Current Status of Endolysin-Based Treatments against Gram-Negative Bacteria
Abstract
:1. Introduction
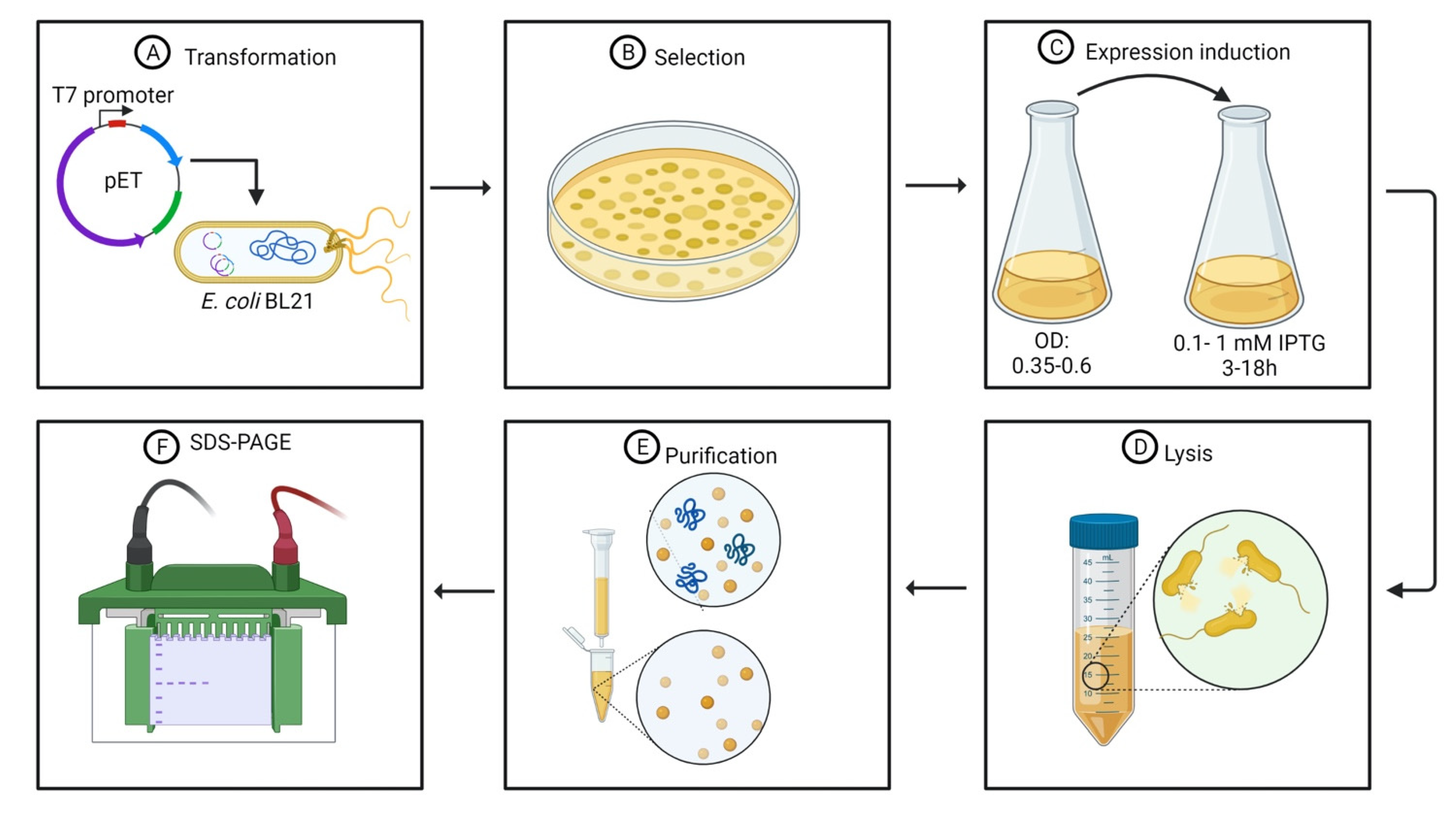
2. Availability of tRNAs and Protein Toxicity Limit Endolysin Heterologous Expression
3. Recombinant His-Tagged Endolysins Are Purified Using Affinity Chromatography
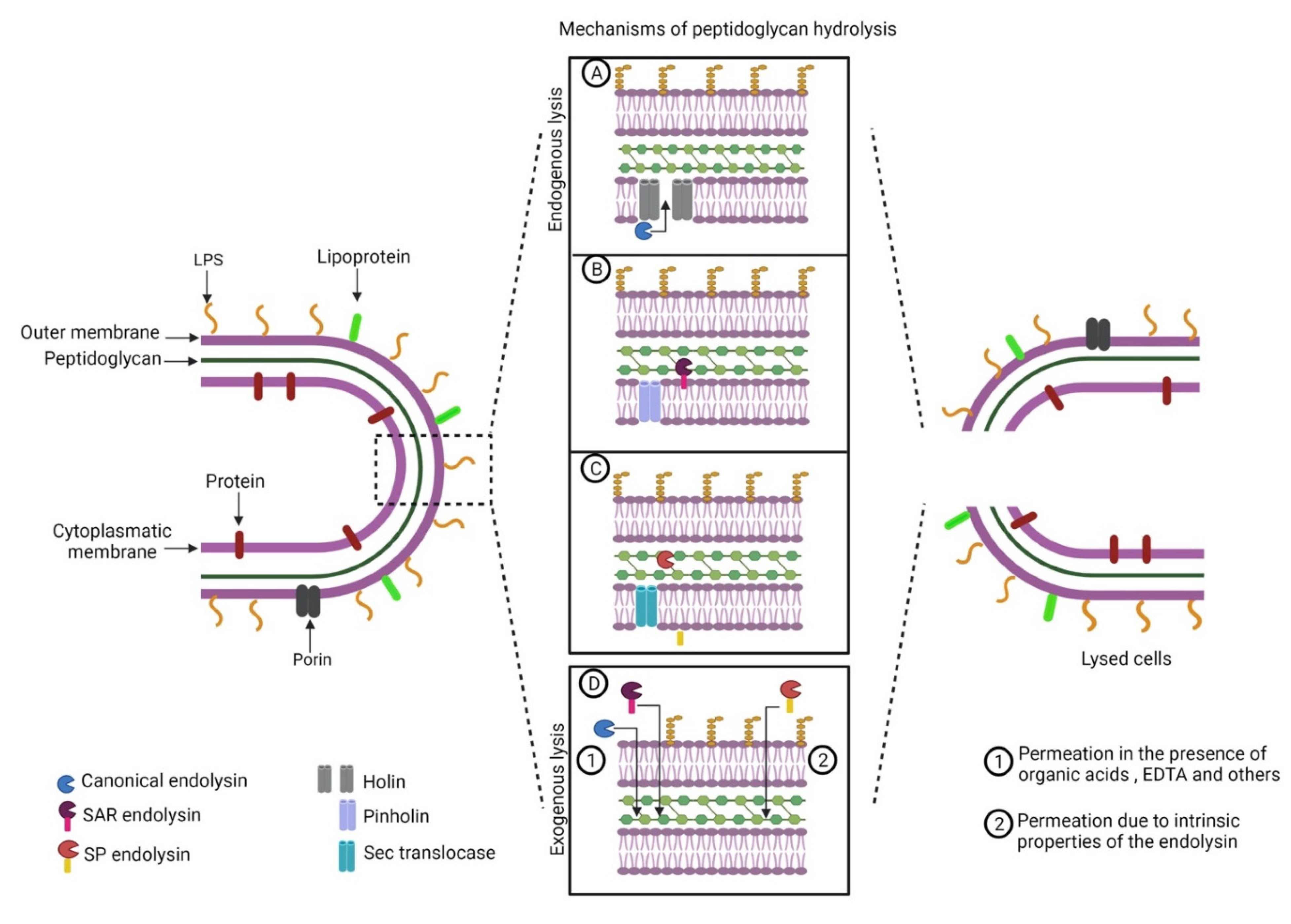
4. Endolysins as Therapeutic Agents against Gram-Negative Bacteria
4.1. Some Native Endolysins Reach the Peptidoglycan Layer Per Se
4.2. Outer Membrane Permeabilizers Improve Native Endolysin Diffusion
4.3. Protein Design Favors Endolysin Permeation through the Outer Membrane
5. Thermally Resistant Endolysins Are Promising Candidates for Drug Development
6. Bacteriophage Depolymerases Might Act Synergistically with Endolysins
7. Metagenomics in the Discovery of New Endolysins
8. Limitations of Endolysin Therapy
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.; Muzammil, S.; Rasool, M.H.; Nisar, M.A.; Alvi, R.F.; Aslam, M.A.; Qamar, M.U.; et al. Antibiotic resistance: A rundown of a global crisis. Infect. Drug Resist. 2018, 11, 1645–1658. [Google Scholar] [CrossRef] [Green Version]
- Ventola, C.L. The Antibiotic Resistance Crisis: Part 1: Causes and threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- Boggione, D.M.; Batalha, L.S.; Gontijo, M.T.; Lopez, M.E.; Teixeira, A.V.; Santos, I.J.; Mendonça, R.C. Evaluation of microencapsulation of the UFV-AREG1 bacteriophage in alginate-Ca microcapsules using microfluidic devices. Colloids Surfaces B Biointerfaces 2017, 158, 182–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gould, K. Antibiotics: From prehistory to the present day. J. Antimicrob. Chemother. 2016, 71, 572–575. [Google Scholar] [CrossRef]
- Pepper, J.W. The evolution of bacterial social life: From the ivory tower to the front lines of public health. Evol. Med. Public Health 2014, 2014, 65–68. [Google Scholar] [CrossRef]
- Zgurskaya, H.I.; López, C.A.; Gnanakaran, S. Permeability Barrier of Gram-Negative Cell Envelopes and Approaches to Bypass It. ACS Infect. Dis. 2015, 1, 512–522. [Google Scholar] [CrossRef] [Green Version]
- Bello, A.; Dingle, T. What’s That Resistance Mechanism? Understanding Genetic Determinants of Gram-Negative Bacterial Resistance. Clin. Microbiol. Newsl. 2018, 40, 165–174. [Google Scholar] [CrossRef]
- Eichenberger, E.; Thaden, J.T. Epidemiology and Mechanisms of Resistance of Extensively Drug Resistant Gram-Negative Bacteria. Antibiotics 2019, 8, 37. [Google Scholar] [CrossRef] [Green Version]
- Breijyeh, Z.; Jubeh, B.; Karaman, R. Resistance of Gram-Negative Bacteria to Current Antibacterial Agents and Approaches to Resolve It. Molecules 2020, 25, 1340. [Google Scholar] [CrossRef] [Green Version]
- Étienne, R.; Woerther, P.-L.; Barbier, F. Mechanisms of antimicrobial resistance in Gram-negative bacilli. Ann. Intensiv. Care 2015, 5, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Exner, M.; Bhattacharya, S.; Christiansen, B.; Gebel, J.; Goroncy-Bermes, P.; Hartemann, P.; Heeg, P.; Ilschner, C.; Kramer, A.; Larson, E.; et al. Antibiotic resistance: What is so special about multidrug-resistant Gram-negative bacteria? GMS Hyg. Infect. Control 2017, 12. [Google Scholar] [CrossRef]
