Development of a Fish Cell Biosensor System for Genotoxicity Detection Based on DNA Damage-Induced Trans-Activation of p21 Gene Expression
Abstract
:Abbreviations
| p53 | a tumor suppressor |
| p21 | cyclin-dependent kinase 1A inhibitor |
| CMV | cytomegalovirus |
| GFP | green fluorescent protein |
| eGFP | enhanced green fluorescent protein |
| HPRT | hypoxanthine-guanine phosphoribosyltransferase |
| TK | thymidine kinase |
| GADD | growth arrest and DNA damage |
| FG | flounder gill |
| G418 | geneticin |
| DMSO | dimethyl sulfoxide |
| MTT | thiazolyl blue tetrazolium bromide |
| DEHP | di(2-ethylhexyl) phthalate |
| S9 | induced rat liver microsomal activation system |
| MEM | minimal essential medium |
| BCS | bovine calf serum |
| FBS | fetal bovine serum |
| PBS | phosphate-buffered saline |
| MEF | mouse embryo fibroblast |
| LARII | Luciferase Assay Reagent II |
| RLU | relative light units |
| ECVAM | European Center for the Validation of Alternative Methods |
| LEC | lowest effective concentration |
1. Introduction
2. Experimental Section
2.1. Chemicals
2.2. Cell Line and Plasmids
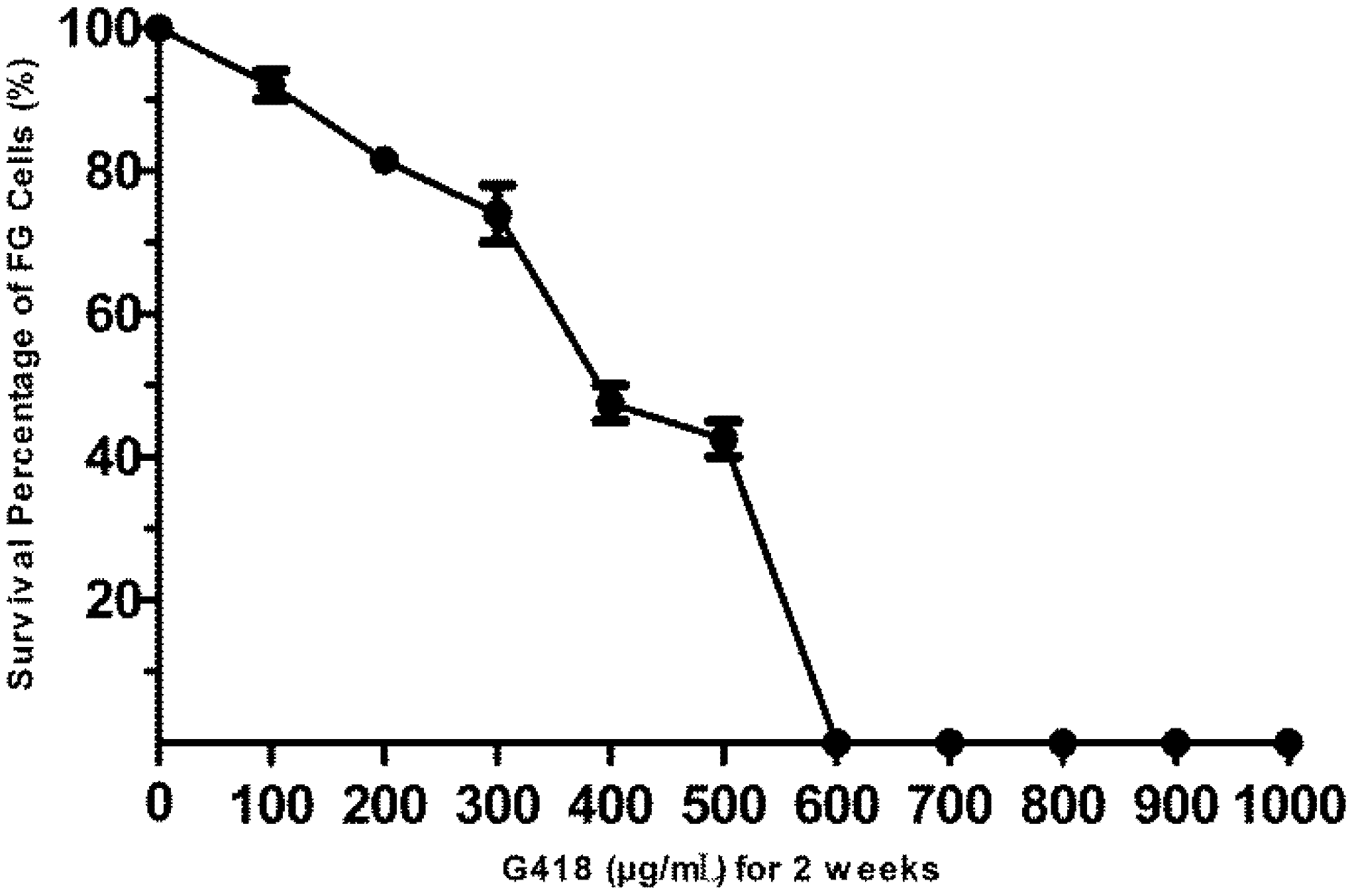
2.3. Analysis of the Sensitivity of FG Cells to G418
2.4. Thiazolyl Blue Tetrazolium Bromide (MTT) Assay
2.5. Validation of the Integrity of the Endogenous p53 Signaling Pathway in FG Cells
2.6. Construction of Stably Transformed FG Cells (p21FGLuc)
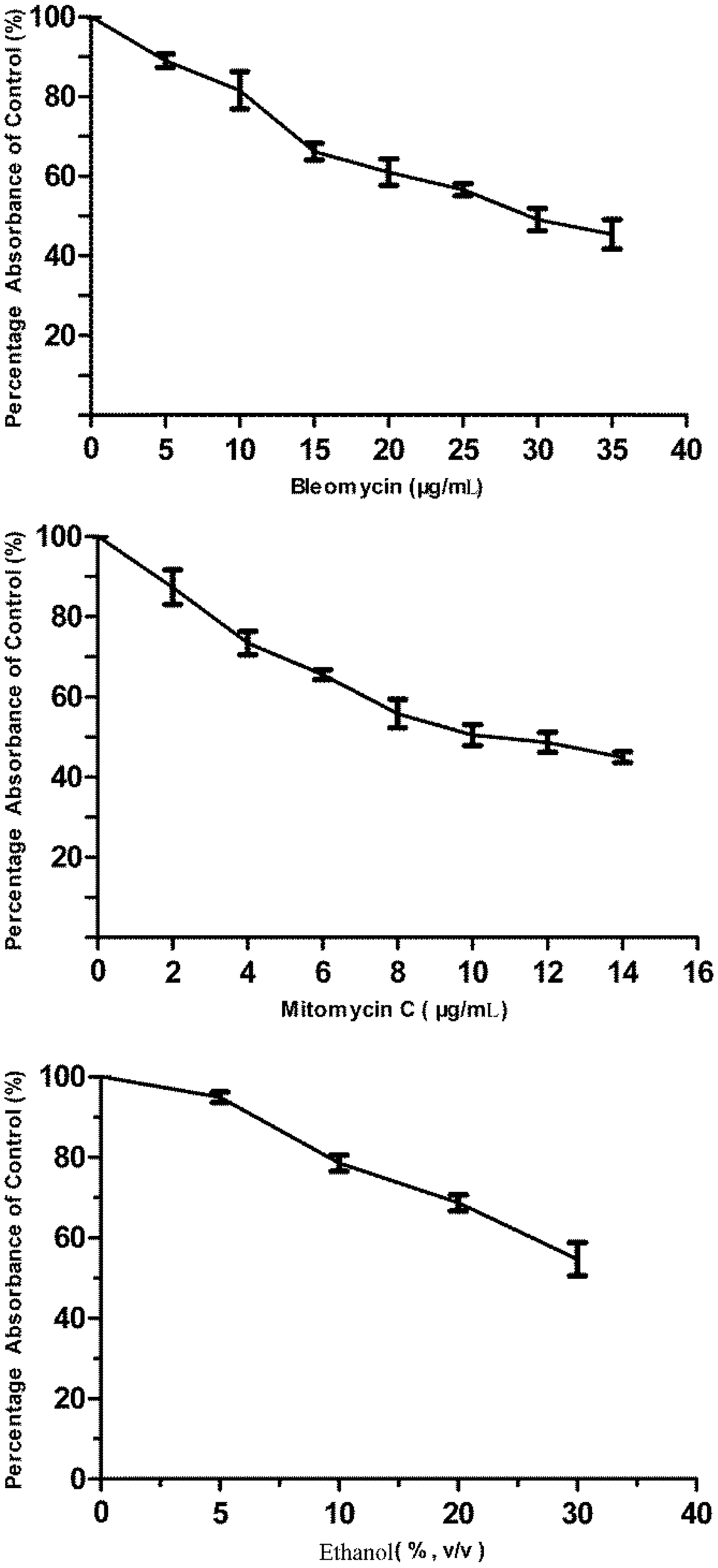
2.7. Challenging the p21FGLuc Cells with Model Genotoxins of Bleomycin and Mitomycin C and Non-Genotoxin of Ethanol
2.8. Dual Luciferase Reporter Gene Assay
2.9. Challenging the p21FGLuc Cells with Varied Doses of Cyclophosphamide, Di(2-ethylhexyl) Phthalate (DEHP), D-Mannitol and Sodium Butyrate
2.10. Statistical Analysis
3. Results
3.1. Cytotoxicity of Bleomycin, Mitomycin C and Ethanol to FG Cells
3.2. The Sensitivity of FG Cells to G418
3.3. FG Cells Have Wild-Type p53 Signaling Pathway
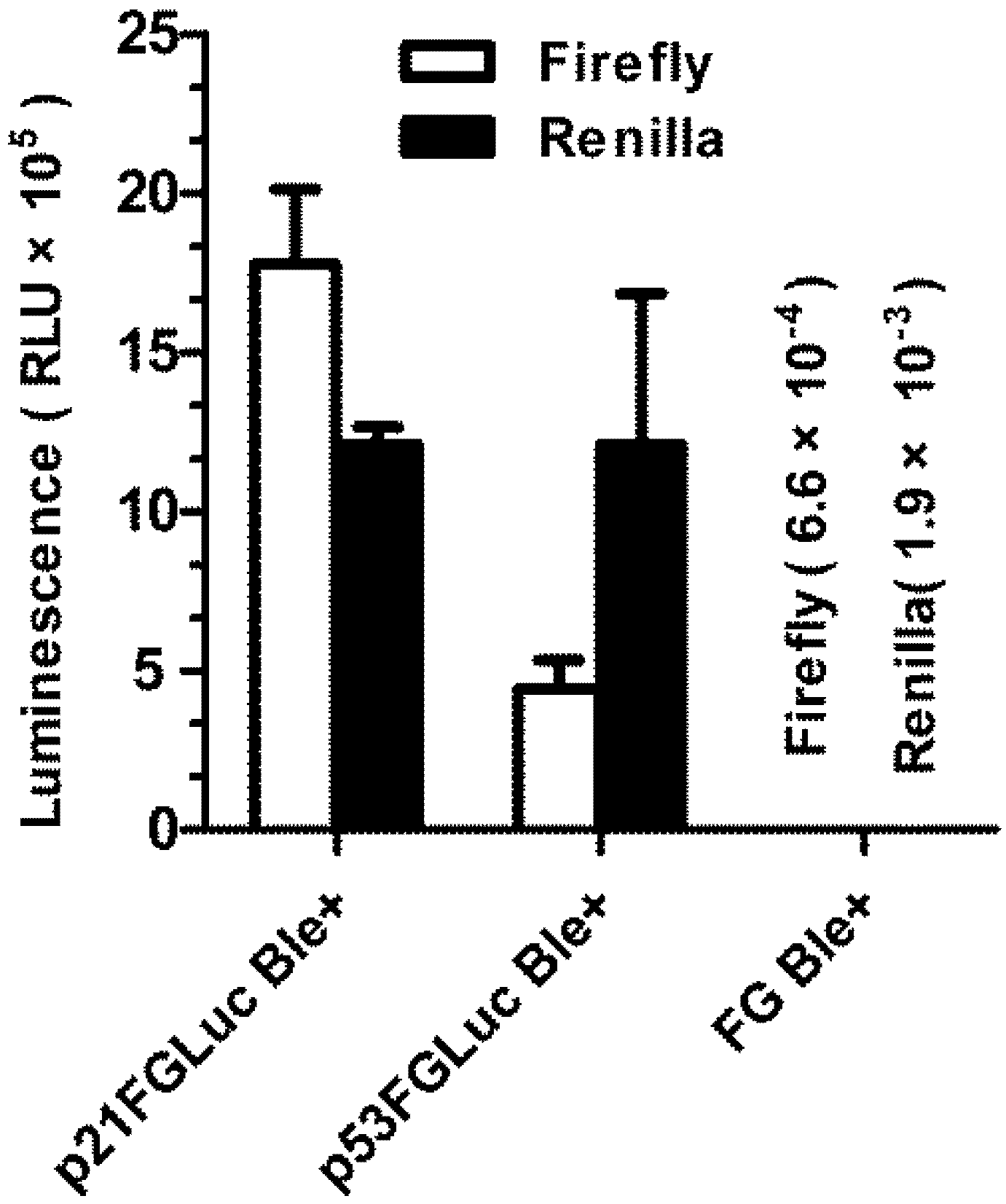
3.4. Construction of Stably Transformed FG Cells (p21FGLuc)
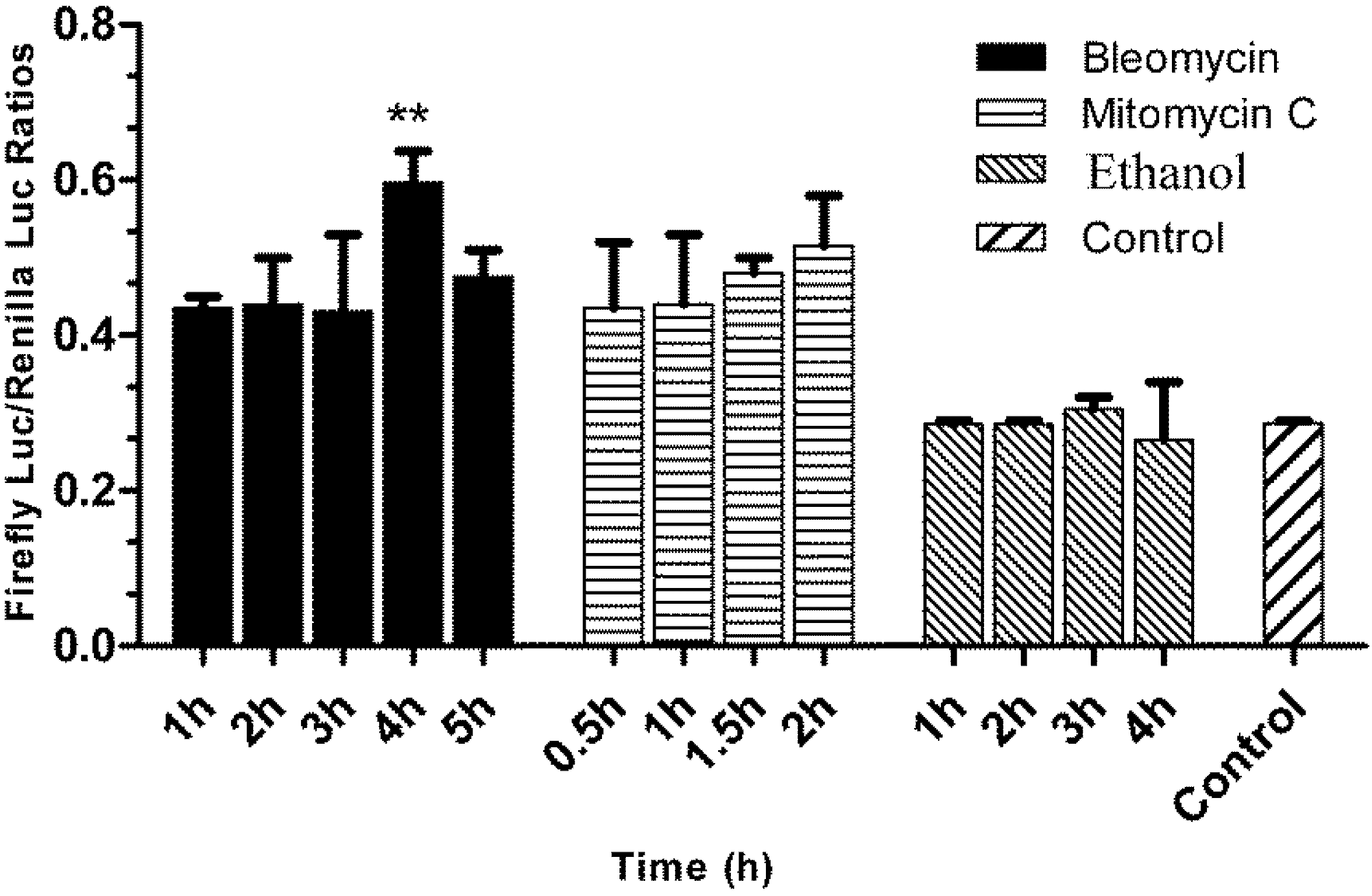
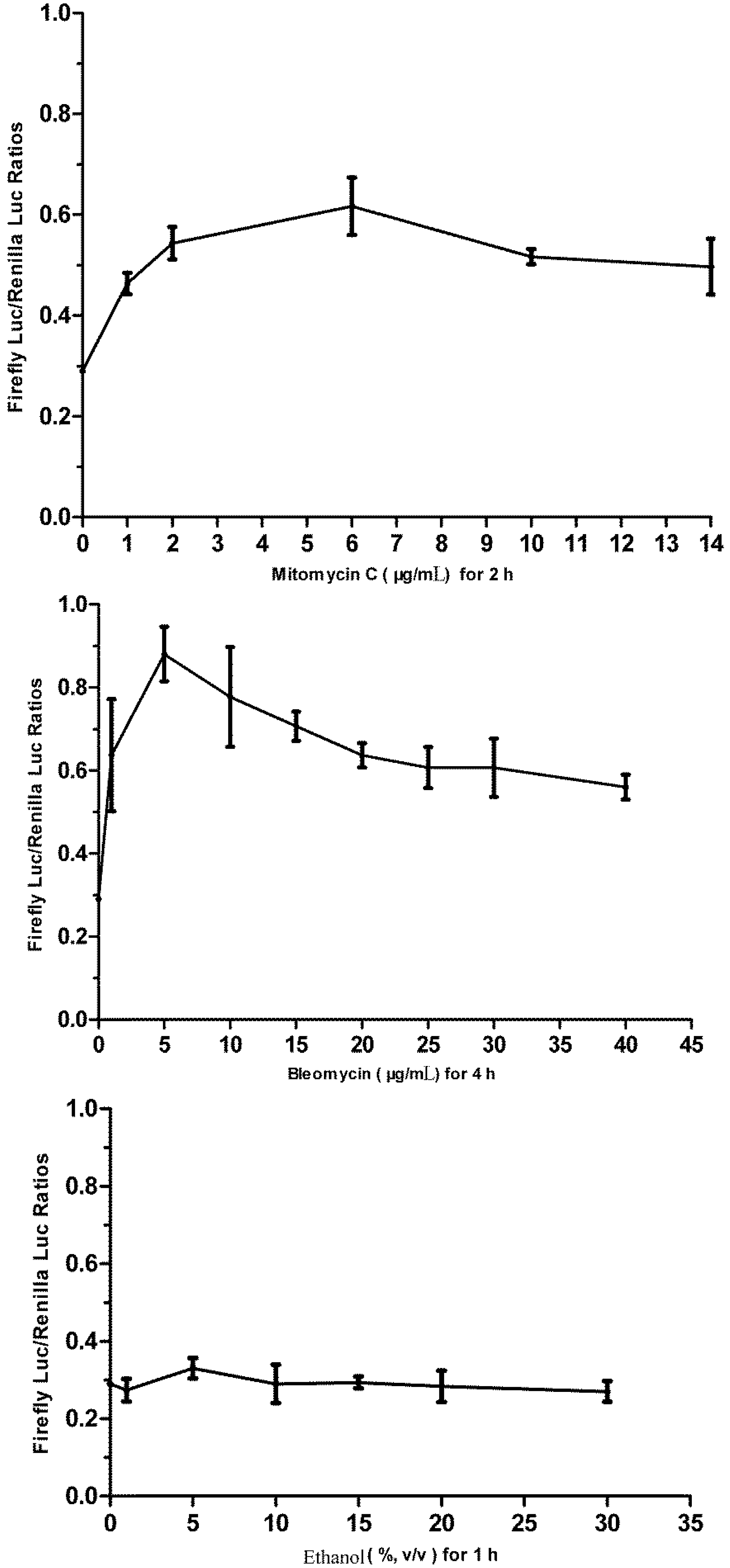
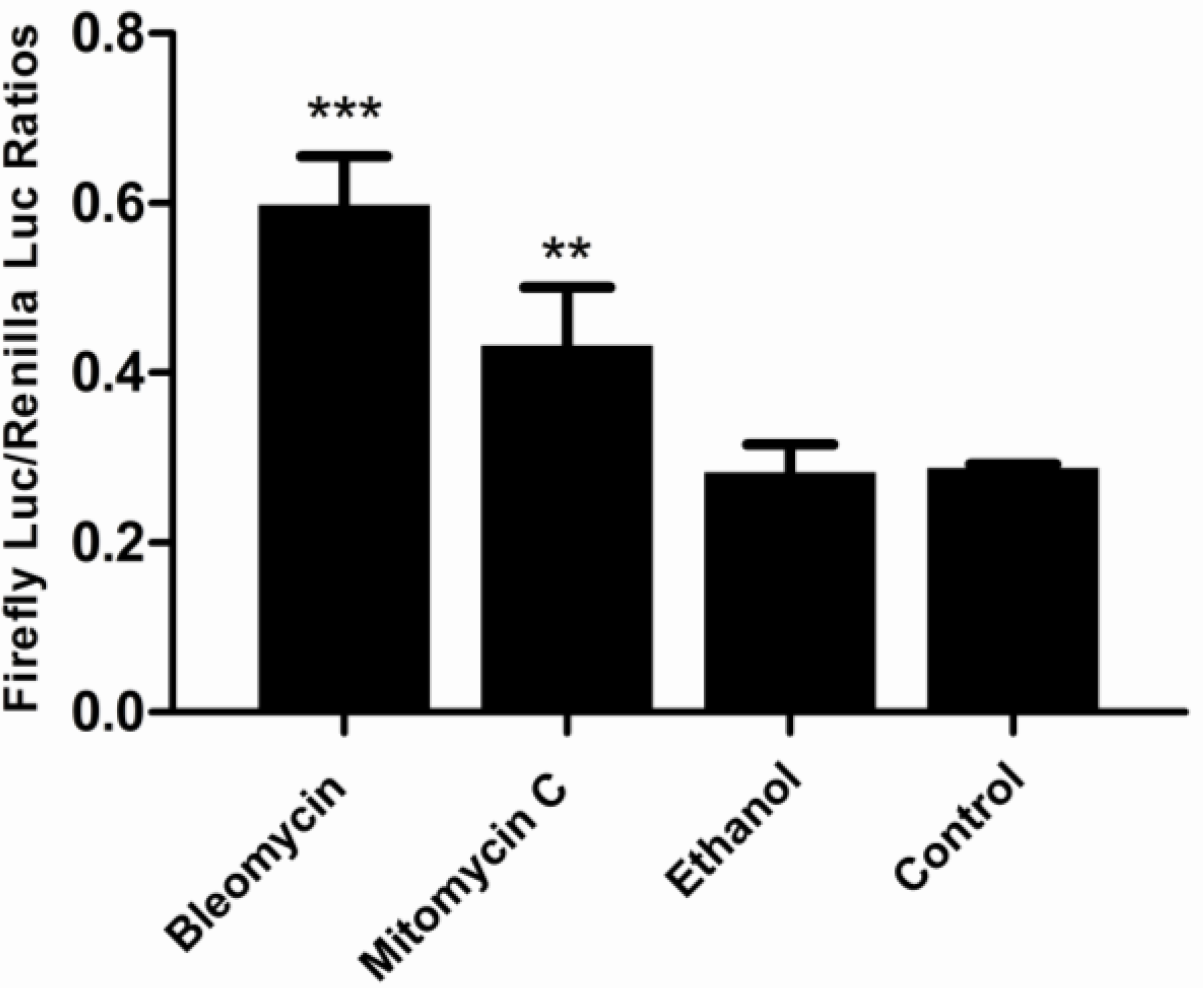
3.5. Responses of the p21FGLuc Cells to the Exposure of Model Genotoxic and Non-Genotoxic Agents
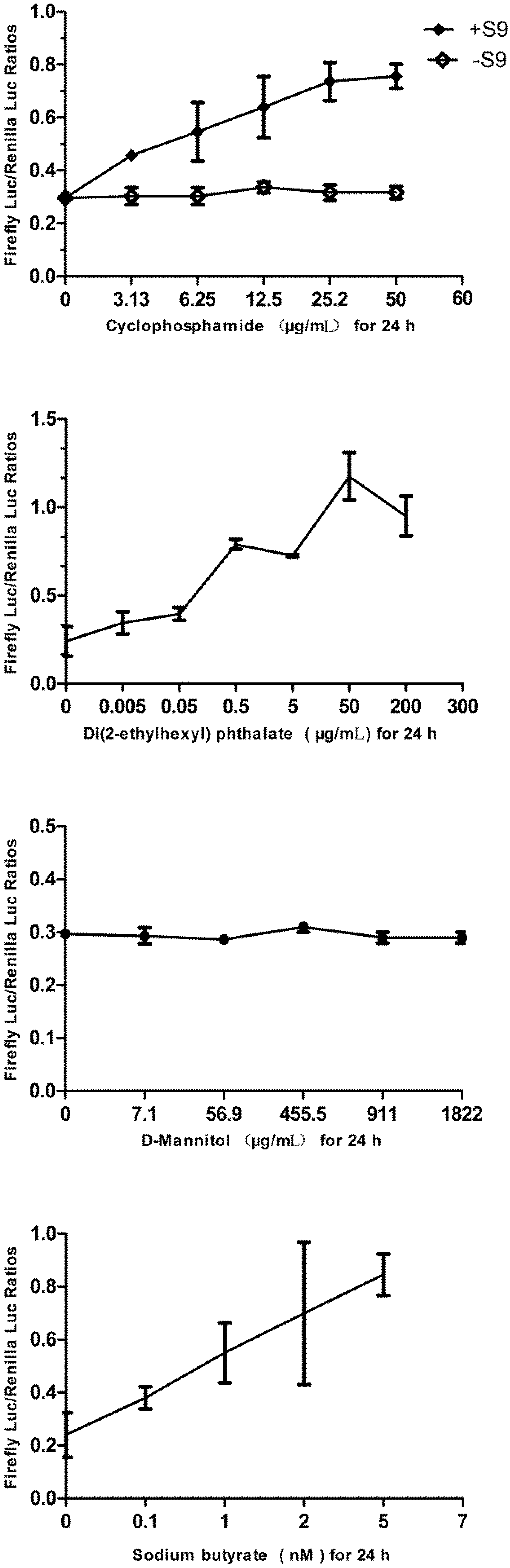
3.6. Responses of the p21FGLuc Cells to Varied Doses of Cyclophosphamide, Di(2-ethylhexyl) Phthalate (DEHP), D-Mannitol and Sodium Butyrate
4. Discussion
4.1. A Prerequisite for FG Cells to Be Engineered into a Genotoxicity Biosensor System Is to Hold a Wild-Type p53 Signaling Pathway
