Microbial Species Isolated from Infected Wounds and Antimicrobial Resistance Analysis: Data Emerging from a Three-Years Retrospective Study
Abstract
1. Introduction
2. Results
3. Discussion
4. Conclusions
5. Material and Methods
5.1. Patients
5.2. Sample Collection
5.3. Species Identification and Antimicrobial Susceptibility Pattern Determination
5.4. Statistical Analysis
5.5. Ethical Considerations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Maillard, J.Y.; Kampf, G.; Cooper, R. Antimicrobial stewardship of antiseptics that are pertinent to wounds: The need for a united approach. JAC Antimicrob. Resist. 2021, 3, dlab027. [Google Scholar] [CrossRef]
- Simões, D.; Miguel, S.P.; Ribeiro, M.P.; Coutinho, P.; Mendonça, A.G.; Correia, I.J. Recent advances on antimicrobial wound dressing: A review. Eur. J. Pharm. Biopharm. 2018, 127, 130–141. [Google Scholar] [CrossRef]
- Van Koppen, C.J.; Hartmann, R.W. Advances in the treatment of chronic wounds: A patent review. Expert Opin. Ther. Pat. 2015, 8, 931–937. [Google Scholar] [CrossRef] [PubMed]
- Negut, I.; Grumezescu, V.; Grumezescu, A.M. Treatment Strategies for Infected Wounds. Molecules 2018, 9, 2392. [Google Scholar] [CrossRef]
- Pallavali, R.R.; Degati, V.L.; Lomada, D.; Reddy, M.C.; Durbaka, V.R.P. Isolation and in vitro evaluation of bacteriophages against MDR-bacterial isolates from septic wound infections. PLoS ONE 2017, 12, e0179245. [Google Scholar] [CrossRef] [PubMed]
- Anguzu, J.R.; Olila, D. Drug sensitivity patterns of bacterial isolates from septic post-operative wounds in a regional referral hospital in Uganda. Afr. Health Sci. 2007, 7, 148–154. [Google Scholar] [PubMed]
- Mehta, R.L.; Kellum, J.A.; Shah, S.V.; Molitoris, B.A.; Ronco, C.; Warnock, D.G.; Levin, A. Acute Kidney Injury Network. Acute Kidney Injury Network: Report of an initiative to improve outcomes in acute kidney injury. Crit. Care 2007, 11, R31. [Google Scholar] [CrossRef]
- Agnihotri, N.; Gupta, V.; Joshi, R.M. Aerobic bacterial isolates from burn wound infections and their antibiograms--a five-year study. Burns 2004, 30, 241–243. [Google Scholar] [CrossRef]
- Puca, V.; Traini, T.; Guarnieri, S.; Carradori, S.; Sisto, F.; Macchione, N.; Muraro, R.; Mincione, G.; Grande, R. The Antibiofilm Effect of a Medical Device Containing TIAB on Microorganisms Associated with Surgical Site Infection. Molecules 2019, 24, 2280. [Google Scholar] [CrossRef]
- Taati Moghadam, M.; Khoshbayan, A.; Chegini, Z.; Farahani, I.; Shariati, A. Bacteriophages, a New Therapeutic Solution for Inhibiting Multidrug-Resistant Bacteria Causing Wound Infection: Lesson from Animal Models and Clinical Trials. Drug Des. Dev. Ther. 2020, 14, 1867. [Google Scholar] [CrossRef]
- Khan, H.A.; Baig, K.F.; Mehboob, R. Nosocomial infections: Epidemiology, prevention, control and surveillance. Asian Pac. J. Trop. Biomed. 2017, 7, 478–482. [Google Scholar] [CrossRef]
- Jeschke, M.G.; Phelan, H.A.; Wolf, S.; Romanowski, K.; Rehou, S.; Saetamal, A.; Weber, J.; Schulz, J.; New, C.; Wiktor, A.; et al. State of the Science Burn Research: Burns in the Elderly. J. Burn Care Res. 2020, 41, 65–83. [Google Scholar] [CrossRef] [PubMed]
- Han, G.; Ceilley, R. Chronic Wound Healing: A Review of Current Management and Treatments. Adv. Ther. 2017, 34, 599–610. [Google Scholar] [CrossRef] [PubMed]
- Dhar, S.; Saraf, R.; Singh, K.; Raina, B. Microbiological profile of chronic burn wounds among patients admitted in burn unit. JK Sci. 2007, 9, 182–185. [Google Scholar]