- Twort, F. An Investigation on the Nature of Ultra-Microscopic Viruses. Lancet 1915, 186, 1241–1243. [Google Scholar] [CrossRef] [Green Version]
- D’Herelle, F. Bacteriophage as a Treatment in Acute Medical and Surgical Infections. Bull. N. Y. Acad. Med. 1931, 7, 329–348. [Google Scholar]
- Jacob, F.; Monod, J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 1961, 3, 318–356. [Google Scholar] [CrossRef]
- Ishino, Y.; Krupovic, M.; Forterre, P. History of CRISPR-Cas from Encounter with a Mysterious Repeated Sequence to Genome Editing Technology. J. Bacteriol. 2018, 200, e00580-17. [Google Scholar] [CrossRef] [Green Version]
- Lopez, M.E.S.; Batalha, L.S.; Vidigal, P.M.P.; Albino, L.A.A.; Boggione, D.M.G.; Gontijo, M.T.P.; Bazzolli, D.M.S.; Mendonca, R.C.S. Genome Sequence of the Enterohemorrhagic Escherichia coli Bacteriophage UFV-AREG1. Genome Announc. 2016, 4, e00412-16. [Google Scholar] [CrossRef] [Green Version]
- Gontijo, M.T.; Batalha, L.S.; Lopez, M.E.; Albino, L.A. Bacteriophage Genome Sequencing: A New Alternative to Understand Biochemical Interactions between Prokaryotic Cells and Phages. J. Microb. Biochem. Technol. 2017, 9, 169–173. [Google Scholar] [CrossRef] [Green Version]
- Lopez, M.; Gontijo, M.; Batalha, L.; Mendonca, R. Bio-Sanitization Using Specific Bacteriophages to Control Escherichia coli O157:H7 in Cherry Tomatoes. Adv. J. Food Sci. Technol. 2018, 16, 92–101. [Google Scholar] [CrossRef]
- Batalha, L.S.; Gontijo, M.T.P.; Teixeira, A.V.N.D.C.; Boggione, D.M.G.; Lopez, M.E.S.; Eller, M.R.; Mendonça, R.C.S. Encapsulation in alginate-polymers improves stability and allows controlled release of the UFV-AREG1 bacteriophage. Food Res. Int. 2021, 139, 109947. [Google Scholar] [CrossRef]
- Young, R.; Wang, I.-N.; Roof, W.D. Phages will out: Strategies of host cell lysis. Trends Microbiol. 2000, 8, 120–128. [Google Scholar] [CrossRef]
- Matamp, N.; Bhat, S.G. Phage Endolysins as Potential Antimicrobials against Multidrug Resistant Vibrio alginolyticus and Vibrio parahaemolyticus: Current Status of Research and Challenges Ahead. Microorganisms 2019, 7, 84. [Google Scholar] [CrossRef] [Green Version]
- Schmelcher, M.; Donovan, D.M.; Loessner, M.J. Bacteriophage endolysins as novel antimicrobials. Future Microbiol. 2012, 7, 1147–1171. [Google Scholar] [CrossRef] [Green Version]
- Young, R. Phage lysis: Three steps, three choices, one outcome. J. Microbiol. 2014, 52, 243–258. [Google Scholar] [CrossRef]
- Guo, M.; Feng, C.; Ren, J.; Zhuang, X.; Zhang, Y.; Zhu, Y.; Dong, K.; He, P.; Guo, X.-K.; Qin, J. A Novel Antimicrobial Endolysin, LysPA26, against Pseudomonas aeruginosa. Front. Microbiol. 2017, 8, 293. [Google Scholar] [CrossRef] [Green Version]
- Saier, M.H.; Reddy, B.L. Holins in Bacteria, Eukaryotes, and Archaea: Multifunctional Xenologues with Potential Biotechnological and Biomedical Applications. J. Bacteriol. 2015, 197, 7–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, R. Phage lysis: Do we have the hole story yet? Curr. Opin. Microbiol. 2013, 16, 790–797. [Google Scholar] [CrossRef] [Green Version]