4.2. Flounder p53 Protein Can Effectively Recognize and Trans-Activate the Expression of Human p21 Gene
4.3. Initial Validation of the Fish Cell Biosensor System for Genotoxicity Detection
| Chemicals | Genotoxicity Results | Concentration Tested | Exposure Time | |
|---|---|---|---|---|
| Direct activators | ||||
| Bleomycin | positive | 1–40 μg/mL | 4 h | |
| Mitomycin C | positive | 1–14 μg/mL | 2 h | |
| Indirect activators | ||||
| Cyclophosphamide | Negative (without S9) | 3.13–50 μg/mL | 24 h | |
| Positive (with S9) | 3.13–50 μg/mL | 24 h | ||
| Alternate toxicants | ||||
| Di(2-ethylhexyl) phthalate (DEHP) | positive | 0.005–200 μg/mL | 24 h | |
| Unreactive activators | ||||
| Ethanol | negative | 1%–30% | 1 h | |
| D-mannitol | negative | 7.1–1,822 μg/mL | 24 h | |
| P21 direct activators | ||||
| Sodium butyrate | false positive | 0.1–5 nM | 24 h | |
Acknowledgements
References
- Al-Sabti, K.; Metcalfe, C.D. Fish micronuclei for assessing genotoxicity in water. Mutat. Res. 1995, 343, 121–135. [Google Scholar] [CrossRef]
- Cambier, S.; Gonzalez, P.; Durrieu, G.; Bourdineaud, J.-P. Cadmium-Induced genotoxicity in zebrafish at environmentally relevant doses. Ecotoxicol. Environ. Saf. 2010, 73, 312–319. [Google Scholar] [CrossRef]
- Diekmann, M.; Hultsch, V.; Nagel, R. On the relevance of genotoxicity for fish populations I: Effects of a model genotoxicant on zebrafish (Danio rerio) in a complete life-cycle test. Aquat. Toxicol. 2004, 68, 13–26. [Google Scholar] [CrossRef]
- De Flora, S.; Viganò, L.; D’Agostini, F.; Camoirano, A.; Bagnasco, M.; Bennicelli, C.; Melodia, F.; Arillo, A. Multiple genotoxicity biomarkers in fish exposed in situ to polluted river water. Mutat. Res. 1993, 319, 167–177. [Google Scholar] [CrossRef]
- Na, N.; Guo, H.R.; Zhang, S.C.; Li, Z.J.; Yin, L.C. In vitro and in vivo acute toxicity of fenpyroximate to flounder Paralichthys olivaceus and its gill cell line FG. Aquat. Toxicol. 2009, 92, 76–85. [Google Scholar] [CrossRef]
- Yang, F.; Zhang, Q.Q.; Guo, H.R.; Zhang, S.C. Evaluation of cytotoxicity, genotoxicity and teratogenicity of marine sediments from Qingdao coastal areas using in vitro fish cell assay, comet assay and zebrafish embryo test. Toxicol. Vitr. 2010, 24, 2003–2011. [Google Scholar] [CrossRef]
- Babich, H.; Borenfreund, E. Cytotoxicity and genotoxicity assays with cultured fish cells: A review. Toxicol. Vitr. 1991, 5, 91–100. [Google Scholar] [CrossRef]