- Vaez, H.; Beigi, F. Antibiotic susceptibility patterns of aerobic bacterial strains isolated from patients with burn wound infections. Germs 2016, 6, 34–36. [Google Scholar] [CrossRef][Green Version]
- Glik, J.; Kawecki, M.; Gaździk, T.; Nowak, M. The impact of the types of microorganisms isolated from blood and wounds on the results of treatment in burn patients with sepsis. Pol. Przegl. Chir. 2012, 84, 6–16. [Google Scholar] [CrossRef]
- Huszczynski, S.M.; Lam, J.S.; Khursigara, C.M. The Role of Pseudomonas aeruginosa Lipopolysaccharide in Bacterial Pathogenesis and Physiology. Pathogens 2019, 9, 6. [Google Scholar] [CrossRef]
- Ch’ng, J.-H.; Chong, K.K.L.; Lam, L.N.; Wong, J.J.; Kline, K.A. Biofilm-associated infection by enterococci. Nat. Rev. Microbiol. 2018, 17, 82–94. [Google Scholar]
- Pinto, R.M.; Soares, F.A.; Reis, S.; Nunes, C.; Van Dijck, P. Innovative Strategies Toward the Disassembly of the EPS Matrix in Bacterial Biofilms. Front. Microbiol. 2020, 11, 952. [Google Scholar] [CrossRef]
- Grande, R.; Puca, V.; Muraro, R. Antibiotic resistance and bacterial biofilm. Expert Opin. Ther. Pat. 2020, 30, 897–900. [Google Scholar] [CrossRef]
- Koo, H.; Allan, R.N.; Howlin, R.P.; Stoodley, P.; Hall-Stoodley, L. Targeting microbial biofilms: Current and prospective therapeutic strategies. Nat. Rev. Microbiol. 2017, 15, 740–755. [Google Scholar] [CrossRef]
- Anderl, J.N.; Franklin, M.J.; Stewart, P.S. Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob. Agents Chemother. 2000, 44, 1818–1824. [Google Scholar] [CrossRef]
- Neut, D.; Tijdens-Creusen, E.J.; Bulstra, S.K.; Van Der Mei, H.C.; Busscher, H.J. Biofilms in chronic diabetic foot ulcers--a study of 2 cases. Acta Orthop. 2011, 82, 383–385. [Google Scholar] [CrossRef]
- Bjarnsholt, T.; Kirketerp-Møller, K.; Jensen, P.Ø.; Madsen, K.G.; Phipps, R.; Krogfelt, K.; Høiby, N.; Givskov, M. Why chronic wounds will not heal: A novel hypothesis. Wound Repair Regen. 2008, 16, 2–10. [Google Scholar] [CrossRef]
- James, G.A.; Swogger, E.; Wolcott, R.; Pulcini, E.D.; Secor, P.; Sestrich, J.; Costerton, J.W.; Stewart, P.S. Biofilms in chronic wounds. Wound Repair Regen. 2008, 16, 37–44. [Google Scholar] [CrossRef]
- Kennedy, P.; Brammah, S.; Wills, E. Burns, biofilm and a new appraisal of burn wound sepsis. Burns 2010, 36, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Fromantin, I.; Watson, S.; Baffie, A.; Rivat, A.; Falcou, M.C.; Kriegel, I.; De Rycke Ingenior, Y.A. A prospective, descriptive cohort study of malignant wound characteristics and wound care strategies in patients with breast cancer. Ostomy Wound Manag. 2014, 60, 38–48. [Google Scholar]
- Kathju, S.; Nistico, L.; Hall-Stoodley, L.; Post, J.C.; Ehrlich, G.D.; Stoodley, P. Chronic surgical site infection due to suture-associated polymicrobial biofilm. Surg. Infect. (Larchmt) 2009, 10, 457–461. [Google Scholar] [CrossRef] [PubMed]
- Howell-Jones, R.S.; Wilson, M.J.; Hill, K.E.; Howard, A.J.; Price, P.E.; Thomas, D.W. A review of the microbiology, antibiotic usage and resistance in chronic skin wounds. J. Antimicrob. Chemother. 2005, 55, 143–149. [Google Scholar] [CrossRef]
- World Heath Organitazion. Ten Threats to Global Health in 2019. Available online: https://www.who.int/news-room/spotlight/ten-threats-to-global-health-in-2019 (accessed on 25 August 2021).
