmitted Infection. Am. J. Public Health 2007, 97, 1118–1125. [Google Scholar] [CrossRef] [PubMed]
- Anderson, T.; Coughlan, E.; Werno, A. Mycoplasma genitalium Macrolide and Fluoroquinolone Resistance Detection and Clinical Implications in a Selected Cohort in New Zealand. J. Clin. Microbiol. 2017, 55, 3242–3248. [Google Scholar] [CrossRef] [Green Version]
- Sonnenberg, P.; Clifton, S.; Beddows, S.; Field, N.; Soldan, K.; Tanton, C.; Mercer, C.; da Silva, F.C.; Alexander, S.; Copas, A.; et al. Prevalence, risk factors, and uptake of interventions for sexually transmitted infections in Britain: Findings from the National Surveys of Sexual Attitudes and Lifestyles (Natsal). Lancet 2013, 382, 1795–1806. [Google Scholar] [CrossRef] [Green Version]
- Sonnenberg, P.; Ison, C.A.; Clifton, S.; Field, N.P.; Tanton, C.; Soldan, K.; Beddows, S.; Alexander, S.; Khanom, R.; Saunders, P.; et al. Epidemiology of Mycoplasma genitalium in British men and women aged 16–44 years: Evidence from the third National Survey of Sexual Attitudes and Lifestyles (Natsal-3). Int. J. Epidemiol. 2015, 44, 1982–1994. [Google Scholar] [CrossRef] [Green Version]
- Baumann, L.; Cina, M.; Egli-Gany, D.; Goutaki, M.; Halbeisen, F.S.; Lohrer, G.-R.; Ali, H.; Scott, P.; Low, N. Prevalence of Mycoplasma genitalium in different population groups: Systematic review and meta-analysis. Sex. Transm. Infect. 2018, 94, 255–262. [Google Scholar] [CrossRef] [Green Version]
- Cazanave, C.; Manhart, L.; Bébéar, C. Mycoplasma genitalium, an emerging sexually transmitted pathogen. Med. Mal. Infect. 2012, 42, 381–392. [Google Scholar] [CrossRef]
- Manhart, L.E.; Gaydos, C.A.; Taylor, S.N.; Lillis, R.A.; Hook, E.W.; Klausner, J.D.; Remillard, C.V.; Love, M.; McKinney, B.; Getman, D.K. Characteristics of Mycoplasma genitalium urogenital infections in a diverse patient sample from the United States: Results from the Aptima Mycoplasma genitalium evaluation study (AMES). J. Clin. Microbiol. 2020, 58, e00165-20. [Google Scholar] [CrossRef] [Green Version]
- Chrisment, D.; Charron, A.; Cazanave, C.; Pereyre, S.; Bébéar, C. Detection of macrolide resistance in Mycoplasma genitalium in France. J. Antimicrob. Chemother. 2012, 67, 2598–2601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Roy, C.; Hénin, N.; Pereyre, S.; Bébéar, C. Fluoroquinolone-resistant Mycoplasma genitalium, Southwestern France. Emerg. Infect. Dis. 2016, 22, 1677–1679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereyre, S.; Laurier Nadalie, C.; Le Roy, C.; Henin, N.; Gardette, M.; Bébéar, C. Significant difference in macrolide and fluoroquinolone resistance in Mycoplasma genitalium in metropolitan and overseas France in 2018 and 2019. Sex. Transm. Infect. 2021, 97, A81. [Google Scholar]
- Ducours, M.; Alleman, L.; Puges, M.; Deborde, M.; Hessamfar, M.; Le-Marec, F.; Dabis, F.; Pereyre, S.; Bébéar, C.; Descaux, A.; et al. Incidence of sexually transmitted infections during pre-exposure prophylaxis for HIV: A worrying outcome at 2 years! Sex. Transm. Infect. 2019, 95, 552. [Google Scholar] [CrossRef]
- Bradley, I.; Varma, R.; Knight, V.; Iliakis, D.; McNally, L.; Jalocon, D.; Jeoffreys, N.; Chen, S.; McNulty, A. Prevalence of rectal Mycoplasma genitalium and macrolide resistance in men who have sex with men attending Sydney Sexual Health Centre. Sex. Health 2020, 17, 114. [Google Scholar] [CrossRef]
- Dionne-Odom, J.; Geisler, W.M.; Aaron, K.J.; Waites, K.B.; Westfall, A.O.; Van Der Pol, B.; Xiao, L. High prevalence of multidrug-resistant Mycoplasma genitalium in human immunodeficiency virus-infected men who have sex with men in Alabama. Clin. Infect. Dis. 2017, 66, 796–798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kenyon, C.; Manoharan-Basil, S.S. Macrolide consumption and resistance in Mycoplasma genitalium. Lancet Infect. Dis. 2020, 20, 1235–1236. [Google Scholar] [CrossRef]