- Kamman, U.; Bunke, M.; Steinhart, H.; Theobald, N. A permanent fish cell line (EPC) for genotoxicity testing of marine sediments with the comet assay. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2001, 498, 67–77. [Google Scholar] [CrossRef]
- Mitani, H. Comparisons of the radiosensitivity of three goldfish cell lines using short term endpoints. Int. J. Radiat. Biol. 1984, 45, 637–643. [Google Scholar] [CrossRef]
- Cimino, M.C. Comparative overview of current international strategies and guidelines for genetic toxicology testing for regulatory purposes. Environ. Mol. Mutagen. 2006, 47, 362–390. [Google Scholar] [CrossRef]
- Jena, G.B.; Kaul, C.L.; Ramarao, P. Genotoxicity testing, a regulatory requirement for drug discovery and development: Impact of ICH guidelines. Indian J. Pharmacol. 2002, 34, 86–99. [Google Scholar]
- Ames, B.N.; Mccann, J.; Yamasaki, E. Methods for detecting carcinogens and mutagens with the salmonella/mammalian-microsome mutagenicity test. Mutat. Res. 1975, 31, 347–364. [Google Scholar] [CrossRef]
- Hendriks, G.; Atallaha, M.; Raamsman, M.; Morolli, B.; van der Puttena, H.; Jaadar, H.; Tijdens, I.; Lange, R.E.; Mullenders, L.; van de Water, B. Sensitive DsRed fluorescence-based reporter cell systems for genotoxicity and oxidative stress assessment. Mutat. Res. 2011, 709–710, 49–59. [Google Scholar]
- Vanderlelie, D.; Regniers, L.; Borremans, B.; Provoost, A.; Verschaeve, L. The VITOTOX® test, a SOS bioluminescence Salmonella typhimurium test to measure genotoxicity kinetics. Mutat. Res. 1997, 389, 279–290. [Google Scholar] [CrossRef]
- Reifferscheid, G.; Heil, J.; Oda, Y.; Zahn, R.K. A microplate version of the SOS/umu-test for rapid detection of genotoxins and genotoxic potentials of environmental samples. Mutat. Res. 1991, 253, 215–222. [Google Scholar] [CrossRef]
- Westerink, W.M.A.; Stevenson, J.C.R.; Lauwers, A.; Griffioen, G.; Horbach, G.J.; Schoonen, W.G.E.J. Evaluation of the VitotoxTM and RadarScreen assays for the rapid assessment of genotoxicity in the early research phase of drug development. Mutat. Res. 2009, 676, 113–130. [Google Scholar] [CrossRef]
- Brennan, R.J.; Schiestl, R.H. Detecting carcinogens with the yeast DEL assay. Methods Mol. Biol. 2004, 262, 111–124. [Google Scholar]
- Lichtenberg-Fraté, H.; Schmitta, M.; Gellertb, G.; Ludwigc, J. A yeast-based method for the detection of cyto and genotoxicity. Toxicol. Vitr. 2003, 17, 709–716. [Google Scholar] [CrossRef]
- Schafer, B.; Neffgen, A.; Klinner, U. A novel yeast-bassed tool to detect mutagenic and recombinogenic effects simultaneously. Mutat. Res. 2008, 652, 20–29. [Google Scholar] [CrossRef]
