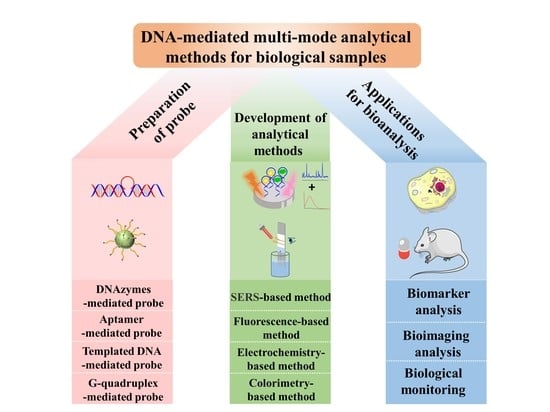
Recent Advances in the DNA-Mediated Multi-Mode Analytical Methods for Biological Samples
Abstract
1. Introduction
2. Preparation of DNA-Mediated Multi-Mode Probes
2.1. DNAzymes-Mediated Multi-Mode Probes
2.2. Aptamer-Mediated Multi-Mode Probes
2.3. Templated DNA-Mediated Multi-Mode Probes
2.4. G-Quadruplex-Mediated Multi-Mode Probes
3. Development of DNA-Mediated Multi-Mode Analytical Methods
3.1. SERS-Based Multi-Mode Methods
3.2. FL-Based Multi-Mode Methods
3.3. Electrochemistry-Based Multi-Mode Methods
3.4. Colorimetry-Based Multi-Mode Methods
4. Applications for Bioanalysis
4.1. Biomarker Analysis
4.2. Bioimaging Analysis
4.3. Biological Monitoring
5. Conclusions and Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ebrahimi, A.; Ravan, H.; Khajouei, S. DNA nanotechnology and bioassay development. Trends Anal. Chem. 2019, 114, 126–142. [Google Scholar] [CrossRef]
- Ma, Y.; Yang, X.; Wei, Y.; Yuan, Q. Applications of DNA nanotechnology in synthesis and assembly of inorganic nanomaterials. Chin. J. Chem. 2016, 34, 291–298. [Google Scholar] [CrossRef]
- Guo, Y.; Hu, Y.; Deng, Z. DNA Directed self-assembly of fluorescent colloidal semiconductor quantum dots and plasmonic metal nanoparticles heterogeneous nanomaterials. Chin. J. Chem. 2016, 34, 259–264. [Google Scholar] [CrossRef]
- Zhu, X.; Yan, X.; Yang, S.; Wang, Y.; Wang, S.; Tian, Y. DNA-mediated assembly of carbon nanomaterials. ChemPlusChem 2022, 87, e202200089. [Google Scholar] [CrossRef]
- Xia, L.; Tang, Y.; Zhang, J.; Dong, T.; Zhou, R. Advances in the DNA nanotechnology for the cancer biomarkers analysis: Attributes and applications. Semin. Cancer Biol. 2022, 86, 1105–1119. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Qiu, L.; Zhang, T.; Tan, W. Integrating DNA nanotechnology with aptamers for biological and biomedical applications. Matter 2021, 4, 461–489. [Google Scholar] [CrossRef]
- Iwe, I.; Li, W.; Li, Z.; Huang, J. Hairpin DNA-Mediated isothermal amplification (HDMIA) techniques for nucleic acid testing. Talanta 2021, 226, 122146. [Google Scholar] [CrossRef]
- Huang, R.; Wang, L.; Gai, Q.; Wang, D.; Qian, L. DNA-mediated assembly of carbon nanotubes for enhancing electrochemiluminescence and its application. Sens. Actuators B Chem. 2018, 256, 953–961. [Google Scholar] [CrossRef]
- Belotserkovskii, B.; Veselkov, A.; Filippov, S.; Dobrynin, V.; Mirkin, S.; Frank-Kamenetskii, M. Formation of intramolecular triplex in homopurine-homopyrimidine mirror repeats with point substitutions. Nucleic Acids Res. 1990, 18, 6621–6624. [Google Scholar] [CrossRef]
- Pestov, D.; Dayn, A.; Siyanova, E.; George, D.; Mirkin, S. H-DNA and Z-DNA in the mouse c-Ki-ras promoter. Nucleic Acids Res. 1991, 19, 6527–6532. [Google Scholar] [CrossRef]
- Walker, G.; Fraiser, M.; Schram, J.; Little, M.; Nadeau, J.; Malinowski, D. Strand displacement amplification—An isothermal, in vitro DNA amplification technique. Nucleic Acids Res. 1992, 2, 1691–1696. [Google Scholar] [CrossRef] [PubMed]
- Petty, J.; Zheng, J.; Hud, N.; Dickson, R. DNA-templated Ag nanocluster formation. J. Am. Chem. Soc. 2004, 126, 5207–5212. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Sun, Z.; Qing, M.; Yu, L.; Yan, H.; Luo, H.; Li, N. A “signal on” photoelectrochemical biosensor for miRNA-21 detection based on layer-by-layer assembled nn type BiS3@CdIn2S4 heterojunction. Sens. Actuators B Chem. 2022, 373, 132702. [Google Scholar] [CrossRef]
- Guerrini, L.; Alvarez-Puebla, R. Multiplex SERS chemosensing of metal ions via DNA-mediated recognition. Anal. Chem. 2019, 91, 11778–11784. [Google Scholar] [CrossRef]
- Yan, J.; Tan, Y.; Lin, M.; Xing, H.; Jiang, J. A DNA-mediated crosslinking strategy to enhance cellular delivery and sensor performance of protein spherical nucleic acids. Chem. Sci. 2021, 12, 1803–1809. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, C.; Zhang, L.; Tan, C.; Yang, J.; Chen, B.; Wang, L.; Zhang, H. DNA-templated silver nanoclusters for multiplexed fluorescent DNA detection. Small 2015, 11, 1385–1389. [Google Scholar] [CrossRef]
- Xu, X.; Wang, L.; Cui, W.; Jiang, W. An integrated and restructive probe mediated strand displacement amplification strategy for sensitive and specific DNA methyltransferase activity detection. Sens. Actuators B Chem. 2018, 266, 124–130. [Google Scholar] [CrossRef]
- Wen, C.; Zhao, L.; Wang, Y.; Wang, K.; Li, H.; Li, X.; Zi, M.; Zeng, J. Colorimetric and photothermal dual-mode lateral flow immunoassay based on Au-Fe3O4 multifunctional nanoparticles for detection of Salmonella typhimurium. Microchim. Acta 2023, 190, 57. [Google Scholar] [CrossRef]
- Li, Z.; Sun, W.; Duan, W.; Jiang, Y.; Chen, M.; Lin, G.; Wang, Q.; Fan, Z.; Tong, Y.; Chen Luo Li, J.; et al. Guiding epilepsy surgery with an LRP1-targeted SPECT/SERRS dual-mode imaging probe. ACS Appl. Mater. Interfaces 2023, 15, 14–25. [Google Scholar] [CrossRef]
- Zha, R.; Wu, R.; Zong, Y.; Wang, Z.; Wu, T.; Zhong, Y.; Liang, H.; Chen, L.; Li, C.; Wang, Y. A high performance dual-mode biosensor based on Nd-MOF nanosheets functionalized with ionic liquid and gold nanoparticles for sensing of ctDNA. Talanta 2023, 258, 124377. [Google Scholar] [CrossRef]