- World Health Organization. Antimicrobial Resistance Global Report on Surveillance: 2014 Summary; World Health Organization: Geneva, Switzerland, 2014; p. 6. [Google Scholar]
- Rice, L.B. Federal Funding for the Study of Antimicrobial Resistance in Nosocomial Pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef]
- Boucher, H.W.; Talbot, G.H.; Bradley, J.S.; Edwards, J.E.; Gilbert, D.; Rice, L.B.; Scheld, M.; Spellberg, B.; Bartlett, J. Bad Bugs, No Drugs: No ESKAPE! An Update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009, 48, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Webster, T.J. Bacteria antibiotic resistance: New challenges and opportunities for implant-associated orthopedic infections. J. Orthop. Res. 2018, 36, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Zimlichman, E.; Henderson, D.; Tamir, O.; Franz, C.; Song, P.; Yamin, C.K.; Keohane, C.; Denham, C.R.; Bates, D.W. Health care-associated infections: A meta-analysis of costs and financial impact on the US healthcare system. JAMA Intern. Med. 2013, 173, 2039–2046. [Google Scholar] [CrossRef] [PubMed]
- Guan, H.; Dong, W.; Lu, Y.; Jiang, M.; Zhang, D.; Aobuliaximu, Y.; Dong, J.; Niu, Y.; Liu, Y.; Guan, B.; et al. Distribution and Antibiotic Resistance Patterns of Pathogenic Bacteria in Patients With Chronic Cutaneous Wounds in China. Front. Med. (Lausanne) 2021, 17, 609584. [Google Scholar] [CrossRef] [PubMed]
- Rahim, K.; Saleha, S.; Zhu, X.; Huo, L.; Basit, A.; Franco, O.L. Bacterial contribution in chronicity of wounds. Microb. Ecol. 2017, 73, 710–721. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.K.; Cheng, N.C.; Cheng, C.M. Biofilms in chronic wounds: Pathogenesis and diagnosis. Trends Biotechnol. 2019, 37, 505–517. [Google Scholar] [CrossRef]
- Mohammed, A.; Seid, M.E.; Gebrecherkos, T.; Tiruneh, M.; Moges, F. Bacterial Isolates and Their Antimicrobial Susceptibility Patterns of Wound Infections among Inpatients and Outpatients Attending the University of Gondar Referral Hospital, Northwest Ethiopia. Int. J. Microbiol. 2017, 2017, 8953829. [Google Scholar] [CrossRef] [PubMed]
- Yeong, E.K.; Sheng, W.H.; Hsueh, P.R.; Hsieh, S.M.; Huang, H.F.; Ko, A.T.; Tai, H.C.; Lai, H.S.; Chang, S.C. The Wound Microbiology and the Outcomes of the Systemic Antibiotic Prophylaxis in a Mass Burn Casualty Incident. J. Burn Care Res. 2020, 41, 95–103. [Google Scholar] [CrossRef]
- Wong, S.Y.; Manikam, R.; Muniandy, S. Prevalence and antibiotic susceptibility of bacteria from acute and chronic wounds in Malaysian subjects. J. Infect. Dev. Ctries. 2015, 9, 936–944. [Google Scholar] [CrossRef]