- Afanassiev, V.; Sefton, M.; Anantachaiyong, T.; Barker, G.; Walmsley, R.; Wölfl, S. Application of yeast cells transformed with GFP expression constructs containing the RAD54 or RNR2 promoter as a test for the genotoxic potential of chemical substances. Mutat. Res. 2000, 464, 297–308. [Google Scholar] [CrossRef]
- Cahill, P.A.; Knight, A.W.; Billinton, N.; Barker, M.G.; Walsh, L.; Keenan, P.O.; Williams, C.V.; Tweats, D.J.; Walmsley, R.M. The GreenScreen genotoxicity assay: A screening validation programme. Mutagenesis 2004, 19, 105–119. [Google Scholar] [CrossRef]
- Walmsley, R.M.; Billinton, N.; Heyer, W.-D. Green fluorescence protein as a reporter for the DNA damage-induced gene RAD54 in Saccharomyces cerevisiae. Yeast 1997, 13, 1535–1545. [Google Scholar] [CrossRef]
- Benton, M.G.; Glasser, N.R.; Palecek, S.P. The utilization of a Saccharomyces cerevisiae HUG1P-GFP promoter-reporter construct for the selective detection of DNA damage. Mutat. Res. 2007, 633, 21–34. [Google Scholar] [CrossRef]
- Billinton, N.; Barker, M.G.; Michel, C.E.; Knight, A.W.; Heyer, W.-D.; Goddard, N.J.; Fielden, P.R.; Walmsley, R.M. Development of a green fluorescent protein reporter for a yeast genotoxicity biosensor. Biosens. Bioelectron. 1998, 13, 831–838. [Google Scholar] [CrossRef]
- Knight, A.W.; Billinton, N.; Cahill, P.A.; Scott, A.; Harvey, J.S.; Roberts, K.J.; Tweats, D.J.; Keenan, P.O.; Walmsley, R.M. An analysis of results from 305 compounds tested with the yeast RAD54-GFP genotoxicity assay (GreenScreen GC)-including relative predictivity of regulatory tests and rodent carcinogenesis and performance with autofluorescent and coloured compounds. Mutagenesis 2007, 22, 409–416. [Google Scholar] [CrossRef]
- Liu, X.; Kramer, J.A.; Swaffield, J.C.; Hu, Y.; Chai, G.; Wilson, A.G.E. Development of a highthroughput yeast-based assay for detection of metabolically activated genotoxins. Mutat. Res. 2008, 653, 63–69. [Google Scholar] [CrossRef]
- Fairbairn, D.W.; Olive, P.L.; O’Neill, K.L. The comet assay: A comprehensive review. Mutat. Res. 1995, 339, 37–59. [Google Scholar] [CrossRef]
- Jha, A.N. Ecotoxicological applications and significance of the comet assay. Mutagenesis 2008, 23, 207–221. [Google Scholar] [CrossRef]
- Singh, N.P.; McCoy, M.T.; Tice, R.R.; Schneider, E.L. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp. Cell Res. 1998, 175, 184–191. [Google Scholar]
- Clements, J. Gene mutation assays in mammalian cells. Methods Mol. Biol. 1995, 43, 277–286. [Google Scholar]