- Berry, J.D.; Rajaure, M.; Young, R. Spanin function requires subunit homodimerization through intermolecular disulfide bonds. Mol. Microbiol. 2013, 88, 35–47. [Google Scholar] [CrossRef]
- Gontijo, M.T.P.; Vidigal, P.M.P.; Lopez, M.E.S.; Brocchi, M. Bacteriophages that infect Gram-negative bacteria as source of signal-arrest-release motif lysins. Res. Microbiol. 2021, 172, 103794. [Google Scholar] [CrossRef]
- Ghose, C.; Euler, C.W. Gram-Negative Bacterial Lysins. Antibiotics 2020, 9, 74. [Google Scholar] [CrossRef] [Green Version]
- Love, M.J.; Abeysekera, G.S.; Muscroft-Taylor, A.C.; Billington, C.; Dobson, R.C. On the catalytic mechanism of bacteriophage endolysins: Opportunities for engineering. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2020, 1868, 140302. [Google Scholar] [CrossRef]
- Oliveira, H.; Boas, D.V.; Emesnage, S.; Kluskens, L.D.; Lavigne, R.; Sillankorva, S.; Esecundo, F.; Eazeredo, J. Structural and Enzymatic Characterization of ABgp46, a Novel Phage Endolysin with Broad Anti-Gram-Negative Bacterial Activity. Front. Microbiol. 2016, 7, 208. [Google Scholar] [CrossRef] [Green Version]
- Altamirano, F.L.G.; Barr, J.J. Phage Therapy in the Postantibiotic Era. Clin. Microbiol. Rev. 2019, 32. [Google Scholar] [CrossRef] [Green Version]
- Brives, C.; Pourraz, J. Phage therapy as a potential solution in the fight against AMR: Obstacles and possible futures. Palgrave Commun. 2020, 6, 1–11. [Google Scholar] [CrossRef]
- Kortright, K.E.; Chan, B.K.; Koff, J.L.; Turner, P.E. Phage Therapy: A Renewed Approach to Combat Antibiotic-Resistant Bacteria. Cell Host Microbe 2019, 25, 219–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Principi, N.; Silvestri, E.; Esposito, S. Advantages and limitations of bacteriophages for the treatment of bacterial infections. Front. Pharmacol. 2019, 10, 513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaur, J.; Kumar, A.; Kaur, J. Strategies for optimization of heterologous protein expression in E. coli: Roadblocks and reinforcements. Int. J. Biol. Macromol. 2018, 106, 803–822. [Google Scholar] [CrossRef]
- Rosano, G.L.; Ceccarelli, E.A. Recombinant protein expression in Escherichia coli: Advances and challenges. Front. Microbiol. 2014, 5, 172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nieuwkoop, T.; Claassens, N.J.; Van Der Oost, J. Improved protein production and codon optimization analyses in Escherichia coli by bicistronic design. Microb. Biotechnol. 2018, 12, 173–179. [Google Scholar] [CrossRef] [Green Version]
- Parret, A.H.; Besir, H.; Meijers, R. Critical reflections on synthetic gene design for recombinant protein expression. Curr. Opin. Struct. Biol. 2016, 38, 155–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gopal, G.J.; Kumar, A. Strategies for the Production of Recombinant Protein in Escherichia coli. Protein J. 2013, 32, 419–425. [Google Scholar] [CrossRef]
- Jia, B.; Jeon, C.O. High-throughput recombinant protein expression in Escherichia coli: Current status and future perspectives. Open Biol. 2016, 6, 6. [Google Scholar] [CrossRef] [Green Version]