- Serra, R.; Grande, R.; Butrico, L.; Rossi, A.; Settimio, U.F.; Caroleo, B.; Amato, B.; Gallelli, L.; de Franciscis, S. Chronic wound infections: The role of Pseudomonas aeruginosa and Staphylococcus aureus. Expert Rev. Anti-Infect. Ther. 2015, 13, 605–613. [Google Scholar] [CrossRef]
- Baffoni, M.; Bessa, L.J.; Grande, R.; Di Giulio, M.; Mongelli, M.; Ciarelli, A.; Cellini, L. Laser irradiation effect on Staphylococcus aureus and Pseudomonas aeruginosa biofilms isolated from venous leg ulcer. Int. Wound J. 2012, 9, 517–524. [Google Scholar] [CrossRef]
- Li, L.; Dai, J.X.; Xu, L.; Chen, Z.H.; Li, X.Y.; Liu, M.; Wen, Y.Q.; Chen, X.D. Antimicrobial resistance and pathogen distribution in hospitalized burn patients: A multicenter study in Southeast China. Medicine 2018, 97, e11977. [Google Scholar] [CrossRef]
- Sisay, M.; Worku, T.; Edessa, D. Microbial epidemiology and antimicrobial resistance patterns of wound infection in Ethiopia: A meta-analysis of laboratory-based cross-sectional studies. BMC Pharmacol. Toxicol. 2019, 20, 35. [Google Scholar] [CrossRef]
- Colsky, A.S.; Kirsner, R.S.; Kerdel, F.A. Analysis of antibiotic susceptibilities of skin wound flora in hospitalized dermatology patients. The crisis of antibiotic resistance has come to the surface. Arch. Dermatol. 1998, 134, 1006–1009. [Google Scholar] [CrossRef]
- Kabanangi, F.; Joachim, A.; Nkuwi, E.J.; Manyahi, J.; Moyo, S.; Majigo, M. High Level of Multidrug-Resistant Gram-Negative Pathogens Causing Burn Wound Infections in Hospitalized Children in Dar es Salaam, Tanzania. Int. J. Microbiol. 2021, 2021, 6644185. [Google Scholar] [CrossRef] [PubMed]
- Abdu, A.; Egbagba, J.; Fente, B. Identification and antimicrobial susceptibility profile of bacterial pathogens isolated from wound infections in a tertiary hospital, Bayelsa South southern, Nigeria. Trop. J. Pathol. Microbiol. 2019, 5, 966–975. [Google Scholar] [CrossRef]
- Kolar, M.; Cermak, P.; Hobzova, L.; Bogdanova, K.; Neradova, K.; Mlynarcik, P.; Bostik, P. Antibiotic Resistance in Nosocomial Bacteria Isolated from Infected Wounds of Hospitalized Patients in Czech Republic. Antibiotics 2020, 9, 342. [Google Scholar] [CrossRef] [PubMed]
- Krumkamp, R.; Oppong, K.; Hogan, B.; Strauss, R.; Frickmann, H.; Wiafe-Akenten, C.; Boahen, K.G.; Rickerts, V.; McCormick Smith, I.; Groß, U.; et al. Spectrum of antibiotic resistant bacteria and fungi isolated from chronically infected wounds in a rural district hospital in Ghana. PLoS ONE 2020, 15, e0237263. [Google Scholar] [CrossRef] [PubMed]