- Ohno, K.; Tanaka-Azuma, Y.; Yoneda, Y.; Yamada, T.G. Genotoxicity test system based on p53R2 gene expression in human cells: Examination with 80 chemicals. Mutat. Res. 2005, 588, 47–57. [Google Scholar] [CrossRef]
- Ohno, K.; Ishihata, K.; Ohno, K.; Tanaka-Azuma, Y.; Yamada, T. A genotoxicity test system based on p53R2 gene expression in human cells: Assessment of its reactivity to various classes of genotoxic chemicals. Mutat. Res. 2008, 656, 27–35. [Google Scholar] [CrossRef]
- Papathanasiou, M.A.; Kerr, N.C.; Robbins, J.H.; McBride, O.W.; Alamo, I., Jr.; Barrett, S.F.; Hickson, I.D.; Fornace, A.J., Jr. Induction by ionizing radiation of the gadd45 gene in cultured human cells: Lack of mediation by protein kinase C. Mol. Cell Biol. 1991, 11, 1009–1016. [Google Scholar]
- Hastwell, P.W.; Chai, L.-L.; Roberts, K.J.; Webster, T.W.; Harvey, J.S.; Rees, R.W.; Walmsley, R.M. High-Specificity and high-sensitivity genotoxicity assessment in a human cell line: Validation of the GreenScreen HC GADD45a-GFP genotoxicity assay. Mutat. Res. 2006, 607, 160–175. [Google Scholar] [CrossRef]
- Hastwell, P.W.; Webster, T.W.; Tate, M.; Billinton, N.; Lynch, A.M.; Harvey, J.S.; Rees, R.W.; Walmsley, R.M. Analysis of 75 marketed pharmaceuticals using the GADD45a-GFP ‘GreenScreen HC’ genotoxicity assay. Mutagenesis 2009, 24, 455–463. [Google Scholar] [CrossRef]
- Knight, A.W.; Birrell, L.; Walmsley, R.M. Development and validation of a higher throughput screening approach to genotoxicity testing using the GADD45a-GFP GreenScreen HC assay. J. Biomol. Screen. 2009, 14, 16–30. [Google Scholar]
- Leonardo, A.D.; Linke, S.P.; Clarkin, K.; Wahl, G.M. DNA damage triggers a prolonged p53-dependent G1 arrest and long-term induction of Cipl in normal human fibroblasts. Genes Dev. 1994, 8, 2540–2551. [Google Scholar] [CrossRef]
- Waldman, T.; Kinzler, K.W.; Vogelstein, B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995, 55, 5187–5190. [Google Scholar]
- Zager, V.; Cemazar, M.; Hreljac, I.; Lah, T.T.; Sersa, G.; Filipic, M. Development of human cell biosensor system for genotoxicity detection based on DNA damage-induced gene expression. Radiol. Oncol. 2010, 44, 42–51. [Google Scholar] [CrossRef]
- Guo, H.R.; Zhang, S.C. Cytotoxicity and genotoxicity of polyethylenimine and nickel chloride in red sea bream(Pagrosomus major)fin cell line RSBF. Chin. J. Oceanol. Limnol. 2002, 20, 323–331. [Google Scholar] [CrossRef]
- Li, H.Y.; Zhang, S.C. In vitro cytotoxicity of the organophosphorus pesticide parathion to FG-9307 cells. Toxicol. Vitr. 2001, 15, 643–647. [Google Scholar] [CrossRef]