- Plotka, M.; Kaczorowska, A.-K.; Stefanska, A.; Morzywolek, A.; Fridjonsson, O.H.; Dunin-Horkawicz, S.; Kozlowski, L.; Hreggvidsson, G.O.; Kristjansson, J.K.; Dabrowski, S.; et al. Novel Highly Thermostable Endolysin from Thermus scotoductus MAT2119 Bacteriophage Ph2119 with Amino Acid Sequence Similarity to Eukaryotic Peptidoglycan Recognition Proteins. Appl. Environ. Microbiol. 2013, 80, 886–895. [Google Scholar] [CrossRef] [Green Version]
- Bailly-Bechet, M.; Danchin, A.; Iqbal, M.; Marsili, M.; Vergassola, M. Codon Usage Domains over Bacterial Chromosomes. PLoS Comput. Biol. 2006, 2, e37. [Google Scholar] [CrossRef]
- Chithambaram, S.; Prabhakaran, R.; Xia, X. The effect of mutation and selection on codon adaptation in Escherichia coli bacteriophage. Genetics 2014, 197, 301–315. [Google Scholar] [CrossRef] [Green Version]
- Sharp, P.M. Codon Usage Bias. In Encyclopedia of Genetics; Brenner, S., Miller, J.H., Eds.; Academic Press: Cambridge, MA, USA, 2001; pp. 402–406. ISBN 978-0-12-227080-2. [Google Scholar]
- Shilling, P.J.; Mirzadeh, K.; Cumming, A.J.; Widesheim, M.; Köck, Z.; Daley, D.O. Improved designs for pET expression plasmids increase protein production yield in Escherichia coli. Commun. Biol. 2020, 3, 1–8. [Google Scholar] [CrossRef]
- Kim, S.; Lee, D.-W.; Jin, J.-S.; Kim, J. Antimicrobial activity of LysSS, a novel phage endolysin, against Acinetobacter baumannii and Pseudomonas aeruginosa. J. Glob. Antimicrob. Resist. 2020, 22, 32–39. [Google Scholar] [CrossRef]
- Oliveira, H.; Thiagarajan, V.; Walmagh, M.; Sillankorva, S.; Lavigne, R.; Neves-Petersen, M.T.; Kluskens, L.; Azeredo, J. A Thermostable Salmonella Phage Endolysin, Lys68, with Broad Bactericidal Properties against Gram-Negative Pathogens in Presence of Weak Acids. PLoS ONE 2014, 9, e108376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, F.M.; Gondil, V.S.; Li, C.; Jiang, M.; Li, J.; Yu, J.; Wei, H.; Yang, H. A Novel Acinetobacter baumannii Bacteriophage Endolysin LysAB54 With High Antibacterial Activity Against Multiple Gram-Negative Microbes. Front. Cell. Infect. Microbiol. 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Yang, E.; Chang, P.-S.; Ryu, S. Preparation and characterization of endolysin-containing liposomes and evaluation of their antimicrobial activities against gram-negative bacteria. Enzym. Microb. Technol. 2019, 128, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.-A.; Shin, H.; Kang, D.-H.; Ryu, S. Characterization of endolysin from a Salmonella Typhimurium-infecting bacteriophage SPN1S. Res. Microbiol. 2012, 163, 233–241. [Google Scholar] [CrossRef]
- Lim, J.-A.; Shin, H.; Heu, S.; Ryu, S. Exogenous Lytic Activity of SPN9CC Endolysin Against Gram-Negative Bacteria. J. Microbiol. Biotechnol. 2014, 24, 803–811. [Google Scholar] [CrossRef]