- Percival, S.L.; Dowd, S.E. Microbiology of wounds. In Microbiology of Wounds; Percival, S.L., Cutting, K., Eds.; CRC Press: Boca Raton, FL, USA, 2010; pp. 187–218. [Google Scholar]
- Percival, S.L. Importance of biofilm formation in surgical infection. Br. J. Surg. 2017, 104, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Stephens, P.; Wall, I.B.; Wilson, M.J.; Hill, K.E.; Davies, C.E.; Hill, C.M.; Harding, K.G.; Thomas, D.W. Anaerobic cocci populating the deep tissues of chronic wounds impair cellular wound healing responses in vitro. Br. J. Dermatol. 2003, 148, 456–466. [Google Scholar] [CrossRef]
- Charles, P.G.; Uçkay, I.; Kressmann, B.; Emonet, S.; Lipsky, B.A. The role of anaerobes in diabetic foot infections. Anaerobe 2015, 34, 8–13. [Google Scholar] [CrossRef] [PubMed]
- EUCAST. European Committee on Antimicrobial Susceptibility Testing. In EUCAST Guidelines for Detection of Resistance Mechanisms and Specific Resistances of Clinical and/or Epidemiological Importance, Version 2.0, EUCAST Development Laboratory for Antimicrobial Susceptibility Testing of bacteria c/o Clinical Microbiology, Central Hospital Sweden. 2017. Available online: https://www.eucast.org/resistance_mechanisms/ (accessed on 12 March 2021).
| Characteristics | 2017 N (%) | 2018 N (%) | 2019 N (%) | Total N (%) |
|---|---|---|---|---|
| Sample | 62 (25.9) | 81 (33.9) | 96 (40.2) | 239 (100.0) |
| Gender | ||||
| F | 41 (31.8) | 47 (36.4) | 41 (31.8) | 129 (54.0) |
| M | 21 (19.1) | 34 (30.9) | 55 (50.0) | 110 (46.0) |
| Age, years median (IQ) | 70.5 (51–80) | 75 (58.5–86) | 73 (55–84) | 73 (55–84) |
| Setting | ||||
| community-based | 34 (23.8) | 54 (37.8) | 55 (38.4) | 143 (59.8) |
| hospital-based | 28 (29.2) | 27 (28.1) | 41 (42.7) | 96 (40.2) |
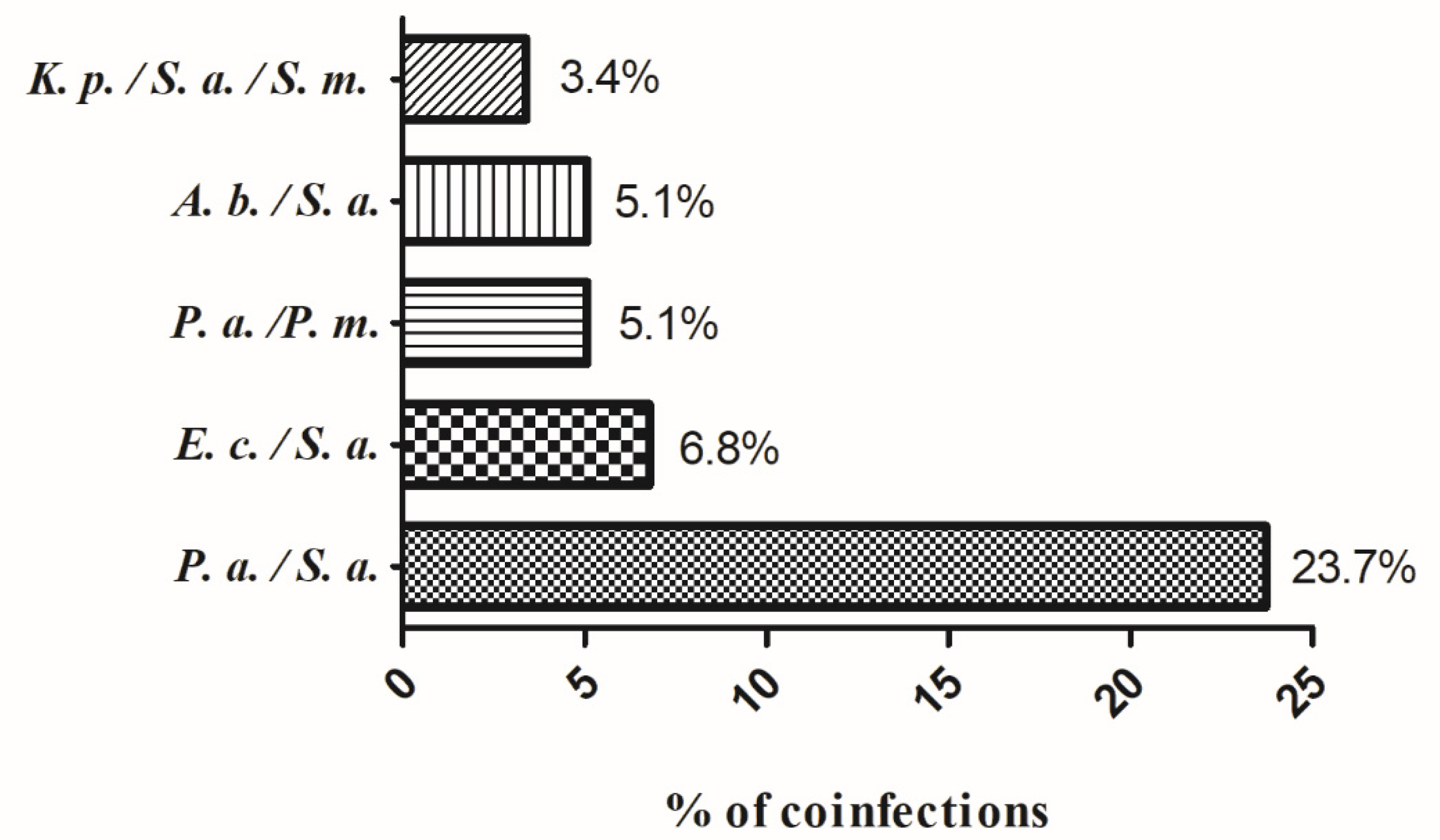
| Co-infection | ||||
| no | 50 (27.8) | 59 (32.8) | 71 (39.4) | 180 (75.3) |
| yes | 12 (20.3) | 22 (37.3) | 25 (42.4) | 59 (24.7) |
| Resistance Profile | 2017 N (%) | 2018 N (%) | 2019 N (%) | Total N (%) |
|---|---|---|---|---|
| Resistance | ||||
| no | 8 (28.6) | 11 (39.3) | 9 (32.1) | 28 (11.8) |
| yes | 54 (25.6) | 70 (33.2) | 87 (41.2) | 211 (88.2) |
| Multi-resistance | ||||
| no | 7 (26.9) | 5 (19.2) | 14 (53.9) | 26 (12.2) |
| 2 antimicrobials | 17 (38.6) | 14 (31.8) | 13 (29.6) | 44 (20.8) |
| 3 antimicrobials | 3 (12.5) | 10 (41.7) | 11 (45.8) | 24 (11.6) |
| 4 antimicrobials | 5 (16.1) | 11 (35.5) | 15 (48.4) | 31 (14.6) |
| 5 antimicrobials | 5 (20.8) | 6 (25.0) | 13 (54.2) | 24 (11.6) |
| ≥6 antimicrobials | 17 (27.4) | 24 (38.7) | 21 (33.9) | 62 (29.2) |