- Yin, L.C.; Guo, H.R.; Zhang, S.C.; Wang, J. Study on the acute toxicity and genotoxicity of herbicide butachlor in flounder, Paralichihys olivaceus, and flounder gill (FG) cells. J. Ocean Univ. China 2007, 37, 167–171. [Google Scholar]
- Xu, Y.Y.; Guo, H.R.; Qin, X.; SU, F.; Yin, L.C. In vitro acute cytotoxicity of abamectin to the gill cell line of flounder Paralichthy olivaceus. J. Ocean Univ. China 2007, 6, 369–372. [Google Scholar] [CrossRef]
- Su, F.; Zhang, S.C.; Li, H.Y.; Guo, H.R. In vitro acute cytotoxicity of neonicotinoid insecticide imidacloprid to gill cell line of flounder Paralichthy olivaceus. Chin. J. Oceanol. Limnol. 2007, 25, 209–214. [Google Scholar] [CrossRef]
- Xiao, Q.; Zhang, S.C.; Guo, H.R.; Su, F.; Xu, Y.Y. Nonylphenol causes decrease in antioxidant enzyme activities, increase in O2− content, and alteration in ultrastructures of FG cells, a flounder (Paralichthy olivaceus) gill cell line. Toxicol. Mech. Methods 2007, 17, 127–134. [Google Scholar] [CrossRef]
- Tong, S.L.; Li, H.; Miao, H.Z. The establishment and partial characterization of a continuous fish cell line FG-9307 from the gill of flounder Paralichthys olivaceus. Aquaculture 1997, 156, 327–333. [Google Scholar] [CrossRef]
- Taylor, A.M.R.; Rosney, C.M.; Campbell, J.B. Unusual sensitivity of ataxia telangiectasia cells to bleomycin. Cancer Res. 1979, 39, 1046–1050. [Google Scholar]
- Nelson, W.G.; Kastan, M.B. The DNA strand breaks: The DNA template alterations that trigger p53-dependent DNA damage response pathways. Mol. Cell Biol. 1994, 14, 1815–1823. [Google Scholar]
- Donner, E.M.; Preston, R.J. The relationship between p53 status, DNA repair and chromatid aberration induction in G2 mouse embryo fibroblast cells treated with bleomycin. Carcinogenesis 1996, 17, 1161–1165. [Google Scholar] [CrossRef]
- Conner, D.A. Mouse embryo fibroblast (MEF) feeder cell preparation. Curr. Protoc. Mol. Biol. 2000. [Google Scholar] [CrossRef]
- Kirkland, D.; Kasper, P.; Müller, L.; Corvi, R.; Speit, G. Recommended lists of genotoxic and non-genotoxic chemicals for assessment of the performance of new or improved genotoxicity tests: A follow-up to an ECVAM workshop. Mutat. Res. 2008, 653, 99–108. [Google Scholar] [CrossRef]
- Birrell, L.; Cahill, P.; Hughesa, C.; Tatea, M.; Walmsley, R.M. GADD45a-GFP GreenScreen HC assay results for the ECVAM recommended lists of genotoxic and non-genotoxic chemicals for assessment of new genotoxicity tests. Mutat. Res. 2010, 695, 87–95. [Google Scholar] [CrossRef]