- Yan, G.; Yang, R.; Fan, K.; Dong, H.; Gao, C.; Wang, S.; Yu, L.; Cheng, Z.; Lei, L. External lysis of Escherichia coli by a bacteriophage endolysin modified with hydrophobic amino acids. AMB Express 2019, 9, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antonova, N.P.; Vasina, D.V.; Lendel, A.M.; Usachev, E.V.; Makarov, V.V.; Gintsburg, A.L.; Tkachuk, A.P.; Gushchin, V.A. Broad Bactericidal Activity of the Myoviridae Bacteriophage Lysins LysAm24, LysECD7, and LysSi3 against Gram-Negative ESKAPE Pathogens. Viruses 2019, 11, 284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antonova, N.P.; Vasina, D.V.; Rubalsky, E.O.; Fursov, M.V.; Savinova, A.S.; Grigoriev, I.V.; Usachev, E.V.; Shevlyagina, N.V.; Zhukhovitsky, V.G.; Balabanyan, V.U.; et al. Modulation of Endolysin LysECD7 Bactericidal Activity by Different Peptide Tag Fusion. Biomolecules 2020, 10, 440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blasco, L.; Ambroa, A.; Trastoy, R.; Bleriot, I.; Moscoso, M.; Fernandez-Garcia, L.; Perez-Nadales, E.; Fernández-Cuenca, F.; Torre-Cisneros, J.; Oteo-Iglesias, J.; et al. In vitro and in vivo efficacy of combinations of colistin and different endolysins against clinical strains of multi-drug resistant pathogens. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- Ramos, C.; Abreu, P.; Nascimento, A.; Ho, P. A high-copy T7 Escherichia coli expression vector for the production of recombinant proteins with a minimal N-terminal His-tagged fusion peptide. Braz. J. Med Biol. Res. 2004, 37, 1103–1109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, P.-C.; Liu, Z.-Q. Construction of pET-32 α (+) vector for protein expression and purification. N. Am. J. Med. Sci. 2012, 4, 651–655. [Google Scholar] [CrossRef] [Green Version]
- Bornhorst, J.A.; Falke, J.J. Purification of proteins using polyhistidine affinity tags. Methods Enzymol. 2000, 326, 245–254. [Google Scholar] [CrossRef] [Green Version]
- Mondal, S.I.; Draper, L.A.; Ross, R.P.; Hill, C. Bacteriophage endolysins as a potential weapon to combat Clostridioides difficile infection. Gut Microbes 2020, 12, 1813533. [Google Scholar] [CrossRef] [PubMed]
- Briers, Y.; Walmagh, M.; Grymonprez, B.; Biebl, M.; Pirnay, J.-P.; Defraine, V.; Michiels, J.; Cenens, W.; Aertsen, A.; Miller, S.; et al. Art-175 Is a Highly Efficient Antibacterial against Multidrug-Resistant Strains and Persisters of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2014, 58, 3774–3784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutiérrez, D.; Fernández, L.; Rodríguez, A.; García, P. Are Phage Lytic Proteins the Secret Weapon to Kill Staphylococcus aureus? mBio 2018, 9, e01923-17. [Google Scholar] [CrossRef] [Green Version]
- Loeffler, J.M.; Nelson, D.; Fischetti, V.A. Rapid Killing of Streptococcus pneumoniae with a Bacteriophage Cell Wall Hydrolase. Science 2001, 294, 2170–2172. [Google Scholar] [CrossRef]
- Gilmer, D.B.; Schmitz, J.E.; Euler, C.W.; Fischetti, V.A. Novel Bacteriophage Lysin with Broad Lytic Activity Protects against Mixed Infection by Streptococcus pyogenes and Methicillin-Resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2013, 57, 2743–2750. [Google Scholar] [CrossRef] [Green Version]
- Pastagia, M.; Euler, C.; Chahales, P.; Fuentes-Duculan, J.; Krueger, J.G.; Fischetti, V.A. A Novel Chimeric Lysin Shows Superiority to Mupirocin for Skin Decolonization of Methicillin-Resistant and -Sensitive Staphylococcus aureus Strains. Antimicrob. Agents Chemother. 2010, 55, 738–744. [Google Scholar] [CrossRef] [Green Version]