| Species | 2017 N (%) | 2018 N (%) | 2019 N (%) | Total N (%) |
|---|---|---|---|---|
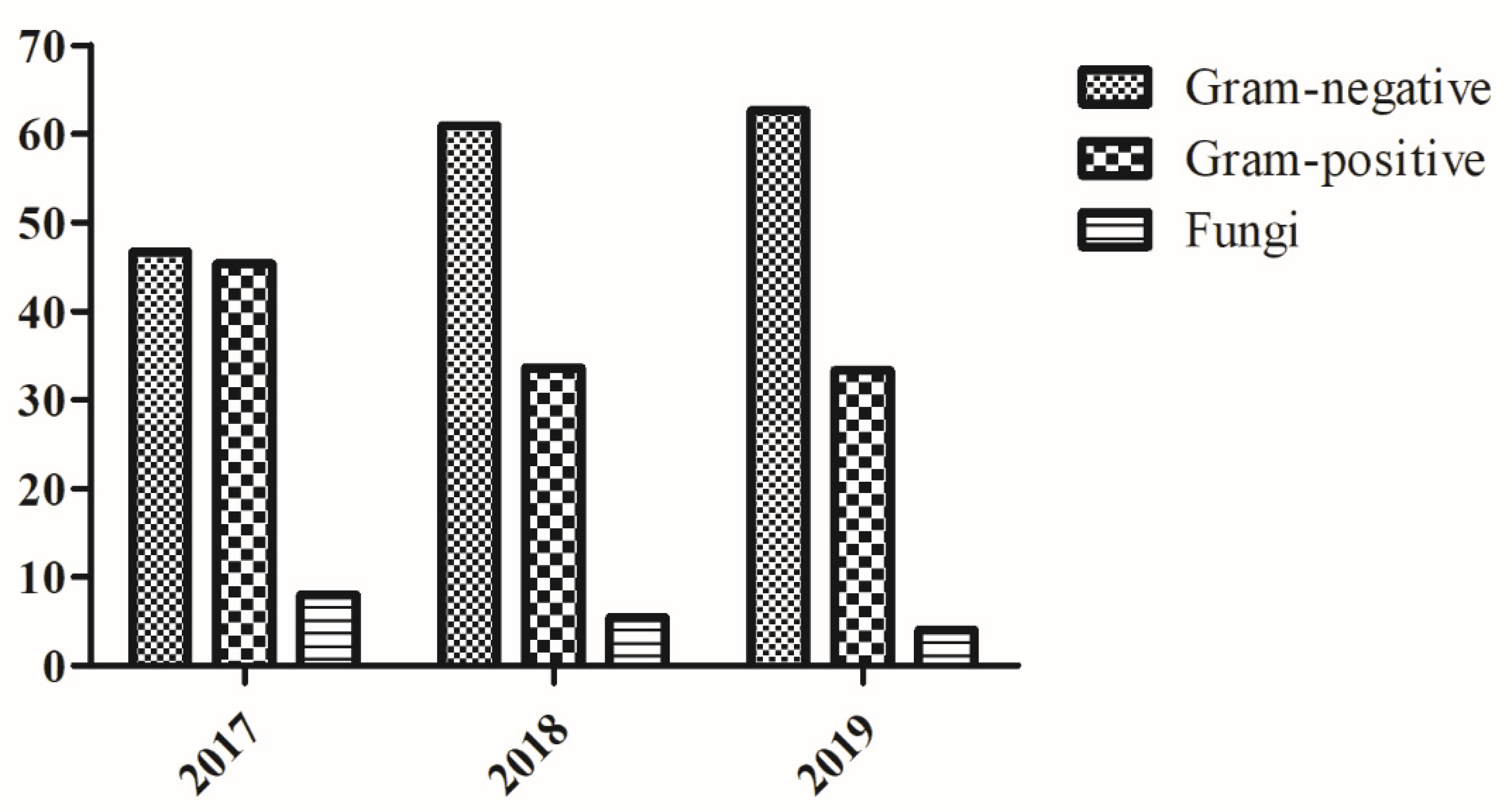
| Gram-negative | 40 (22.3) | 63 (35.2) | 76 (42.5) | 179 (57.9) |
| Acinetobacter baumannii/haemolyticus | 4 (23.6) | 5 (29.4) | 8 (47.0) | 17 (9.5) |
| Alcaligenes sp. | 3 (100.0) | - | - | 3 (1.7) |
| Citrobacter freundii | 1 (50.0) | - | 1 (50.0) | 2 (1.1) |
| Enterobacter cloacae | 2 (33.3) | 2 (33.3) | 2 (33.3) | 6 (3.4) |
| Escherichia coli | 9 (24.4) | 11 (29.7) | 17 (45.9) | 37 (20.7) |
| Klebsiella ornithinolytica | - | - | 1 (100.0) | 1 (0.6) |
| Klebsiella pneumoniae | 1 (20.0) | 3 (60.0) | 1 (20.0) | 5 (2.8) |
| Morganella morganii | - | 3 (60.0) | 2 (40.0) | 5 (2.8) |
| Proteus mirabilis | 5 (25.0) | 6 (30.0) | 9 (45.0) | 20 (11.2) |
| Proteus vulgaris | - | 1 (100.0) | - | 1 (0.6) |
| Providencia sp. | - | 2 (100.0) | - | 2 (1.1) |
| Pseudomonas aeruginosa | 14 (19.4) | 25 (34.8) | 33 (45.8) | 72 (40.2) |
| Pseudomonas fluorescens/putida | - | 1 (100.0) | - | 1 (0.6) |
| Serratia marcescens | 1 (14.3) | 4 (57.1) | 2 (28.6) | 7 (3.9) |
| Gram-positive | 40 (35.5) | 37 (32.7) | 36 (31.8) | 113 (36.6) |
| Enterococcus avium | 1 (100.0) | - | - | 1 (0.9) |
| Enterococcus faecalis | 2 (66.7) | 1 (33.3) | - | 3 (2.7) |
| Staphylococcus aureus | 29 (32.2) | 32 (35.6) | 29 (32.2) | 90 (79.4) |
| Staphylococcus auricularis | 1 (50.0) | - | 1 (50.0) | 2 (1.8) |
| Staphylococcus epidermidis | 3 (100.0) | - | - | 3 (2.7) |
| Staphylococcus haemolyticus | 2 (40.0) | 1 (20.0) | 2 (40.0) | 5 (4.4) |
| Staphylococcus lugdunensis | 1 (50.0) | - | 1 (50.0) | 2 (1.8) |
| Staphylococcus schleiferi subsp. coagulans | - | - | 1 (100.0) | 1 (0.9) |
| Staphylococcus sciuri | - | - | 1 (100.0) | 1 (0.9) |
| Staphylococcus simulans | 1 (50.0) | 1 (50.0) | - | 2 (1.8) |
| Streptococcus agalactiae | - | - | 1 (100.0) | 1 (0.9) |
| Streptococcus pyogenes | - | 1 (100.0) | - | 1 (0.9) |
| Streptococcus salivarius | - | 1 (100.0) | - | 1 (0.9) |
| Fungi | 8 (47.0) | 5 (29.4) | 4 (23.6) | 17 (5.5) |
| Candida albicans | 2 (28.6) | 3 (42.8) | 2 (28.6) | 7 (41.2) |
| Candida glabrata | 2 (66.7) | 1 (33.3) | - | 3 (17.6) |
| Candida guilliermondii | - | - | 1 (100.0) | 1 (5.9) |
| Candida parapsilosis | 2 (66.7) | 1 (33.3) | - | 3 (17.6) |
| Candida stellatoidea | 1 (100.0) | - | - | 1 (5.9) |
| Candida tropicalis | - | - | 1 (100.0) | 1 (5.9) |
| Candida sp. | 1 (100.0) | - | - | 1 (5.9) |
| Infection | p | |||
|---|---|---|---|---|
| 1 Species N (%) | 2 Species N (%) | 3 Species N (%) | ||
| age | ||||
| younger median age | 83 (46.1) | 25 (51.0) | 6 (60.0) | 0.605 |