- Blundell, R.A. The biology of p21Waf1/Cip1. Am. J. Biochem. Biotechnol. 2006, 2, 33–40. [Google Scholar] [CrossRef]
- Nakano, K.; Mizuno, T.; Sowa, Y.; Orita, T.; Yoshino, T.; Okuyama, Y.; Fujita, T.; Ohtani-Fujita, N.; Matsukawa, Y.; Tokinoi, T.; et al. Butyrate activates the WAF1/Cip1 gene promoter through Sp1 sites in a p53-negative human colon cancer cell line. J. Biol. Chem. 1997, 272, 22199–22206. [Google Scholar]
- Enoch, T.; Norbury, C. Cellular responses to DNA damage: Cell-Cycle checkpoints, apoptosis and the roles of p53 and ATM. Trends Biochem. Sci. 1995, 20, 426–430. [Google Scholar] [CrossRef]
- Vousden, K.H.; Lu, X. Live or let die: The cell response to p53. Nat. Rev. Cancer 2002, 2, 594–604. [Google Scholar] [CrossRef]
- Kuerbitz, S.J.; Plunkett, B.S.; Walsh, W.V.; Kastan, M.B. Wild-Type p53 is a cell cycle checkpoint determinant following irradiation. Proc. Natl. Acad. Sci. USA 1992, 89, 7491–7495. [Google Scholar]
- Lane, D.P. Cancer. p53, guardian of the genome. Nature 1992, 358, 15–16. [Google Scholar] [CrossRef]
- Levine, A.J. p53, the cellular gatekeeper for growth and division. Cell 1997, 88, 323–331. [Google Scholar] [CrossRef]
- Guo, H.R.; Zhang, S.C.; Li, H.Y. Spontaneous neoplastic transformation of the cell line FG-9307 from Olive flounder. N. Am. J. Aquac. 2003, 65, 44–48. [Google Scholar] [CrossRef]
- Mohanty, B.P. A p53-like protein from a freshwater mollusc Lamellidens corrianus. Indian J. Biochem. Biophys. 2006, 43, 247–250. [Google Scholar]
- Van Beneden, R.J.; Walker, C.W.; Laughner, E.S. Characterization of gene expression of a p53 homologue in the soft-shell clam (Mya arenaria). Mol. Mar. Biol. Biotechnol. 1997, 6, 116–122. [Google Scholar]
- Ishioka, C.; Englert, C.; Winge, P.; Yan, Y.X.; Engelstein, M.; Friend, S.H. Mutational analysis of the carboxy-terminal portion of p53 using both yeast and mammalian cell assays in vivo. Oncogene 1995, 10, 1485–1492. [Google Scholar]
- Jin, S.; Martinek, S.; Joo, W.S.; Wortman, J.R.; Mirkovic, N.; Sali, A.; Yandell, M.D.; Pavletich, N.P.; Young, M.W.; Levine, A.J. Identification and characterization of a p53 homologue in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2000, 97, 7301–7306. [Google Scholar]
- Soussi, T.; Caron de Fromentel, C.; Méchali, M.; May, P.; Kress, M. Cloning and characterization of a cDNA from Xenopus laevis coding for a protein homologous to human and murine p53. Oncogene 1987, 1, 71–78. [Google Scholar]