- Wang, J.; Liu, Y.; Li, X.; Chen, H.; Xu, J. Core-shell plasmonic nanomaterials toward: Dual-mode imaging analysis of glutathione and enhanced chemodynamic therapy. Anal. Chem. 2021, 93, 10317–10325. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Yuan, X.; Huang, Z.; Zhang, W.; Huang, F.; Ren, L. Dual-mode fluorescence and magnetic resonance imaging byperylene diimide-based Gd-containing magnetic ionic liquids. ACS Biomater. Sci. Eng. 2020, 6, 6405–6414. [Google Scholar] [CrossRef]
- Chen, X.; Lin, C.; Chen, Y.; Luo, F.; Wang, Y.; Chen, X. Terminal protection of a small molecule-linked loop DNA probe for turn-on label-free fluorescence detection of proteins. Biosens. Bioelectron. 2016, 83, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Viehrig, M.; Rajendran, S.; Sanger, K.; Schmidt, M.; Alstrøm, T.S.; Rindzevicius, T.; Zór, K.; Boisen, A. Quantitative SERS assay on a single chip enabled by electrochemically assisted regeneration: A method for detection of melamine in milk. Anal. Chem. 2020, 92, 4317–4325. [Google Scholar] [CrossRef]
- Sun, Y.; Li, S.; Chen, R.; Wu, P.; Liang, J. Ultrasensitive and rapid detection of T-2 toxin using a target-responsive DNA hydrogel. Sens. Actuators B Chem. 2020, 311, 127912. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, X.; Li, N.; Xu, Y.; Ma, Y.; Huang, Z.; Luo, H.; Huo, D. Typing of cancer cells by microswimmer based on Co-Fe-MOF for one-step simultaneously detect multiple biomarkers. Biosens. Bioelectron. 2023, 230, 115263. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Sun, D.W.; Ma, J.; Wang, Z.; Qin, A.; Tang, B.Z. Simultaneous sensing of ammonia and temperatures using a dual-mode freshness indicator based on Au/Cu nanoclusters for packaged seafood. Food Chem. 2023, 418, 135929. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Li, X.; Zhang, Y.; Zhou, K.; Yuan, L.; Shi, R.; Zhang, K.; Fu, Q. Sustainable and green synthesis of waste-biomass-derivedcarbon dots for parallel and semi-quantitative visual detection of Cr(VI) and Fe3+. Molecules 2022, 27, 1258. [Google Scholar] [CrossRef] [PubMed]
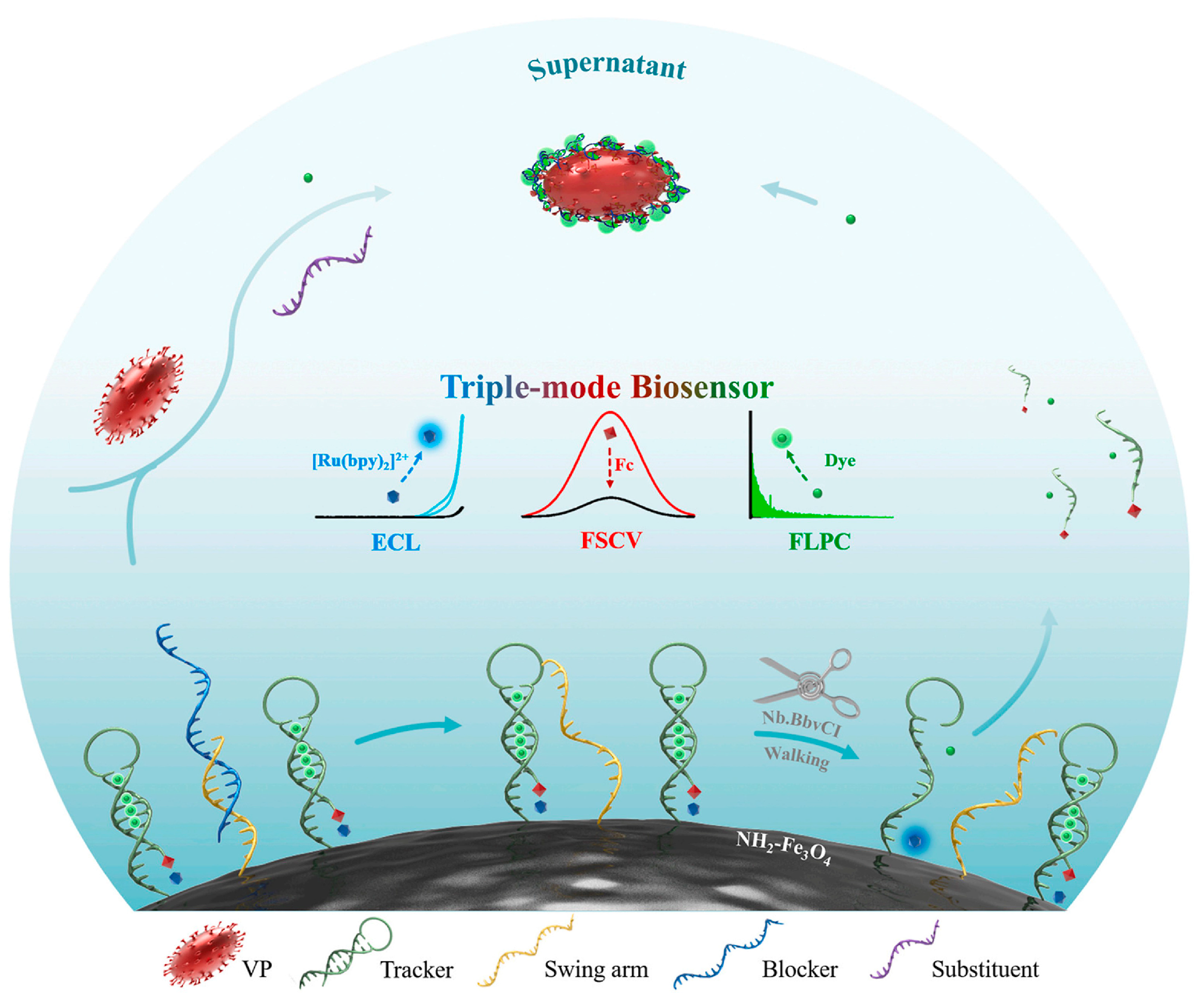
- Wang, Y.; Zhai, H.; Guo, Q.; Zhang, Y.; Gao, X.; Yang, Q.; Sun, X.; Guo, Y.; Zhang, Y. A dual-modal electrochemical aptasensor based on intelligent DNA walker with cascade signal amplification powered by Nb. BbvCI for Pb2+. Sci. Total Environ. 2023, 863, 160910. [Google Scholar] [CrossRef]
- Wang, H.; Zhao, B.; Ye, Y.; Qi, X.; Zhang, Y.; Xia, X.; Wang, X.; Zhou, N. A fluorescence and surface-enhanced Raman scattering dual-mode aptasensor for rapid and sensitive detection of ochratoxin A. Biosens. Bioelectron. 2022, 207, 114164. [Google Scholar] [CrossRef]
- Liu, S.; Huo, Y.; Deng, S.; Li, G.; Li, S.; Huang, L.; Ren, S.; Gao, Z. A facile dual-mode aptasensor based on AuNPs@MIL-101 nanohybrids for ultrasensitive fluorescence and surface-enhanced Raman spectroscopy detection of tetrodotoxin. Biosens. Bioelectron. 2022, 201, 113891. [Google Scholar] [CrossRef]
- Li, M.; Yan, M.; Xu, B.; Zhao, C.; Wang, D.; Wang, Y.; Chen, H. A dual-mode optical fiber sensor for SERS and fluorescence detection in liquid. Spectrochim. Acta A 2023, 290, 122267. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zong, S.; Zhang, Y.; Wang, Z.; Wang, Y.; Zhu, K.; Yang, K.; Wang, Z.; Chen, B.; Cui, Y. A SERS-colorimetric dual-mode aptasensor for the detection of cancer biomarker MUC1. Anal. Bioanal. Chem. 2020, 412, 5707–5718. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Zhang, S.; Ouyang, Y.; Zhang, C.; Liu, M.; Zhang, Y.; Deng, L. Aeromonas salmonicida aptamer selection and construction for colorimetric and ratiometric fluorescence dual-model aptasensor combined with g-C3N4 and G-quadruplex. Talanta 2023, 252, 123857. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zhao, W.; Zhou, H.; Zhang, Q.; Zhang, S. ATP-triggered intracellular in situ aggregation of a gold-nanoparticle-equipped triple-helix molecular switch for fluorescence imaging and photothermal tumor therapy. Langmuir 2022, 38, 3755–3764. [Google Scholar] [CrossRef]
- Kim, G.; Kim, J.; Kim, S.; Kato, T.; Yoon, J.; Noh, S.; Park, E.; Park, C.; Lee, T.; Choi, J. Fabrication of MERS-nanovesicle biosensor composed of multi-functional DNA aptamer/graphene-MoS2 nanocomposite based on electrochemical and surface-enhanced Raman spectroscopy. Sens. Actuators B Chem. 2022, 352, 131060. [Google Scholar] [CrossRef]
- Gao, Y.; Han, Y.; Wang, C.; Qiang, L.; Gao, J.; Wang, Y.; Liu, H.; Han, L.; Zhang, Y. Rapid and sensitive triple-mode detection of causative SARS-CoV-2 virus specific genes through interaction between genes and nanoparticles. Anal. Chim. Acta 2021, 1154, 338330. [Google Scholar] [CrossRef]
- Li, C.; Li, J.; Liang, A.; Wen, G.; Jiang, Z. Aptamer turn-on SERS/RRS/fluorescence tri-mode platform for ultra-trace urea determination using Fe/N-doped carbon dots. Front. Chem. 2021, 9, 613083. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Song, C.; Wang, L. DNA-mediated dynamic plasmonic nanostructures: Assembly, actuation, optical properties, and biological applications. Phys. Chem. Chem. Phys. 2022, 24, 23959–23979. [Google Scholar] [CrossRef]
- Le, T.; Van Park, J.S.; Lee, S. Synchronous Fe3+-enhanced colorimetric and liquid SERS-based detection of dopamine using core-shell plasmonic nanoparticles. ACS Appl. Nano Mater. 2023, 6, 6726–6738. [Google Scholar] [CrossRef]
- Sun, C.; Dong, W.; Peng, J.; Wan, X.; Sun, Z.; Li, D.; Wang, S. Dual-mode fluorescence–SERS sensor for sensitive and selective detection of uranyl ions based on satellite Fe3O4-Au@CdTe nanostructure. Sens. Actuators B Chem. 2020, 325, 128644. [Google Scholar] [CrossRef]