| older median age | 97 (53.9) | 24 (49.0) | 4 (40.0) | |
| gender | ||||
| F | 97 (53.9) | 27 (55.1) | 5 (50.0) | 0.956 |
| M | 83 (46.1) | 22 (44.9) | 5 (50.0) | |
| Microorganisms | N (%) |
|---|---|
| Acinetobacter baumannii/haemolyticus | 6 (9.7) |
| Citrobacter freundii | 1 (1.6) |
| Enterobacter cloacae | 1 (1.6) |
| Escherichia coli | 8 (12.9) |
| Klebsiella pneumoniae | 4 (6.5) |
| Morganella morganii | 2 (3.2) |
| Proteus mirabilis | 13 (21.0) |
| Pseudomonas aeruginosa | 8 (12.9) |
| Staphylococcus aureus | 12 (19.4) |
| Staphylococcus auricularis | 2 (3.2) |
| Staphylococcus epidermidis | 3 (4.8) |
| Staphylococcus haemolyticus | 2 (3.2) |
| Microorganisms | N. of Resistances | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 N (%) | 2 N (%) | 3 N (%) | 4 N (%) | 5 N (%) | 6 N (%) | 7 N (%) | 8 N (%) | 9 N (%) | 10 N (%) | 12 N (%) | |
| Acinetobacter baumannii/haemolyticus | 1 (3.8) | 1 (2.3) | 4 (16.7) | 2 (6.5) | 3 (12.4) | 3 (14.3) | 3 (17.7) | - | - | - | - |
| Citrobacter freundii | - | 1 (2.3) | - | - | - | - | - | - | - | - | 1 (25.0) |
| Enterobacter cloacae | - | 3 (6.8) | - | 2 (6.5) | - | 1 (4.8) | - | - | - | - | - |
| Entrococcus faecalis | 1 (3.8) | - | - | - | - | - | - | - | - | - | - |
| Escherichia coli | 4 (15.5) | 3 (6.8) | 3 (12.5) | 6 (19.3) | 4 (16.6) | 4 (19.0) | 1 (5.8) | 1 (11.1) | 1 (10.0) | - | 1 (25.0) |
| Klebsiella ornithinolytica | - | - | - | - | 1 (4.2) | - | - | - | - | - | - |
| Klebsiella pneumoniae | - | - | - | - | 1 (4.2) | - | 2 (11.8) | - | 1 (10.0) | - | 1 (25.0) |
| Morganella morganii | - | - | - | 2 (6.5) | 1 (4.2) | 1 (4.8) | 1 (5.8) | - | - | - | - |
| Proteus mirabilis | 1 (3.8) | 2 (4.5) | 2 (8.3) | 1 (3.2) | 1 (4.2) | 3 (14.3) | 2 (11.8) | 2 (22.2) | 5 (50.0) | 1 (100.0) | - |
| Proteus vulgaris | - | - | - | - | 1 (4.2) | - | - | - | - | - | - |
| Providencia sp. | - | 1 (2.3) | - | 1 (3.2) | - | - | - | - | - | - | - |
| Pseudomonas aeruginosa | 12 (46.2) | 6 (13.7) | 3 (12.5) | 3 (9.7) | 6 (25.0) | 2 (9.5) | 5 (29.5) | 1 (11.1) | - | - | - |
| Serratia marcescens | 1 (3.8) | 2 (4.5) | 2 (8.3) | 1 (3.2) | 1 (4.2) | - | - | - | - | - | - |
| Staphylococcus aureus | 4 (15.5) | 24 (54.5) | 6 (25.0) | 12 (38.7) | 4 (16.6) | 5 (23.8) | 2 (11.8) | 2 (22.2) | 2 (20.0) | - | 1 (25.0) |
| Staphylococcus auricularis | - | - | - | - | - | - | - | 2 (22.2) | - | - | - |
| Staphylococcus epidermidis | - | - | - | - | - | 2 (9.5) | - | - | 1 (10.0) | - | - |
| Staphylococcus haemolyticus | 1 (3.8) | - | 1 (4.2) | 1 (3.2) | - | - | 1 (5.8) | 1 (11.1) | - | - | - |
