Complementary DNA Significantly Enhancing Signal Response and Sensitivity of a Molecular Beacon Probe to Aflatoxin B1
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Analysis Procedures for AFB1 Detection
2.3. Specificity Tests
2.4. Detection of AFB1 Spiked in Complex Matrix
3. Results and Discussions
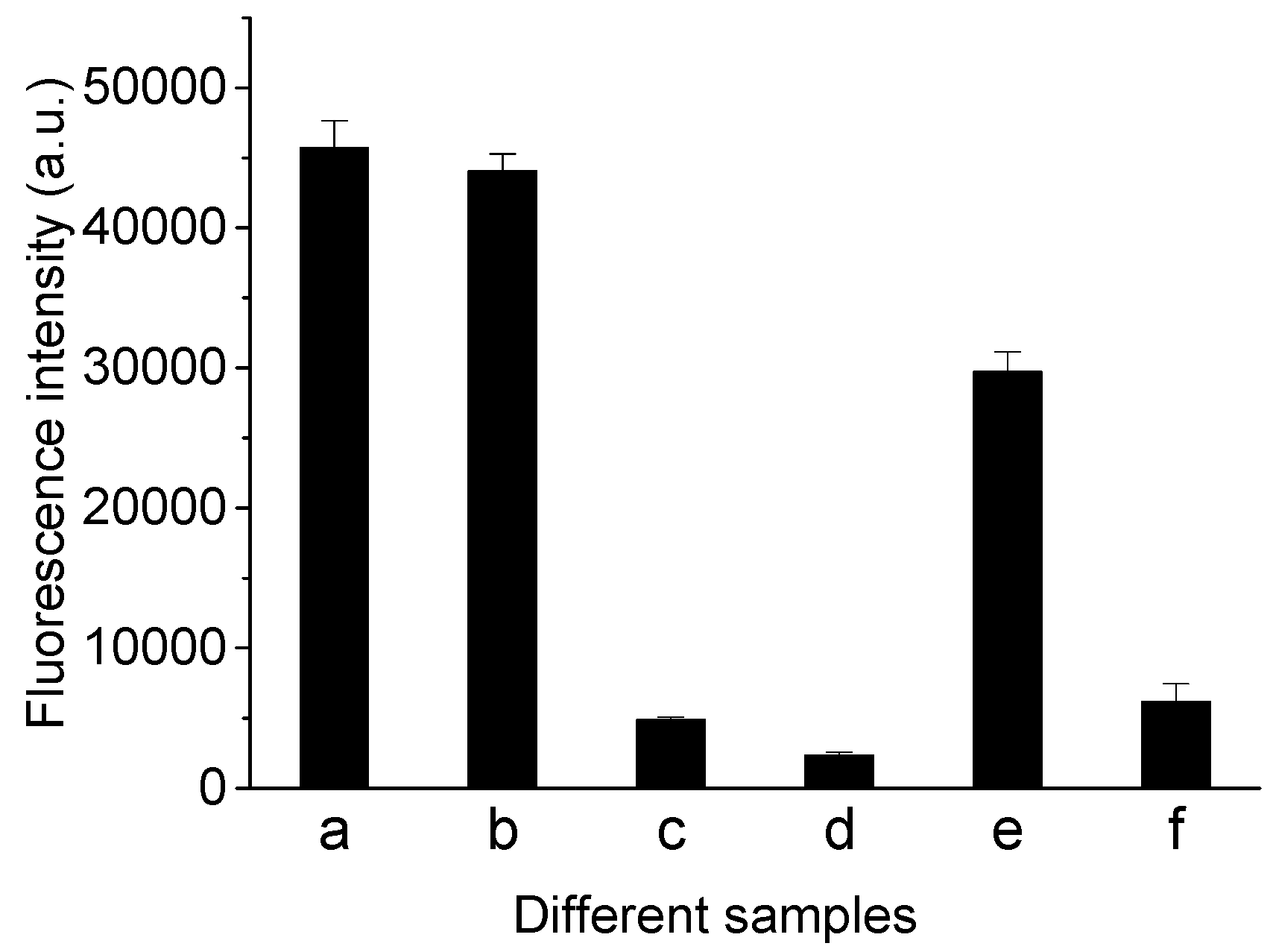
3.1. Inspiration and Feasibility of This Proposal
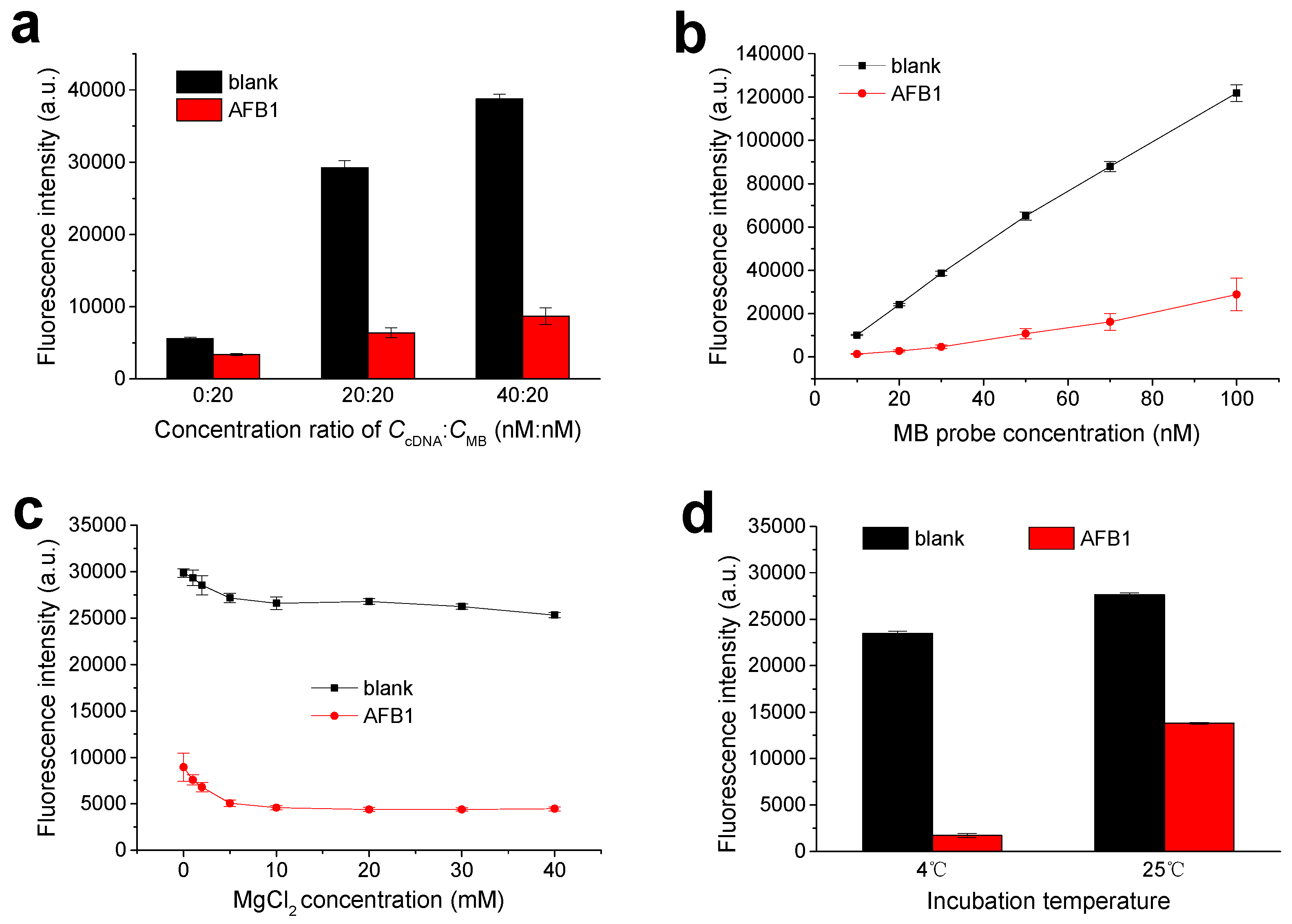
3.2. Optimization of Experimental Conditions
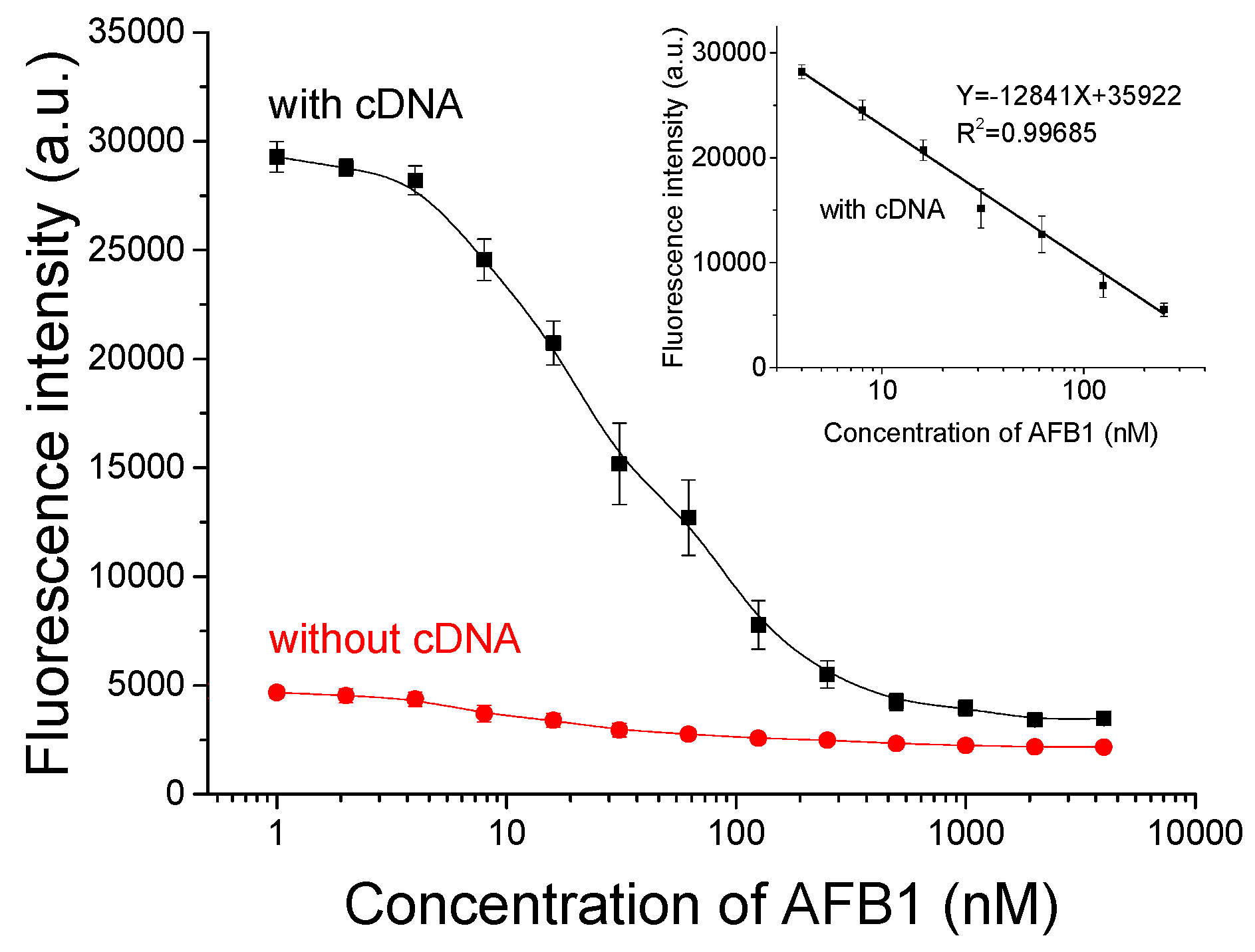
3.3. Sensitivity Analysis of Novel Aptameric Sensor against AFB1
3.4. Specificity of This Detection Method
3.5. Complex Matrix Interference Tests
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Rushing, B.R.; Selim, M.I. Aflatoxin B1: A review on metabolism, toxicity, occurrence in food, occupational exposure, and detoxification methods. Food Chem. Toxicol. 2019, 124, 81–100. [Google Scholar] [CrossRef] [PubMed]