- Wu, Z.; Cui, B. Simultaneous fluorometric and chirality based aptasensing of sulfamethazine by using upconversion nanoparticles and Au@Ag@Au core-shell nanoparticles. Microchim. Acta 2019, 186, 555. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Fu, X.; Kong, G.; Yin, Y.; Meng, H.M.; Ke, G.; Zhang, X. DNAzyme-gold nanoparticle-based probes for biosensing and bioimaging. J. Mater. Chem. B 2020, 8, 9449–9465. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Liu, R.; Bai, T.; Wei, M.; He, B.; Suo, Z. A low-noise ratiometric fluorescence biosensor for detection of Pb2+ based on DNAzyme and exonuclease III–assisted cascade signal amplification. Anal. Bioanal. Chem. 2022, 414, 1899–1907. [Google Scholar] [CrossRef]
- Li, D.; Ling, S.; Cheng, X.; Yang, Z.; Lv, B. Development of a DNAzyme-based colorimetric biosensor assay for dual detection of Cd2+ and Hg2+. Anal. Bioanal. Chem. 2021, 413, 7081–7091. [Google Scholar] [CrossRef]
- Lian, K.; Chen, G.; Wang, X.; Zhang, W.; Hu, X.; Wang, H.; Li, Y.; Xi, D.; Wang, Y. Fluorescent detection of brown spot of tobacco caused by Alternaria alternata based on lambda exonuclease-induced DNAzyme amplification. RSC Adv. 2023, 13, 1587–1593. [Google Scholar] [CrossRef]
- Lee, C.Y.; Liao, C.H.; Fang, N.M.; Hsieh, Y.Z. DNAzyme-amplified label-free biosensor for the simple and sensitive detection of pyrophosphatase. Biosensors 2021, 11, 422. [Google Scholar] [CrossRef]
- Liu, L.; Chu, H.; Yang, J.; Sun, Y.; Ma, P.; Song, D. Construction of a magnetic-fluorescent-plasmonic nanosensor for the determination of MMP-2 activity based on SERS-fluorescence dual-mode signals. Biosens. Bioelectron. 2022, 212, 114389. [Google Scholar] [CrossRef]
- Li, C.; Chen, P.; Wang, Z.; Ma, X. A DNAzyme-gold nanostar probe for SERS-fluorescence dual-mode detection and imaging of calcium ions in living cells. Sens. Actuators B Chem. 2021, 347, 130596. [Google Scholar] [CrossRef]
- Huang, W.; Zhan, D.; Xie, Y.; Li, X.; Lai, G. Dual CHA-mediated high-efficient formation of a tripedal DNA walker for constructing a novel proteinase-free dual-mode biosensing strategy. Biosens. Bioelectron. 2022, 197, 113708. [Google Scholar] [CrossRef]
- Xue, Y.; Xie, H.; Wang, Y.; Feng, S.; Sun, J.; Huang, J.; Yang, X. Novel and sensitive electrochemical/fluorescent dual-mode biosensing platform based on the cascaded cyclic amplification of enzyme-free DDSA and functional nucleic acids. Biosens. Bioelectron. 2022, 218, 114762. [Google Scholar] [CrossRef]
- Hu, Z.; Zhang, J.; Li, Y.; Song, S.; Feng, C.; Wang, J.; Zhang, F.; Wang, J.; Liu, X. G-quadruplex-deficient precursor hairpin probes for ultra-low background dual-mode detection of miRNAs. Talanta 2023, 253, 123954. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Gálvez, L.; Sulleiro, M.; Gutiérrez-Sánchez, C.; García-Nieto, D.; Luna, M.; Pérez, E.; García-Mendiola, T.; Lorenzo, E. MoS2-carbon nanodots as a new electrochemiluminescence platform for breast cancer biomarker detection. Biosensors 2023, 13, 348. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Wang, M.; Lai, W.; Song, X.; Li, J.; Liu, D.; Wei, Z.; Hong, C. Construction of electrochemical and electrochemiluminescent dual-mode aptamer sensors based on ferrocene dual-functional signal probes for the sensitive detection of Alternariol. Microchim. Acta 2023, 190, 57. [Google Scholar] [CrossRef]
- Shahdost-Fard, F.; Faridfar, S.; Keihan, A.H.; Aghaei, M.; Petrenko, I.; Ahmadi, F.; Ehrlich, H.; Rahimi-Nasrabadi, M. Applicability of a green nanocomposite consisted of spongin decorated Cu2WO4(OH)2 and AgNPs as a high-performance aptasensing platform in Staphylococcus aureus detection. Biosensors 2023, 13, 271. [Google Scholar] [CrossRef]
- Liu, Y.; Cai, Z.; Sheng, L.; Ma, M.; Wang, X. A magnetic relaxation switching and visual dual-mode sensor for selective detection of Hg2+ based on aptamers modified Au@Fe3O4 nanoparticles. J. Hazard. Mater. 2020, 388, 121728. [Google Scholar] [CrossRef]
- Duan, W.; Wang, X.; Wang, H.; Li, F. Fluorescent and colorimetric dual-mode aptasensor for thrombin detection based on target-induced conjunction of split aptamer fragments. Talanta 2018, 180, 76–80. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, Y.; Yi, H.C.; Gu, H.W.; Yin, X.L.; Xiang, D.L.; Zou, P. An electrochemical and colorimetric dual-mode aptasensor for Staphylococcus aureus based on a multifunctional MOF and magnetic separation technique. Microchem. J. 2023, 190, 108681. [Google Scholar] [CrossRef]
- Zhang, D.; Chu, S.; Wang, L.; Zhan, X.; Zhou, P.; Zhang, D. Dual-mode colorimetric determination of As (III) based on negatively-charged aptamer-mediated aggregation of positively-charged AuNPs. Anal. Chim. Acta 2022, 1221, 340111. [Google Scholar] [CrossRef]
- Yu, W.; Lin, X.; Duan, N.; Wang, Z.; Wu, S. A fluorescence and surface-enhanced Raman scattering dual-mode aptasensor for sensitive detection of deoxynivalenol based on gold nanoclusters and silver nanoparticles modified metal-polydopamine framework. Anal. Chim. Acta 2023, 1244, 340846. [Google Scholar] [CrossRef]
- Yu, J.; Wu, H.; He, L.; Tan, L.; Jia, Z.; Gan, N. The universal dual-mode aptasensor for simultaneous determination of different bacteria based on naked eyes and microfluidic-chip together with magnetic DNA encoded probes. Talanta 2021, 225, 122062. [Google Scholar] [CrossRef]