- Gerstmans, H.; Rodriguez-Rubio, L.; Lavigne, R.; Briers, Y. From endolysins to Artilysin®s: Novel enzyme-based approaches to kill drug-resistant bacteria. Biochem. Soc. Trans. 2016, 44, 123–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischetti, V.A. Bacteriophage endolysins: A novel anti-infective to control Gram-positive pathogens. Int. J. Med. Microbiol. 2010, 300, 357–362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischetti, V.A. Using phage Lytic Enzymes to Control Pathogenic Bacteria. BMC Oral Health 2006, 6, S16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, D.; Loomis, L.; Fischetti, V.A. Prevention and elimination of upper respiratory colonization of mice by group A streptococci by using a bacteriophage lytic enzyme. Proc. Natl. Acad. Sci. USA 2001, 98, 4107–4112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gondil, V.S.; Harjai, K.; Chhibber, S. Endolysins as emerging alternative therapeutic agents to counter drug-resistant infections. Int. J. Antimicrob. Agents 2020, 55, 105844. [Google Scholar] [CrossRef] [PubMed]
- Ciepluch, K.; Skrzyniarz, K.; Barrios-Gumiel, A.; Quintana, S.; Sánchez-Nieves, J.; De La Mata, F.J.; Maciejewska, B.; Drulis-Kawa, Z.; Arabski, M. Dendronized Silver Nanoparticles as Bacterial Membrane Permeabilizers and Their Interactions with P aeruginosa Lipopolysaccharides, Lysozymes, and Phage-Derived Endolysins. Front. Microbiol. 2019, 10, 2771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finnegan, S.; Percival, S. EDTA: An Antimicrobial and Antibiofilm Agent for Use in Wound Care. Adv. Wound Care 2015, 4, 415–421. [Google Scholar] [CrossRef] [Green Version]
- Bollenbach, T. Antimicrobial interactions: Mechanisms and implications for drug discovery and resistance evolution. Curr. Opin. Microbiol. 2015, 27, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Sercombe, L.; Veerati, T.; Moheimani, F.; Wu, S.; Sood, A.K.; Hua, S. Advances and Challenges of Liposome Assisted Drug Delivery. Front. Pharmacol. 2015, 6, 286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zampara, A.; Sørensen, M.C.H.; Grimon, D.; Antenucci, F.; Vitt, A.R.; Bortolaia, V.; Briers, Y.; Brøndsted, L. Exploiting phage receptor binding proteins to enable endolysins to kill Gram-negative bacteria. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Lukacik, P.; Barnard, T.J.; Keller, P.W.; Chaturvedi, K.S.; Seddiki, N.; Fairman, J.W.; Noinaj, N.; Kirby, T.L.; Henderson, J.P.; Steven, A.C.; et al. Structural engineering of a phage lysin that targets Gram-negative pathogens. Proc. Natl. Acad. Sci. USA 2012, 109, 9857–9862. [Google Scholar] [CrossRef] [Green Version]
- Briers, Y.; Walmagh, M.; Van Puyenbroeck, V.; Cornelissen, A.; Cenens, W.; Aertsen, A.; Oliveira, H.; Azeredo, J.; Verween, G.; Pirnay, J.-P.; et al. Engineered Endolysin-Based “Artilysins” To Combat Multidrug-Resistant Gram-Negative Pathogens. mBio 2014, 5, e01379-14. [Google Scholar] [CrossRef] [Green Version]
- Mikoulinskaia, G.V.; Chernyshov, S.V.; Shavrina, M.S.; Molochkov, N.V.; Lysanskaya, V.Y.; Zimin, A.A. Two novel thermally resistant endolysins encoded by pseudo T-even bacteriophages RB43 and RB49. J. Gen. Virol. 2018, 99, 402–415. [Google Scholar] [CrossRef] [PubMed]