- Ostry, V.; Malir, F.; Toman, J.; Grosse, Y. Mycotoxins as human carcinogens-the IARC monographs classification. Mycotoxin Res. 2017, 33, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Marchese, S.; Polo, A.; Ariano, A.; Velotto, S.; Costantini, S.; Severino, L. Aflatoxin B1 and M1: Biological properties and their involvement in cancer development. Toxins 2018, 10, 214. [Google Scholar] [CrossRef] [PubMed]
- Ali, N. Aflatoxins in rice: Worldwide occurrence and public health perspectives. Toxicol. Rep. 2019, 6, 1188–1197. [Google Scholar] [CrossRef] [PubMed]
- Mahato, D.K.; Lee, K.E.; Kamle, M.; Devi, S.; Dewangan, K.N.; Kumar, P.; Kang, S.G. Aflatoxins in food and feed: An overview on prevalence, detection and control strategies. Front. Microbiol. 2019, 10, 2266. [Google Scholar] [CrossRef]
- Yao, H.; Hruska, Z.; Diana Di Mavungu, J. Developments in detection and determination of aflatoxins. World Mycotoxin J. 2015, 8, 181–191. [Google Scholar] [CrossRef]
- Abbas, H.K.; Shier, W.T.; Horn, B.W.; Weaver, M.A. Cultural methods for aflatoxin detection. J. Toxicol. -Toxin. Rev. 2004, 23, 295–315. [Google Scholar] [CrossRef]
- Li, P.; Zhang, Q.; Zhang, W. Immunoassays for aflatoxins. TrAC Trends Anal. Chem. 2009, 28, 1115–1126. [Google Scholar] [CrossRef]