- Ali, G.; Omer, K. Nanozyme and stimulated fluorescent Cu-based metal–organic frameworks (Cu-MOFs) functionalized with engineered aptamers as a molecular recognition element for thrombin detection in the plasma of COVID-19 patients. ACS Omega 2022, 7, 36804–36810. [Google Scholar] [CrossRef]
- Li, J.; Li, C.; Jiang, Z. TbMOF@Au catalytic determination of trace malathion with aptamer SERS/RRS/Abs assay. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2023, 294, 122581. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Wang, J.; Wang, Y.; Du, Y.; Huang, Y.; Tang, A.; Cui, Y.; Kong, D. DNA nanostructure-based nucleic acid probes: Construction and biological applications. Chem. Sci. 2021, 12, 7602–7622. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Wu, J.; Li, Z. Biosensing using hairpin DNA probes. Rev. Anal. Chem. 2015, 34, 1–27. [Google Scholar] [CrossRef]
- Chandler, M.; Shevchenko, O.; Vivero-Escoto, J.; Striplin, C.; Afonin, K. DNA-templated synthesis of fluorescent silver nanoclusters. J. Chem. Educ. 2020, 97, 1992–1996. [Google Scholar] [CrossRef]
- Rolband, L.; Yourston, L.; Chandler, M.; Beasock, D.; Danai, L.; Kozlov, S.; Marshall, N.; Shevchenko, O.; Krasnoslobodtsev, A.; Afonin, K. DNA-templated fluorescent silver nanoclusters inhibit bacterial growth while being non-toxic to mammalian cells. Molecules 2021, 26, 4045. [Google Scholar] [CrossRef]
- Yourston, L.; Dhoqina, P.; Marshall, N.; Mahmud, R.; Kuether, E.; Krasnoslobodtsev, A. Hg2+ Detection with rational design of DNA-templated fluorescent silver nanoclusters. Processes 2021, 9, 1699. [Google Scholar] [CrossRef]
- Gupta, A.; Krasnoslobodtsev, A. DNA-templated silver nanoclusters as dual-mode sensitive probes for self-powered biosensor fueled by glucose. Nanomaterials 2023, 13, 1299. [Google Scholar] [CrossRef]
- Li, H.; Lu, Y.; Pang, J.; Sun, J.; Yang, F.; Wang, Z.; Liu, Y. DNA-scaffold copper nanoclusters integrated into a cerium (III)-triggered Fenton-like reaction for the fluorometric and colorimetric enzymatic determination of glucose. Microchim. Acta 2019, 186, 862. [Google Scholar] [CrossRef]
- Qing, T.; Long, C.; Wang, X.; Zhang, K.; Zhang, P.; Feng, B. Detection of micrococcal nuclease for identifying Staphylococcus aureus based on DNA templated fluorescent copper nanoclusters. Microchim. Acta 2019, 186, 248. [Google Scholar] [CrossRef]
- Yuan, J.; Shao, W.; Chen, S.; Huang, Z.; Tan, J. Recent advances in fluorescent probes for G-quadruplex nucleic acids. Biochem. Biophys. Res. Commun. 2020, 531, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Liu, Y.; Wan, Q.; Lu, Q.; Liu, J.; Ren, Y.; Tang, J.; Su, Q.; Luo, Y. Label-free, reusable, equipment-free, and visual detection of hydrogen sulfide using a colorimetric and fluorescent dual-mode sensing platform. Anal. Chem. 2023, 95, 5920–5926. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zhou, Y.; Liu, J. G-quadruplex DNA for construction of biosensors. Trends Anal. Chem. 2020, 132, 116060. [Google Scholar] [CrossRef]
- Chen, J.; Wang, M.; Zhou, C.; Zhang, J.; Su, X. Label-free and dual-mode biosensor for HPV DNA based on DNA/silver nanoclusters and G-quadruplex/hemin DNAzyme. Talanta 2022, 247, 123554. [Google Scholar] [CrossRef]
- Li, D.; Li, C.; Liang, A.; Jiang, Z. SERS and fluorescence dual-mode sensing trace hemin and K+ based on G-quarplex/hemin DNAzyme catalytic amplification. Sens. Actuators B Chem. 2019, 297, 126799. [Google Scholar] [CrossRef]
- Li, H.; Yang, Q.; Wang, Z.; Li, F. Iridium complex with specific intercalation in the G-quadruplex: A phosphorescence and electrochemiluminescence dual-mode homogeneous biosensor for enzyme-free and label-free detection of microRNA. ACS Sens. 2023, 8, 1529–1535. [Google Scholar] [CrossRef] [PubMed]
- Dejeu, J.; Van der Heyden, A.; Spinelli, N.; Defrancq, E.; Coche-Guérente, L. Recent progress in the design of G-quadruplex–based electrochemical aptasensors. Curr. Opin. Electrochem. 2021, 30, 100812. [Google Scholar] [CrossRef]
- Zhu, L.; Li, G.; Shao, X.; Huang, K.; Luo, Y.; Xu, W. A colorimetric zinc (II) assay based on the use of hairpin DNAzyme recycling and a hemin/G-quadruplex lighted DNA nanoladder. Microchim. Acta 2020, 187, 26. [Google Scholar]
- Xu, J.; Zhang, B.; Zhang, Y.; Mai, L.; Hu, W.; Chen, C.J.; Liu, J.; Zhu, G. Recent advances in disease diagnosis based on electrochemical-optical dual-mode detection method. Talanta 2022, 253, 124037. [Google Scholar] [CrossRef]
- Alvarez-Puebla, R.A.; Pazos-Perez, N.; Guerrini, L. SERS-fluorescent encoded particles as dual-mode optical probes. Appl. Mater. Today 2018, 13, 1–14. [Google Scholar] [CrossRef]
- Subasinghe, S.; Pautler, R.; Samee, M.A.; Yustein, J.; Allen, M. Dual-mode tumor imaging using probes that are responsive to hypoxia-induced pathological conditions. Biosensors 2022, 12, 478. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Chen, L.; Wei, J.; Cui, X.; Cheng, Z.; Wang, T.; Chao, I.; Zhao, Y.; Gao, H.; Li, P. A two-step strategy for simultaneous dual-mode detection of methyl-paraoxon and Ni (II). Ecotoxicol. Environ. Saf. 2022, 239, 113668. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Jia, Y.; Song, X.; Lu, J.; Lu, X.; Liu, B.; Han, J.; Huang, Y.; Zhang, J.; Chen, T. Giant gold nanowire vesicle-based colorimetric and SERS dual-mode immunosensor for ultrasensitive detection of Vibrio parahemolyticus. Anal. Chem. 2018, 90, 6124–6130. [Google Scholar] [PubMed]