- Plotka, M.; Kaczorowska, A.-K.; Morzywolek, A.; Makowska, J.; Kozlowski, L.; Thorisdottir, A.; Skírnisdottir, S.; Hjörleifsdottir, S.; Fridjonsson, O.H.; Hreggvidsson, G.O.; et al. Biochemical Characterization and Validation of a Catalytic Site of a Highly Thermostable Ts2631 Endolysin from the Thermus scotoductus Phage vB_Tsc2631. PLoS ONE 2015, 10, e0137374. [Google Scholar] [CrossRef] [Green Version]
- Plotka, M.; Kapusta, M.; Dorawa, S.; Kaczorowska, A.-K.; Kaczorowski, T. Ts2631 Endolysin from the Extremophilic Thermus scotoductus Bacteriophage vB_Tsc2631 as an Antimicrobial Agent against Gram-Negative Multidrug-Resistant Bacteria. Viruses 2019, 11, 657. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Xiong, Y.; Xiao, Y.; Han, J.; Deng, X.; Lin, L. MMPphg from the thermophilic Meiothermus bacteriophage MMP17 as a potential antimicrobial agent against both Gram-negative and Gram-positive bacteria. Virol. J. 2020, 17, 1–10. [Google Scholar] [CrossRef]
- Knecht, L.E.; Veljkovic, M.; Fieseler, L. Diversity and Function of Phage Encoded Depolymerases. Front. Microbiol. 2020, 10, 2949. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Li, P.; Zhu, Y.; Huang, Y.; Gao, M.; Yuan, X.; Niu, W.; Liu, H.; Fan, H.; Qin, Y.; et al. Identification of a Novel Acinetobacter baumannii Phage-Derived Depolymerase and Its Therapeutic Application in Mice. Front. Microbiol. 2020, 11, 1407. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.-J.; Lin, T.-L.; Lin, Y.-T.; Su, P.-A.; Chen, C.-T.; Hsieh, P.-F.; Hsu, C.-R.; Chen, C.-C.; Hsieh, Y.-C.; Wang, J.-T. Identification of Capsular Types in Carbapenem-Resistant Klebsiella pneumoniae Strains bywzcSequencing and Implications for Capsule Depolymerase Treatment. Antimicrob. Agents Chemother. 2015, 59, 1038–1047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernández-Ruiz, I.; Coutinho, F.H.; Rodriguez-Valera, F. Thousands of Novel Endolysins Discovered in Uncultured Phage Genomes. Front. Microbiol. 2018, 9, 1033. [Google Scholar] [CrossRef] [Green Version]
- Gondil, V.S.; Chhibber, S. Bacteriophage and Endolysin Encapsulation Systems: A Promising Strategy to Improve Therapeutic Outcomes. Front. Pharmacol. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Barreto, P.D.S.; Vellas, B.; Rolland, Y. Physical activity and exercise in the context of SARS-CoV-2: A perspective from geroscience field. Ageing Res. Rev. 2021, 66, 101258. [Google Scholar] [CrossRef] [PubMed]
- Agu, R.U.; Ugwoke, M.I.; Armand, M.; Kinget, R.; Verbeke, N. The lung as a route for systemic delivery of therapeutic proteins and peptides. Respir. Res. 2001, 2, 198–209. [Google Scholar] [CrossRef] [Green Version]
- Murray, E.; Draper, L.; Ross, R.; Hill, C. The Advantages and Challenges of Using Endolysins in a Clinical Setting. Viruses 2021, 13, 680. [Google Scholar] [CrossRef]
- Furfaro, L.L.; Payne, M.S.; Chang, B.J. Bacteriophage Therapy: Clinical Trials and Regulatory Hurdles. Front. Cell. Infect. Microbiol. 2018, 8, 376. [Google Scholar] [CrossRef] [Green Version]
- Love, M.J.; Bhandari, D.; Dobson, R.C.J.; Billington, C. Potential for Bacteriophage Endolysins to Supplement or Replace Antibiotics in Food Production and Clinical Care. Antibiotics 2018, 7, 17. [Google Scholar] [CrossRef] [Green Version]