| Staphylococcus lugdunensis | 1 (3.8) | 1 (2.3) | - | - | - | - | - | - | - | - | - |
| Staphylococcus sciuri | - | - | - | - | 1 (4.2) | - | - | - | - | - | - |
| Staphylococcus simulans | - | - | 2 (8.3) | - | - | - | - | - | - | - | - |
| Streptococcus salivarius | - | - | 1 (4.2) | - | - | - | - | - | - | - | - |
| N. of Resistances | p | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 N (%) | 2 N (%) | 3 N (%) | 4 N (%) | 5 N (%) | 6 N (%) | 7 N (%) | 8 N (%) | 9 N (%) | 10 N (%) | 12 N (%) | ||
| age | 0.080 | |||||||||||
| younger median age | 14 (53.8) | 29 (65.9) | 13 (54.2) | 12 (38.7) | 8 (33.3) | 9 (29.0) | 10 (59.8) | 5 (55.6) | 2 (20.0) | 1 (100.0) | 1 (25.0) | |
| older median age | 12 (46.2) | 15 (34.1) | 11 (45.8) | 19 (61.3) | 16 (66.7) | 12 (71.0) | 7 (41.2) | 4 (44.5) | 8 (80.0) | 0 (0.0) | 3 (75.0) | |
| gender | 0.625 | |||||||||||
| F | 14 (53.8) | 20 (45.5) | 13 (54.2) | 17 (54.8) | 13 (54.2) | 12 (57.1) | 11 (64.7) | 7 (77.8) | 6 (60.0) | 1 (100.0) | 2 (50.0) | |
| M | 12 (46.2) | 24 (54.5) | 11 (45.8) | 14 (45.2) | 11 (45.8) | 9 (42.9) | 6 (35.3) | 2 (22.2) | 4 (40.0) | 0 (0.0) | 2 (50.0) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Puca, V.; Marulli, R.Z.; Grande, R.; Vitale, I.; Niro, A.; Molinaro, G.; Prezioso, S.; Muraro, R.; Di Giovanni, P. Microbial Species Isolated from Infected Wounds and Antimicrobial Resistance Analysis: Data Emerging from a Three-Years Retrospective Study. Antibiotics 2021, 10, 1162. https://doi.org/10.3390/antibiotics10101162
Puca V, Marulli RZ, Grande R, Vitale I, Niro A, Molinaro G, Prezioso S, Muraro R, Di Giovanni P. Microbial Species Isolated from Infected Wounds and Antimicrobial Resistance Analysis: Data Emerging from a Three-Years Retrospective Study. Antibiotics. 2021; 10(10):1162. https://doi.org/10.3390/antibiotics10101162
Chicago/Turabian StylePuca, Valentina, Roberta Zita Marulli, Rossella Grande, Irene Vitale, Antonietta Niro, Gina Molinaro, Silvia Prezioso, Raffaella Muraro, and Pamela Di Giovanni. 2021. "Microbial Species Isolated from Infected Wounds and Antimicrobial Resistance Analysis: Data Emerging from a Three-Years Retrospective Study" Antibiotics 10, no. 10: 1162. https://doi.org/10.3390/antibiotics10101162
APA StylePuca, V., Marulli, R. Z., Grande, R., Vitale, I., Niro, A., Molinaro, G., Prezioso, S., Muraro, R., & Di Giovanni, P. (2021). Microbial Species Isolated from Infected Wounds and Antimicrobial Resistance Analysis: Data Emerging from a Three-Years Retrospective Study. Antibiotics, 10(10), 1162. https://doi.org/10.3390/antibiotics10101162