- Liu, M.; Wang, Z.; Pan, L.; Cui, Y.; Liu, Y. A SERS/fluorescence dual-mode nanosensor based on the human telomeric G-quadruplex DNA: Application to mercury (II) detection. Biosens. Bioelectron. 2015, 69, 142–147. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Li, C.; He, C.; Zhou, Y.; Wang, Z.; Duan, N.; Wu, S. Upconversion nanoparticles assembled with gold nanourchins as luminescence and surface-enhanced Raman scattering dual-mode aptasensors for detection of ochratoxin A. ACS Appl. Nano Mater. 2021, 4, 8231–8240. [Google Scholar] [CrossRef]
- Zhang, J.; Ma, X.; Wang, Z. Surface-enhanced Raman scattering-fluorescence dual-mode nanosensors for quantitative detection of cytochrome c in living cells. Anal. Chem. 2019, 91, 6600–6607. [Google Scholar] [CrossRef]
- Huang, L.; Huang, H.; Zhang, Z.; Li, G. Contractile hairpin DNA-mediated dual-mode strategy for simultaneous quantification of lactoferrin and iron ion by surface-enhanced Raman scattering and fluorescence analysis. Anal. Chem. 2023, 95, 5946–5954. [Google Scholar] [CrossRef]
- Li, C.; Song, M.; Wu, S.; Wang, Z.; Duan, N. Selection of aptamer targeting levamisole and development of a colorimetric and SERS dual-mode aptasensor based on AuNPs/Cu-TCPP (Fe) nanosheets. Talanta 2023, 251, 123739. [Google Scholar] [CrossRef]
- Liu, J.; Chen, J.; Wu, D.; Huang, M.; Chen, J.; Pan, R.; Wu, Y.; Li, G. CRISPR-/Cas12a-mediated liposome-amplified strategy for the surface-enhanced Raman scattering and naked-eye detection of nucleic acid and application to food authenticity screening. Anal. Chem. 2021, 93, 10167–10174. [Google Scholar] [CrossRef]
- Joyce, C.; Fothergill, S.M.; Xie, F. Recent advances in gold-based metal enhanced fluorescence platforms for diagnosis and imaging in the near-infrared. Mater. Today Adv. 2020, 7, 100073. [Google Scholar] [CrossRef]
- Huang, L.; Zhang, Z.; Li, G. DNA strand displacement based surface-enhanced Raman scattering-fluorescence dual-mode nanoprobes for quantification and imaging of vascular endothelial growth factor in living cells. Biosens. Bioelectron. 2022, 204, 114069. [Google Scholar] [PubMed]
- Wang, Y.; Huo, T.; Du, Y.; Qian, M.; Lin, C.; Nie, H.; Li, W.; Hao, T.; Zhang, X.; Lin, N. Huang, R. Sensitive CTC analysis and dual-mode MRI/FL diagnosis based on a magnetic core-shell aptasensor. Biosens. Bioelectron. 2022, 215, 114530. [Google Scholar] [CrossRef] [PubMed]
- Shin, B.; Park, J.S.; Chun, H.S.; Yoon, S.; Kim, W.K.; Lee, J. A fluorescence/colorimetric dual-mode sensing strategy for miRNA based on graphene oxide. Anal. Bioanal. Chem. 2020, 412, 233–242. [Google Scholar] [CrossRef]
- Singh, R.; Kurian, A.; Patel, K.; Mandakhbayar, N.; Lee, N.; Knowles, J.; Lee, J.; Kim, H.W. Label-free fluorescent mesoporous bioglass for drug delivery, optical triple-mode imaging, and photothermal/photodynamic synergistic cancer therapy. ACS Appl. Bio Mater. 2020, 3, 2218–2229. [Google Scholar] [CrossRef]
- Shen, H.; Qileng, A.; Yang, H.; Liang, H.; Zhu, H.; Liu, Y.; Lei, H.; Liu, W. “Dual-signal-on” integrated-type biosensor for portable detection of miRNA: Cas12a-induced photoelectron chemistry and fluorescence strategy. Anal. Chem. 2021, 93, 11816–11825. [Google Scholar] [CrossRef]
- Yu, Q.; Guo, Q.; Zhou, J.; Yuan, X.; Huang, K.; Chen, P. Filter-assisted smartphone colorimetry/ICP-MS dual-mode biosensor of butyrylcholinesterase in clinical samples. Sens. Actuators B Chem. 2022, 370, 132472. [Google Scholar] [CrossRef]
- Wang, X.; Liao, X.; Mei, L.; Zhang, M.; Chen, S.; Qiao, X.; Hong, C. An immunosensor using functionalized Cu2O/Pt NPs as the signal probe for rapid and highly sensitive CEA detection with colorimetry and electrochemistry dual modes. Sens. Actuators B Chem. 2021, 341, 130032. [Google Scholar] [CrossRef]
- Yu, J.; Xiao, S.; Yu, Z.; Hui, Y.; Li, T.; Wu, D.; Bi, W.; Gan, N.; Jia, Z. On-site and dual-mode detection of live Vibrio parahaemolyticus in waters: A universal pathogen sensing platform based on a smart hydrogel aptasensor imbedded with gold nanoclusters. Sens. Actuators B Chem. 2022, 366, 131947. [Google Scholar] [CrossRef]
- Li, S.; Zhang, H.; Huang, Z.; Jia, Q. Spatially confining copper nanoclusters in porous ZrO2 for fluorescence/colorimetry/smartphone triple-mode detection of metoprolol tartrate. Biosens. Bioelectron. 2023, 231, 115290. [Google Scholar] [CrossRef]
- Wang, L.; Zheng, S.; Lu, L.; Li, C.; Wang, F. A dual-mode ratiometric fluorescence and smartphone-assisted colorimetric sensing platform based on bifunctional Fe, Co-CQD for glucose analysis at physiological pH. Anal. Chim. Acta 2023, 1239, 340701. [Google Scholar] [CrossRef]
- Cui, W.; Hu, Z.; Unocic, R.; Van Tendeloo, G.; Sang, X. Atomic defects, functional groups and properties in MXenes. Chin. Chem. Lett. 2021, 32, 339–344. [Google Scholar] [CrossRef]
- Wu, Z.; Sun, D.W.; Pu, H.; Wei, Q. A dual signal-on biosensor based on dual-gated locked mesoporous silica nanoparticles for the detection of Aflatoxin B1. Talanta 2023, 253, 124027. [Google Scholar] [CrossRef]
- Yu, M.; Liu, S.; Su, D.; Jiang, S.; Zhang, G.; Qin, Y.; Li, M. Controllable MXene nano-sheet/Au nanostructure architectures for the ultra-sensitive molecule Raman detection. Nanoscale 2019, 11, 22230–22236. [Google Scholar] [CrossRef] [PubMed]