| Endolysin | Vector | System | Induction | Reference |
|---|---|---|---|---|
| LysAm24 | pET42b | BL21(DE3) pLysS | LB broth (37 °C, 240 rpm) to an OD600 of 0.55–0.65 and 1 mM IPTG at 37 °C for 3 h | [54] |
| LysECD7 | ||||
| LysSi3 | ||||
| BSP16Lys | pET28a | BL21(DE3) | LB broth (37 °C) to an OD600 of 0.5 and 0.5 mM IPTG at 37 °C for 3 h | [55] |
| LysAB54 | pET28a | BL21(DE3) | LB broth (37 °C) to an OD600 of 0.6 and 1.0 mM IPTG at 16 °C for 16 h | [49] |
| ElyA1 | pET28a | Rosetta (DE3) pLysS | LB broth (37 °C, 180 rpm) and 1 mM IPTG at 30 °C for 5 h | [56] |
| ElyA2 | ||||
| LysPA26 | pET28b | BL21(DE3) | LB broth (37 °C) and 1 mM IPTG at 25 °C for 5 h | [24] |
| LysSS | pET21 | BL21(DE3) | LB broth (37 °C) to an OD600 of 0.5 and 0.1 mM IPTG at 18 °C for 16 h | [47] |
| LysSPN1S | pET15 | BL21(DE3) | LB broth to an OD600 of 0.6 and 1 mM IPTG for 3 h | [51] |
| LysSPN9CC | pET29b | BL21(DE3) | LB broth to an OD600 of 0.6 and 1 mM IPTG for 4 h | [52] |
| Lys68 | pET28a | BL21(DE3) | LB broth (37 °C, 120 rpm) to an OD600 of 0.6 and 0.5 mM IPTG at 16 °C for 18 h | [48] |
| Abgp46 | pET15b | BL21(DE3) | [31] | |
| Lysep3 | pET28a | BL21(DE3) | LB broth (37 °C, 180 rpm) to an OD600 of 0.35 and 0.1 mM IPTG at 37 °C for 3 h | [53] |
| Type of Study | Main Findings |
|---|---|
| Native endolysins | Polyhistidine tags influence outer membrane permeation. Transmembrane regions in the N-terminal region of endolysins comprising signal–arrest–release domains could be responsible for outer membrane permeation. The mechanisms of outer membrane permeation by endolysins remain inconclusive, but both in vitro and in vivo research revealed that some native endolysins can inhibit Gram-negative bacteria. |
| Native endolysins combined with outer membrane permeabilizers | Organic acids improve the antimicrobial activity of endolysins or facilitate the permeation of endolysins. Liposomes, silver nanoparticles, and polymyxins facilitate outer membrane permeation of endolysins. |
| Engineered endolysins | Hybrids of endolysins with outer membrane receptors, bacteriocins, and outer membrane-destabilizing peptides enhance endolysin diffusion. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gontijo, M.T.P.; Jorge, G.P.; Brocchi, M. Current Status of Endolysin-Based Treatments against Gram-Negative Bacteria. Antibiotics 2021, 10, 1143. https://doi.org/10.3390/antibiotics10101143
Gontijo MTP, Jorge GP, Brocchi M. Current Status of Endolysin-Based Treatments against Gram-Negative Bacteria. Antibiotics. 2021; 10(10):1143. https://doi.org/10.3390/antibiotics10101143
Chicago/Turabian StyleGontijo, Marco Túlio Pardini, Genesy Perez Jorge, and Marcelo Brocchi. 2021. "Current Status of Endolysin-Based Treatments against Gram-Negative Bacteria" Antibiotics 10, no. 10: 1143. https://doi.org/10.3390/antibiotics10101143
APA StyleGontijo, M. T. P., Jorge, G. P., & Brocchi, M. (2021). Current Status of Endolysin-Based Treatments against Gram-Negative Bacteria. Antibiotics, 10(10), 1143. https://doi.org/10.3390/antibiotics10101143