- Cheng, R.; Ford, B.L.; ÓNeal, P.E.; Mathews, C.Z.; Bradford, C.S.; Thongtan, T.; Barnes, D.W.; Hendricks, J.D.; Bailey, G.S. Zebrafish (Danio rerio) p53 tumor suppressor gene: cDNA sequence and expression during embryogenesis. Mol. Mar. Biol. Biotechnol. 1997, 6, 88–97. [Google Scholar]
- Lu, W.-J.; Abrams, J.M. Lessons from p53 in non-mammalian models. Cell Death Differ. 2006, 13, 909–912. [Google Scholar] [CrossRef]
- Langheinrich, U.; Hennen, E.; Stott, G.; Vacun, G. Zebrafish as a model organism for the identification and characterization of drugs and genes affecting p53 signalling. Curr. Biol. 2002, 12, 2023–2028. [Google Scholar] [CrossRef]
- Tomasz, M. Mitomycin C: Small, fast and deadly (but very selective). Chem. Biol. 1995, 2, 575–579. [Google Scholar] [CrossRef]
- Claussen, C.A.; Long, E.C. Nucleic acid recognition by metal complexes of bleomycin. Chem. Rev. 1999, 99, 2797–2816. [Google Scholar] [CrossRef]
- Chastain, G. Alcohol, neurotransmitter systems, and behavior. J. Gen. Psychol. 2006, 133, 329–335. [Google Scholar] [CrossRef]
- Cohen, J.L.; Jao, J.Y. Enzymatic basis of cyclophosphamide activation by hepatic microsomes of the rat. J. Pharmacol. Exp. Ther. 1970, 174, 206–210. [Google Scholar]
- Tan, G.H. Residue levels of phthalate esters in water and sediment samples from the Klang River basin. Bull. Environ. Contam. Toxicol. 1995, 54, 171–176. [Google Scholar]
- Caldwell, J.C. DEHP: Genotoxicity and Potential Carcinogenic Mechanisms—A Review. Available online: http://dx.doi.org/10.1016/j.mrrev.2012.03.001 (accessed on 27 July 2012).
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Geng, D.; Zhang, Z.; Guo, H. Development of a Fish Cell Biosensor System for Genotoxicity Detection Based on DNA Damage-Induced Trans-Activation of p21 Gene Expression. Biosensors 2012, 2, 318-340. https://doi.org/10.3390/bios2030318
Geng D, Zhang Z, Guo H. Development of a Fish Cell Biosensor System for Genotoxicity Detection Based on DNA Damage-Induced Trans-Activation of p21 Gene Expression. Biosensors. 2012; 2(3):318-340. https://doi.org/10.3390/bios2030318
Chicago/Turabian StyleGeng, Deyu, Zhixia Zhang, and Huarong Guo. 2012. "Development of a Fish Cell Biosensor System for Genotoxicity Detection Based on DNA Damage-Induced Trans-Activation of p21 Gene Expression" Biosensors 2, no. 3: 318-340. https://doi.org/10.3390/bios2030318
APA StyleGeng, D., Zhang, Z., & Guo, H. (2012). Development of a Fish Cell Biosensor System for Genotoxicity Detection Based on DNA Damage-Induced Trans-Activation of p21 Gene Expression. Biosensors, 2(3), 318-340. https://doi.org/10.3390/bios2030318