- Kadhom, M.; Kalash, K.; Al-Furaiji, M. Performance of 2D MXene as an adsorbent for malachite green removal. Chemosphere 2022, 290, 133256. [Google Scholar]
- Xu, M.; Chen, K.; Zhu, L.; Zhang, S.; Wang, M.; He, L.; Zhang, Z.; Du, M. MOF@COF heterostructure hybrid for dual-mode photoelectrochemical-electrochemical HIV-1 DNA sensing. Langmuir 2021, 37, 13479–13492. [Google Scholar] [CrossRef]
- Zhai, J.; Li, X.; Zhang, J.; Pan, H.; Peng, Q.; Gan, H.; Su, S.; Yuwen, L.; Song, C. SERS/electrochemical dual-mode biosensor based on multi-functionalized molybdenum disulfide nanosheet probes and SERS-active Ag nanorods array electrodes for reliable detection of cancer-related miRNA. Sens. Actuators B Chem. 2022, 368, 132245. [Google Scholar] [CrossRef]
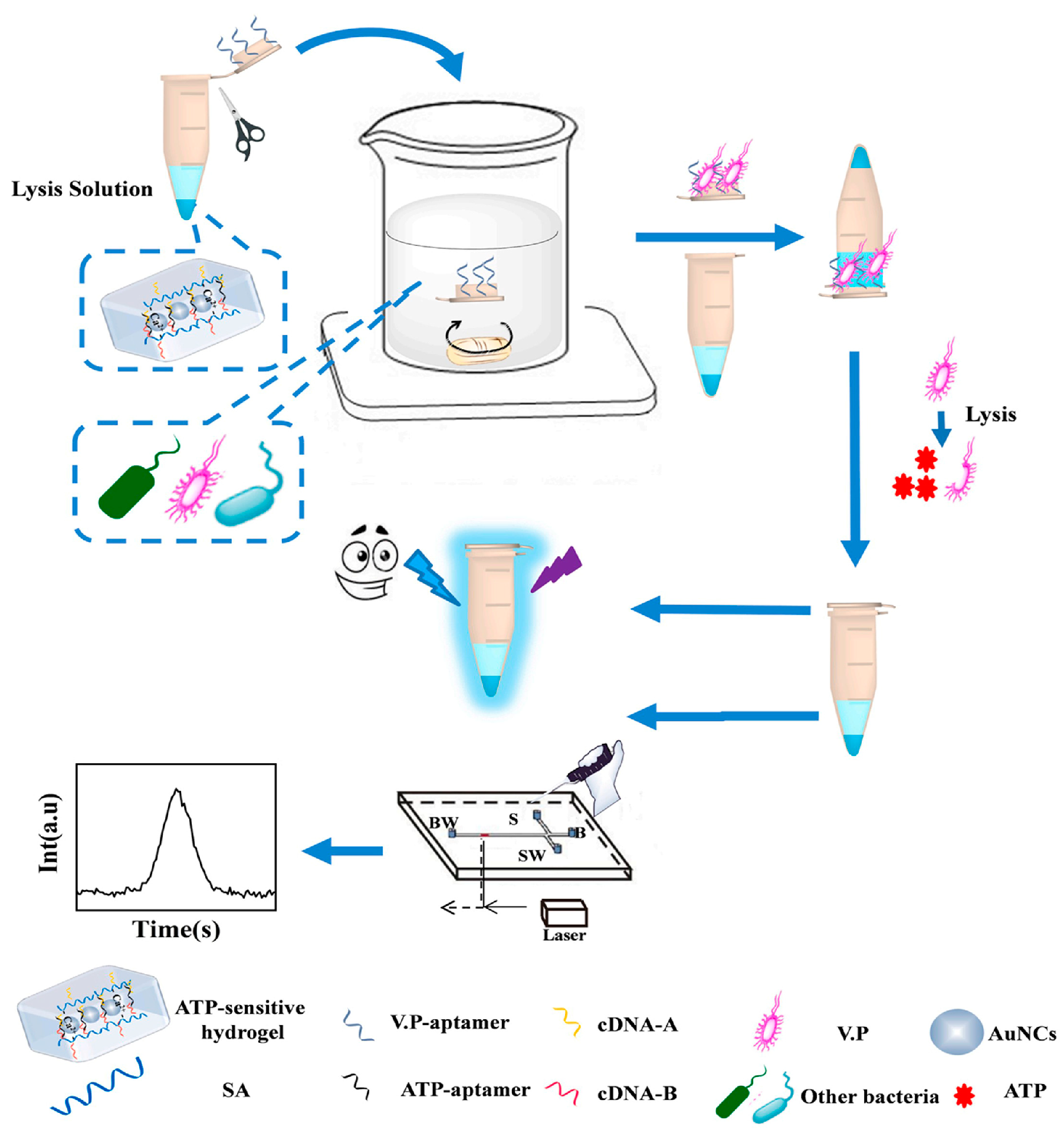
- Wei, W.; Lin, H.; Hao, T.; Wang, S.; Hu, Y.; Guo, Z.; Luo, X. DNA walker-mediated biosensor for target-triggered triple-mode detection of Vibrio parahaemolyticus. Biosens. Bioelectron. 2021, 186, 113305. [Google Scholar] [CrossRef]
- Hu, K.; Qin, L.; Ren, X.; Guo, Z.; Wang, S.; Hu, Y. Deoxyribonucleic acid-guided dual-mode electro-chemical/chemiluminescent platform for sensitive and selective examination of Pb2+. J. Electroanal. Chem. 2022, 922, 116757. [Google Scholar] [CrossRef]
- Zhang, X.; Zhi, H.; Wang, F.; Zhu, M.; Meng, H.; Wan, P.; Feng, L. Target-responsive smart nanomaterials via a Au–S binding encapsulation strategy for electrochemical/colorimetric dual-mode paper-based analytical devices. Anal. Chem. 2022, 94, 2569–2577. [Google Scholar] [CrossRef]
- Liu, T.; Li, N.; Dong, J.; Zhang, Y.; Fan, Y.; Lin, S.; Luo, H.; Li, N. A colorimetric and fluorometric dual-signal sensor for arginine detection by inhibiting the growth of gold nanoparticles/carbon quantum dots composite. Biosens. Bioelectron. 2017, 87, 772–778. [Google Scholar] [CrossRef]
- Dinh, V.; Lee, N. Dual-mode visual detection strategies of viable pathogens for point-of-care testing. Biosens. Bioelectron. 2023, 221, 114904. [Google Scholar] [CrossRef]
- Song, C.; Li, J.; Sun, Y.; Jiang, X.; Zhang, J.; Dong, C.; Wang, L. Colorimetric/SERS dual-mode detection of mercury ion via SERS-Active peroxidase-like Au@AgPt NPs. Sens. Actuators B Chem. 2020, 310, 127849. [Google Scholar] [CrossRef]
- Shen, Y.; Gao, X.; Zhang, Y.; Chen, H.; Ye, Y.; Wu, Y. Polydopamine-based nanozyme with dual-recognition strategy-driven fluorescence-colorimetric dual-mode platform for Listeria monocytogenes detection. J. Hazard. Mater. 2022, 439, 129582. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Bai, L.; Hou, T.; Zhang, L.; Gai, P.; Li, F. Dual-mode colorimetric and homogeneous electrochemical detection of intracellular/extracellular H2O2 based on FeSx/SiO2 nanoparticles with high peroxidase-like activity. Anal. Chim. Acta 2023, 1265, 341332. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Luo, F.; Guo, L.; Qiu, B.; Lin, Z. Target-triggered aggregation of gold nanoparticles for photothermal quantitative detection of adenosine using a thermometer as readout. Anal. Chim. Acta 2020, 110, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Chen, R.; Hou, Y.; Qin, Y.; Li, S.; Yang, S.; Gao, Z. DNA hydrogels combined with microfluidic chips for melamine detection. Anal. Chim. Acta 2022, 1228, 340312. [Google Scholar] [CrossRef]
- Si, Q.; Li, Y.; Huang, Z.; Liu, C.; Chen, X.; Wei, J.; Wang, F. Construction of a simple dual-mode ATP-sensing system for reliable fish freshness evaluation. Anal. Chim. Acta 2023, 1252, 341048. [Google Scholar] [CrossRef]
- Mousavizadeh, F.; Sarlak, N. A sensitive dual mode turn-on fluorescence and colorimetric nanosensor for ultrasensitive detection of trace amount of gluten proteins in bread products based on crystalline nano cellulose and gold nanoparticles. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2023, 287, 122095. [Google Scholar] [CrossRef]
- Panikar, S.; Cialla-May, D.; De la Rosa, E.; Salas, P.; Popp, J. Towards translation of surface-enhanced Raman spectroscopy (SERS) to clinical practice: Progress and trends. Trends Anal. Chem. 2021, 134, 116122. [Google Scholar] [CrossRef]
- Zhong, Y.; Yu, X.; Fu, W.; Chen, Y.; Shan, G.; Liu, Y. Colorimetric and Raman spectroscopic array for detection of hydrogen peroxide and glucose based on etching the silver shell of Au@Ag core-shell nanoparticles. Microchim. Acta 2019, 186, 802. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhu, X.; Li, Y.; Hai, X.; Bi, S. A colorimetric and photothermal dual-mode biosensing platform based on nanozyme-functionalized flower-like DNA structures for tumor-derived exosome detection. Talanta 2023, 258, 124456. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Lv, H.; Wang, J.; Yang, Z.; Ding, Y.; Zhao, B.; Tian, Y. An aptamer-based colorimetric/SERS dual-mode sensing strategy for the detection of sulfadimethoxine residues in animal-derived foods. Anal. Methods 2023, 15, 1047–1053. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Li, D.; Fodjo, E.K.; Deng, W. Colorimetric/fluorescent/SERS triple-channel sensing of Cu2+ in real systems based on chelation-triggered self-aggregation. Chem. Eng. J. 2022, 399, 125840. [Google Scholar] [CrossRef]
- Liu, X.; Wang, X.; Ye, S.; Li, R.; Li, H. A one–two–three multifunctional system for enhanced imaging and detection of intracellular microRNA and chemo genetherapy. ACS Appl. Mater. Interfaces 2021, 13, 27825–27835. [Google Scholar] [CrossRef]