- Soni, S.; Horner, P. Launch of the BASHH guideline for the management of M. genitalium in adults. Sex. Transm. Infect. 2019, 95, 237. [Google Scholar] [CrossRef] [PubMed]
- Murray, G.L.; Bodiyabadu, K.; Danielewski, J.; Garland, S.M.; Machalek, D.A.; Fairley, C.K.; Jensen, J.S.; Williamson, D.A.; Tan, L.Y.; Mokany, E.; et al. The parC mutation G248T (S83I), and concurrent gyrA mutations, are associated with moxifloxacin and sitafloxacin treatment failure for Mycoplasma genitalium. J. Infect. Dis. 2019, 221, 1017–1024. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Takei, M.; Kishii, R.; Yasuda, M.; Deguchi, T. Contribution of Topoisomerase IV Mutation to Quinolone Resistance in Mycoplasma genitalium. Antimicrob. Agents Chemother. 2013, 57, 1772–1776. [Google Scholar] [CrossRef] [Green Version]
- Hamasuna, R.; Le, P.T.; Kutsuna, S.; Furubayashi, K.; Matsumoto, M.; Ohmagari, N.; Fujimoto, N.; Matsumoto, T.; Jensen, J.S. Mutations in ParC and GyrA of moxifloxacin-resistant and susceptible Mycoplasma genitalium strains. PLoS ONE 2018, 13, e0198355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Unemo, M.; Salado-Rasmussen, K.; Hansen, M.; Olsen, A.; Falk, M.; Golparian, D.; Aasterød, M.; Ringlander, J.; Nilsson, C.S.; Sundqvist, M.; et al. Clinical and analytical evaluation of the new Aptima Mycoplasma genitalium assay, with data on M. genitalium prevalence and antimicrobial resistance in M. genitalium in Denmark, Norway and Sweden in 2016. Clin. Microbiol. Infect. 2017, 24, 533–539. [Google Scholar] [CrossRef] [Green Version]
- Guiraud, J.; Lounnas, M.; Boissière, A.; Le Roy, C.; Elguero, E.; Banuls, A.L.; Bébéar, C.; Godreuil, S.; Pereyre, S. Lower mgpB diversity in macrolide-resistant Mycoplasma genitalium infecting men visiting two sexually transmitted infection clinics in Montpellier, France. J. Antimicrob. Chemother. 2020, 76, 43–47. [Google Scholar] [CrossRef] [PubMed]
- Dumke, R.; Rust, M.; Glaunsinger, T. MgpB Types among Mycoplasma genitalium Strains from Men Who Have Sex with Men in Berlin, Germany, 2016–2018. Pathogens 2019, 9, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernández-Huerta, M.; Serra-Pladevall, J.; Esperalba, J.; Moreno-Mingorance, A.; Fernández-Naval, C.; Barberá, M.-J.; Aparicio, D.; Pich, O.Q.; Pumarola, T.; Jensen, J.S.; et al. Single-Locus-Sequence-Based Typing of the mgpB Gene Reveals Transmission Dynamics in Mycoplasma genitalium. J. Clin. Microbiol. 2020, 58, e01886-19. [Google Scholar] [CrossRef] [PubMed]
- Deguchi, T.; Kikuchi, M.; Yasuda, M.; Ito, S. Multidrug-Resistant Mycoplasma genitalium Is Increasing: Table 1. Clin. Infect. Dis. 2015, 62, 405–406. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Su, X.; Le, W.; Li, S.; Yang, Z.; Chaisson, C.; Madico, G.; Gong, X.; Reed, G.W.; Wang, B.; et al. Mycoplasma genitalium in Symptomatic Male Urethritis: Macrolide Use Is Associated with Increased Resistance. Clin. Infect. Dis. 2019, 70, 805–810. [Google Scholar] [CrossRef]
- Read, T.R.H.; Fairley, C.K.; Murray, G.; Jensen, J.S.; Danielewski, J.; Worthington, K.; Doyle, M.; Mokany, E.; Tan, L.; Chow, E.P.F.; et al. Outcomes of Resistance-guided Sequential Treatment of Mycoplasma genitalium Infections: A Prospective Evaluation. Clin. Infect. Dis. 2018, 68, 554–560. [Google Scholar] [CrossRef] [Green Version]
- Horner, P.; Ingle, S.M.; Garrett, F.; Blee, K.; Kong, F.; Muir, P.; Moi, H. Which azithromycin regimen should be used for treating Mycoplasma genitalium? A meta-analysis. Sex. Transm. Infect. 2017, 94, 14–20. [Google Scholar] [CrossRef] [Green Version]
- Durukan, D.; Doyle, M.; Murray, G.; Bodiyabadu, K.; Vodstrcil, L.; Chow, E.P.F.; Jensen, J.S.; Fairley, C.K.; Aguirre, I.; Bradshaw, C.S. Doxycycline and Sitafloxacin Combination Therapy for Treating Highly Resistant Mycoplasma genitalium. Emerg. Infect. Dis. 2020, 26, 1870–1874. [Google Scholar] [CrossRef]