- Qileng, A.; Huang, S.; He, L.; Qin, W.; Liu, W.; Xu, Z.; Liu, Y. Composite films of CdS nanoparticles, MoS2 nanoflakes, reduced graphene oxide, and carbon nanotubes for ratiometric and modular immunosensing-based detection of toxins in cereals. ACS Appl. Nano Mater. 2020, 3, 2822–2829. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Stoltenburg, R.; Reinemann, C.; Strehlitz, B. SELEX—A (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol. Eng. 2007, 24, 381–403. [Google Scholar] [CrossRef] [PubMed]
- Bognar, Z.; Gyurcsanyi, R.E. Aptamers against immunoglobulins: Design, selection and bioanalytical applications. Int. J. Mol. Sci. 2020, 21, 5748. [Google Scholar] [CrossRef] [PubMed]
- Toh, S.Y.; Citartan, M.; Gopinath, S.C.; Tang, T.H. Aptamers as a replacement for antibodies in enzyme-linked immunosorbent assay. Biosens. Bioelectron. 2015, 64, 392–403. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Zhao, F.; Zhang, Y.; Xiong, Y.; Han, S. Recent advances in aptamer-based optical and electrochemical biosensors for detection of pesticides and veterinary drugs. Food Control 2022, 131, 108399. [Google Scholar] [CrossRef]