- Zhang, W.; Ding, X.; Cheng, H.; Yin, C.; Yan, J.; Mou, Z.; Wang, W.; Cui, D.; Fan, C.; Sun, D. Dual-targeted gold nanoprism for recognition of early apoptosis, dual-model imaging and precise cancer photothermal therapy. Theranostics 2019, 9, 5610. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Cao, H.; Du, X.; Chen, T.; Ma, A.; Pan, J. Nonaqueous miscible liquid–liquid electroextraction for fast exhaustive enrichment of ultratrace analytes by an exponential transfer and deceleration mechanism. Anal. Chem. 2020, 93, 1458–1465. [Google Scholar] [CrossRef]
- Song, C.; Jiang, X.; Yang, Y.; Zhang, J.; Larson, S.; Zhao, Y.; Wang, L. High-sensitive assay of nucleic acid using tetrahedral DNA probes and DNA concatamers with a surface-enhanced Raman scattering/surface plasmon resonance dual-mode biosensor based on a silver nanorod-covered silver nanohole array. ACS Appl. Mater. Interfaces 2020, 12, 31242–31254. [Google Scholar] [CrossRef]
- Li, M.; Liu, Z.; Liu, Y.; Luo, H.; Huang, K.J.; Tan, X. Capacitor-parallel-amplified decoupled photoelectrochemical/electrochromic dual-mode bioassay for sensitive detection of microRNA with high reliability. Biosens. Bioelectron. 2023, 232, 115310. [Google Scholar] [CrossRef]
- He, M.Q.; Ai, Y.; Hu, W.; Jia, X.; Wu, L.; Ding, M.; Liang, Q. Dual-functional capping agent-mediated transformation of silver nanotriangles to silver nanoclusters for dual-mode biosensing. Anal. Chem. 2023, 95, 6130–6137. [Google Scholar] [CrossRef]
- Xu, R.; Ouyang, L.; Chen, H.; Zhang, G.; Zhe, J. Recent advances in biomolecular detection based on aptamers and nanoparticles. Biosensors 2023, 13, 474. [Google Scholar] [CrossRef]
- Li, J.; Li, L.; Bi, X.; Liu, X.; Luo, L.; You, T. Fluorescence/colorimetry dual-mode sensing strategy for mercury ion detection based on the quenching effect and nanozyme activity of porous cerium oxide nanorod. Sens. Actuators B Chem. 2022, 360, 131483. [Google Scholar] [CrossRef]
- Zayani, R.; Rabti, A.; Aoun, S.B.; Raouafi, N. Fluorescent and electrochemical bimodal bioplatform for femtomolar detection of microRNAs in blood sera. Sens. Actuators B Chem. 2021, 327, 128950. [Google Scholar] [CrossRef]
- Dong, Y.; Dong, W.; Liang, X.; Wang, Y.R.; Xu, F.; Li, L.; Han, L.; Jiang, L.R. Construction and application of thrombin-activated fluorescence-SERS dual-mode optical nanoprobes. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2023, 293, 122513. [Google Scholar] [CrossRef]
- Ye, X.; Gao, D.; Mu, X.; Wu, Q.; Ma, P.; Song, D. Dual-signal triple-mode optical sensing platform for assisting in the diagnosis of kidney disorders. Anal. Chem. 2023, 95, 4653–4661. [Google Scholar] [CrossRef]
- Li, C.; Chen, P.; Khan, I.M.; Wang, Z.; Zhang, Y.; Ma, X. Fluorescence-Raman dual-mode quantitative detection and imaging of small-molecule thiols in cell apoptosis with DNA-modified gold nanoflowers. J. Mater. Chem. B 2022, 10, 571–581. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.Y.; Xie, W.Z.; Tan, X.; Huang, K.J.; Xu, J. Superior graphdiyne self-powered biosensing platform with highly sensitivity and reliability for dual-mode detection of MicroRNA by integrating T7 Exonuclease and 3D DNA walker induced rolling circle amplification. Anal. Chim. Acta 2023, 1239, 340696. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, S.; Chen, C.; Sun, J.; Yang, X. Enzyme-free aptamer/AuNPs-based fluorometric and colorimetric dual-mode detection for ATP. Sens. Actuators B Chem. 2018, 265, 67–74. [Google Scholar] [CrossRef]
- Guo, W.J.; Yang, X.Y.; Wu, Z.; Zhang, Z.L. A colorimetric and electrochemical dual-mode biosensor for thrombin using a magnetic separation technique. J. Mater. Chem. B 2020, 8, 3574–3581. [Google Scholar] [CrossRef]
- Shi, J.; Li, J.; Liang, A.; Jiang, Z. Highly catalysis MOFCe supported Ag nanoclusters coupled with specific aptamer for SERS quantitative assay of trace dopamine. Talanta 2022, 245, 123468. [Google Scholar] [CrossRef]
- Li, P.; Mei, L.; Li, H.; Hong, C. Dual-mode immunosensor based on Cu-doped Mo2C nanosheets as signal labels. Bioelectrochemistry 2023, 149, 108280. [Google Scholar] [CrossRef]
- Liang, P.; Guo, Q.; Zhao, T.; Wen, C.Y.; Tian, Z.; Shang, Y.; Xing, J.; Jiang, Y.; Zeng, J. Ag nanoparticles with ultrathin Au shell-based lateral flow immunoassay for colorimetric and SERS dual-mode detection of SARS-CoV-2 IgG. Anal. Chem. 2022, 94, 8466–8473. [Google Scholar] [CrossRef]
- Mao, G.; Luo, X.; Ye, S.; Wang, X.; He, J.; Kong, J.; Dai, J.; Yin, W.; Ma, Y. Fluorescence and colorimetric analysis of African swine fever virus based on the RPA-assisted CRISPR/Cas12a strategy. Anal. Chem. 2023, 95, 8063–8069. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Xiao, S.; Zeng, M.; Xie, H.; Gan, N. Dual-mode colorimetric-electrochemical biosensor for Vibrio parahaemolyticus detection based on CuO2 nanodot-encapsulated metal-organic framework nanozymes. Sens. Actuators B Chem. 2023, 387, 133835. [Google Scholar] [CrossRef]
- Du, S.; Xie, B.; Gao, H.; Zhang, J.; Fu, H.; Liao, F.; Liao, Y. Self-powered DNAzyme walker enables dual-mode biosensor construction for electrochemiluminescence and electrochemical detection of microRNA. Anal. Chem. 2023, 95, 7006–7013. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Chang, S.; Wang, X.; Zhou, Z.; Chen, B.; Qian, R.; Li, D. Live-cell profiling of membrane sialic acids by fluorescence imaging combined with SERS labelling. Sens. Actuators B Chem. 2022, 351, 130877. [Google Scholar] [CrossRef]