- Doyle, M.; Vodstrcil, L.A.; Plummer, E.L.; Aguirre, I.; Fairley, C.K.; Bradshaw, C.S. Nonquinolone Options for the Treatment of Mycoplasma genitalium in the Era of Increased Resistance. Open Forum Infect. Dis. 2020, 7, ofaa291. [Google Scholar] [CrossRef] [PubMed]
- Falk, L.; Jensen, J.S. Successful outcome of macrolide-resistant Mycoplasma genitalium urethritis after spectinomycin treatment: A case report. J. Antimicrob. Chemother. 2016, 72, 624–625. [Google Scholar] [CrossRef] [Green Version]
- Hale, H.H.; Helmboldt, C.F.; Plastridge, W.N.; Stula, E.F. Bovine mastitis caused by a Mycoplasma species. Cornell Vet. 1962, 52, 582–591. [Google Scholar] [PubMed]
- Tardy, F.; Aspan, A.; Autio, T.; Ridley, A.; Tricot, A.; Colin, A.; Pohjanvirta, T.; Smid, B.; Harders, F.; Lindegaard, M.; et al. Mycoplasma bovis in Nordic European Countries: Emergence and Dominance of a New Clone. Pathogens 2020, 9, 875. [Google Scholar] [CrossRef] [PubMed]
- Maunsell, F.; Woolums, A.; Francoz, D.; Rosenbusch, R.; Step, D.; Wilson, D.; Janzen, E. Mycoplasma bovis Infections in Cattle. J. Vet. Intern. Med. 2011, 25, 772–783. [Google Scholar] [CrossRef]
- Nicholas, R.A.J. Bovine mycoplasmosis: Silent and deadly. Vet. Rec. 2011, 168, 459–462. [Google Scholar] [CrossRef]
- Perez-Casal, J.; Prysliak, T.; Maina, T.; Suleman, M.; Jimbo, S. Status of the development of a vaccine against Mycoplasma bovis. Vaccine 2017, 35, 2902–2907. [Google Scholar] [CrossRef]
- Stanford, K.; Zaheer, R.; Klima, C.; McAllister, T.; Peters, D.; Niu, Y.D.; Ralston, B. Antimicrobial Resistance in Members of the Bacterial Bovine Respiratory Disease Complex Isolated from Lung Tissue of Cattle Mortalities Managed with or without the Use of Antimicrobials. Microorganisms 2020, 8, 288. [Google Scholar] [CrossRef] [Green Version]
- Klein, U.; de Jong, A.; Youala, M.; El Garch, F.; Stevenin, C.; Moyaert, H.; Rose, M.; Catania, S.; Gyuranecz, M.; Pridmore, A.; et al. New antimicrobial susceptibility data from monitoring of Mycoplasma bovis isolated in Europe. Vet. Microbiol. 2019, 238, 108432. [Google Scholar] [CrossRef]
- Lysnyansky, I.; Ayling, R.D. Mycoplasma bovis: Mechanisms of Resistance and Trends in Antimicrobial Susceptibility. Front. Microbiol. 2016, 7, 595. [Google Scholar] [CrossRef]
- Lerner, U.; Amram, E.; Ayling, R.D.; Mikula, I.; Gerchman, I.; Harrus, S.; Teff, D.; Yogev, D.; Lysnyansky, I. Acquired resistance to the 16-membered macrolides tylosin and tilmicosin by Mycoplasma bovis. Vet. Microbiol. 2013, 168, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Lysnyansky, I.; Mikula, I.; Gerchman, I.; Levisohn, S. Rapid Detection of a Point Mutation in the parC Gene Associated with Decreased Susceptibility to Fluoroquinolones in Mycoplasma bovis. Antimicrob. Agents Chemother. 2009, 53, 4911–4914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, T.; Okubo, T.; Usui, M.; Higuchi, H.; Tamura, Y. Amino acid substitutions in GyrA and ParC are associated with fluoroquinolone resistance in Mycoplasma bovis isolates from Japanese dairy calves. J. Vet. Med. Sci. Jpn. Soc. Vet. Sci. 2013, 75, 1063–1065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heuvelink, A.; Reugebrink, C.; Mars, J. Antimicrobial susceptibility of Mycoplasma bovis isolates from veal calves and dairy cattle in the Netherlands. Vet. Microbiol. 2016, 189, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Soehnlen, M.K.; Kunze, M.E.; Karunathilake, K.E.; Henwood, B.M.; Kariyawasam, S.; Wolfgang, D.R.; Jayarao, B.M. In vitro antimicrobial inhibition of Mycoplasma bovis isolates submitted to the Pennsylvania Animal Diagnostic Laboratory using flow cytometry and a broth microdilution method. J. Vet. Diagn. Investig. 2011, 23, 547–551. [Google Scholar] [CrossRef] [Green Version]