- Najdian, A.; Amanlou, M.; Beiki, D.; Bitarafan-Rajabi, A.; Mirzaei, M.; Ardestani, M. Amino-modified-silica-coated gadolinium-copper nanoclu sters, conjugated to AS1411 aptamer and radiolabeled with technetium-99 m as a novel multimodal imaging agent. Bioorganic Chem. 2022, 125, 105827. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Gao, M.Y.; Zhu, Q.; Chi, B.; Zeng, L.W.; Hu, J.M.; Shen, A.G. Monodispersed plasmonic Prussian blue nanoparticles for zero-background SERS/MRI-guided phototherapy. Nanoscale 2020, 12, 3292–3301. [Google Scholar] [CrossRef]
- Yan, H.; Gao, X.; Zhang, Y.; Chang, W.; Li, J.; Li, X.; Du, Q.; Li, C. Imaging tiny hepatic tumor xenografts via endoglin-targeted paramagnetic/opticalnanoprobe. ACS Appl. Interfaces 2018, 10, 17047–17057. [Google Scholar] [CrossRef]
- Wang, J.; Zhao, T.; Li, X.; Luo, X.; Chen, H.; Xu, J. Dual-mode scattering nanoprobes for imaging hydrogen sulfide in living cells. ACS Appl. Nano Mater. 2021, 4, 7319–7329. [Google Scholar] [CrossRef]
- Zhong, H.; Wu, Y.X.; Yu, S.; Wang, X.; He, K.; Li, D.; Cao, Y.; Gan, N. Two-photon CQDs-based dual-mode nanoprobe for fluorescence imaging and magnetic resonance imaging of intracellular wide pH. Anal. Chem. 2021, 93, 5691–5699. [Google Scholar] [CrossRef] [PubMed]
- Weng, Y.; Guan, S.; Lu, H.; Meng, X.; Kaassis, A.Y.; Ren, X.; Qu, X.; Sun, C.; Zhou, S. Confinement of carbon dots localizing to the ultrathin layered double hydroxides toward simultaneous triple-mode bioimaging and photothermal therapy. Talanta 2018, 184, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Liu, Y.; Jia, P.; Feng, Y.; Fu, S.; Yang, J.; Xiong, L.; Su, F.; Wu, Y.; Huang, Y. Ag nanoparticle-decorated mesoporous silica as a dual-mode Raman sensing platform for detection of volatile organic compounds. ACS Appl. Mater. Interfaces 2021, 4, 1019–1028. [Google Scholar] [CrossRef]
- Lin, X.; Li, C.; Xu, S.; Wang, Z.; Ma, X. SERS-fluorescence dual-mode nanoprobe for the detection and imaging of Bax mRNA during apoptosis. Microchim. Acta 2023, 190, 130. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, C.; Liu, Z.; Li, S.; Ma, P.; Gao, F. Target-triggered nanomaterial self-assembly induced electromagnetic hot-spot generation for SERS-fluorescence dual-Mode in situ monitoring miRNA-guided phototherapy. Anal. Chem. 2021, 93, 13755–13764. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yue, S.; Wang, Y.; Wang, Y.; Xu, Z. A multicolor-SERS dual-mode pH sensor based on smart nano-in-micro particles. Sens. Actuators B Chem. 2020, 310, 127889. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, H.; Ye, S.; Wang, X.; Ma, L. Fluorescent-Raman binary star ratio probe for microRNA detection and imaging in living cells. Anal. Chem. 2020, 93, 1466–1471. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, B.; Liang, Y.; Shi, L.; Cen, X.; Zheng, L.; Liang, E.; Huang, L.; Cheng, K. Near-infrared fluorescent and photoacoustic dual-mode probe for highly sensitive and selective imaging of cysteine in vivo. Anal. Chem. 2022, 94, 10737–10744. [Google Scholar] [CrossRef]
- Shen, Y.; Tian, Q.; Sun, Y.; Xu, J.; Ye, D.; Chen, H. ATP-activatable photosensitizer enables dual fluorescence imaging and targeted photodynamic therapy of tumor. Anal. Chem. 2017, 89, 13610–13617. [Google Scholar] [CrossRef]










| Biomarkers | DNA-Mediated Strategy | Multi-Mode Analysis | LODs | Samples | Ref. |
|---|---|---|---|---|---|
| Glutathione (GSH) | Aptamer | SERS/FL | 0.913 (SERS)/1.454 (FL) μmol/L | Cell | [136] |
| Glucose | Aptamer | PEC/EC | 5.9 × 10−5 (PEC)/6.2 × 10−4 (EC) U/mL | Serum | [137] |
| ATP | Aptamer | Colorimetry /FL | 61.29 (FL)/122.5 (Colorimetry) nmol/L | Urine | [138] |
| Thrombin | Aptamer | Colorimetry/EC | 0.35 (EC)/10 (Colorimetry) nmol/L | Serum | [139] |
| PSA | G-quadruplexes | FL/SERS | 0.275 (FL)/5.01 (SERS) pg/mL | Serum | [140] |
| Carcino-embryonic antigen (CEA) | Aptamer | EC/EC | * 0.33 (DPV)/1.67 (CV) fg/mL | Serum | [141] |
| Human immunoglobulin G(IgG) | Aptamer | Colorimetry/SERS | 0.22 (SERS)/0.52 (Colorimetry) pg/mL | Serum | [142] |
| African Swine Fever Virus | DNAzyme | Colorimetry/FL | 20 (FL)/20 (Colorimetry) copies/mL | Serum | [143] |
| Pathogen (Vibrio parahaemolyticus) | Aptamer | Colorimetry/EC | 30 (Colorimetry)/ 5 (EC) CFU/mL | milk | [144] |
| microRNA | DNAzymes | ECL/EC | 54.3 (ECL)/78.6 (EC) amol/L | Cell | [145] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, L.; Zhang, Z. Recent Advances in the DNA-Mediated Multi-Mode Analytical Methods for Biological Samples. Biosensors 2023, 13, 693. https://doi.org/10.3390/bios13070693
Huang L, Zhang Z. Recent Advances in the DNA-Mediated Multi-Mode Analytical Methods for Biological Samples. Biosensors. 2023; 13(7):693. https://doi.org/10.3390/bios13070693
Chicago/Turabian StyleHuang, Lu, and Zhuomin Zhang. 2023. "Recent Advances in the DNA-Mediated Multi-Mode Analytical Methods for Biological Samples" Biosensors 13, no. 7: 693. https://doi.org/10.3390/bios13070693
APA StyleHuang, L., & Zhang, Z. (2023). Recent Advances in the DNA-Mediated Multi-Mode Analytical Methods for Biological Samples. Biosensors, 13(7), 693. https://doi.org/10.3390/bios13070693