- Becker, C.A.; Thibault, F.M.; Arcangioli, M.-A.; Tardy, F. Loss of diversity within Mycoplasma bovis isolates collected in France from bovines with respiratory diseases over the last 35 years. Infect. Genet. Evol. 2015, 33, 118–126. [Google Scholar] [CrossRef]
- Gerchman, I.; Levisohn, S.; Mikula, I.; Lysnyansky, I. In vitro antimicrobial susceptibility of Mycoplasma bovis isolated in Israel from local and imported cattle. Vet. Microbiol. 2009, 137, 268–275. [Google Scholar] [CrossRef]
- Godinho, K.S.; Rae, A.; Windsor, G.D.; Tilt, N.; Rowan, T.G.; Sunderland, S.J. Efficacy of tulathromycin in the treatment of bovine respiratory disease associated with induced Mycoplasma bovis infections in young dairy calves. Vet. Ther. Res. Appl. Vet. Med. 2005, 6, 96–112. [Google Scholar]
- Mosquera, R.A.; De Jesus-Rojas, W.; Stark, J.M.; Yadav, A.; Jon, C.K.; Atkins, C.L.; Samuels, C.L.; Gonzales, T.R.; McBeth, K.E.; Hashmi, S.S.; et al. Role of prophylactic azithromycin to reduce airway inflammation and mortality in a RSV mouse infection model. Pediatr. Pulmonol. 2018, 53, 567–574. [Google Scholar] [CrossRef] [PubMed]
| Host | Mycoplasma Species | Clinical Signs or Syndrome | Geographical Prevalence |
|---|---|---|---|
| Humans | M. genitalium | Urethritis, cervicitis, pelvic inflammatory disease | Frequent, worldwide |
| M. pneumoniae | Upper and lower respiratory tract infection | Frequent, worldwide | |
| M. hominis | Commensal of the urogenital tract (opportunistic pathogen) | Frequent, worldwide | |
| U. parvum, U. urealyticum | Commensal of the urogenital tract (opportunistic pathogen) | Frequent, worldwide | |
| Cattle | M. bovis | Infectious enzootic bronchopneumonia, mastitis, arthritis, otitis | Frequent, worldwide |
| M. mycoides subsp. mycoides | Contagious bovine pleuropneumonia | Scarce, Africa and Asia | |
| Small ruminants | M. putrefaciens | Contagious agalactia | Regularly reported in Europe, particularly in Mediterranean regions, as well as the Middle East, Asia, North Africa and South America |
| M. agalactiae | |||
| M. mycoides subsp. capri | |||
| M. capricolum subsp. capricolum | |||
| M. capricolum subsp. capripneumoniae | Contagious caprine pleuropneumonia | Scarce, Africa and Asia | |
| M. ovipeumoniae | Atypical pneumonia (facultative pathogen) | Infrequent, worldwide | |
| Chickens, turkeys | M. gallisepticum | Chronic respiratory disease, infectious sinusitis | Frequent, worldwide |
| M. synoviae | Subclinical respiratory tract infections, infectious synovitis, eggshell apex abnormality syndrome in laying-hen flocks (facultative pathogen) | Frequent, worldwide | |
| Swine | M. hyopneumoniae | Enzootic pneumonia | Frequent, worldwide |
| M. hyorhinis | Polyserositis, arthritis (facultative pathogen) | Frequent, worldwide | |
| M. hyosynoviae | Arthritis, polyarthritis (facultative pathogen) | Frequent, worldwide | |
| M. suis | Infectious Anaemia in Pigs, chronic immunosuppression | Frequent, worldwide |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pereyre, S.; Tardy, F. Integrating the Human and Animal Sides of Mycoplasmas Resistance to Antimicrobials. Antibiotics 2021, 10, 1216. https://doi.org/10.3390/antibiotics10101216
Pereyre S, Tardy F. Integrating the Human and Animal Sides of Mycoplasmas Resistance to Antimicrobials. Antibiotics. 2021; 10(10):1216. https://doi.org/10.3390/antibiotics10101216
Chicago/Turabian StylePereyre, Sabine, and Florence Tardy. 2021. "Integrating the Human and Animal Sides of Mycoplasmas Resistance to Antimicrobials" Antibiotics 10, no. 10: 1216. https://doi.org/10.3390/antibiotics10101216
APA StylePereyre, S., & Tardy, F. (2021). Integrating the Human and Animal Sides of Mycoplasmas Resistance to Antimicrobials. Antibiotics, 10(10), 1216. https://doi.org/10.3390/antibiotics10101216