- Liu, L.S.; Wang, F.; Ge, Y.; Lo, P.K. Recent Developments in Aptasensors for Diagnostic Applications. ACS Appl. Mater. Interfaces 2021, 13, 9329–9358. [Google Scholar] [CrossRef]
- Lv, X.; Zhu, L.; Rong, Y.; Shi, H.; Zhang, L.; Yu, J.; Zhang, Y. Ratiometric electrochemiluminescence lab-on-paper device for DNA methylation determination based on highly conductive copper paper electrode. Biosens. Bioelectron. 2022, 214, 114522. [Google Scholar] [CrossRef]
- Zhou, Y.; Lv, S.; Wang, X.Y.; Kong, L.; Bi, S. Biometric photoelectrochemical-visual multimodal biosensor based on 3D hollow HCdS@Au nanospheres coupled with target induced ion exchange reaction for antigen detection. Anal. Chem. 2022, 94, 14492–14501. [Google Scholar] [CrossRef]
- Shen, Y.; Guan, J.; Ma, C.; Shu, Y.; Xu, Q.; Hu, X. Competitive displacement triggering DBP photoelectrochemical aptasensor via cetyltrimethylammonium bromide bridging aptamer and perovskite. Anal. Chem. 2022, 94, 1742–1751. [Google Scholar] [CrossRef]
- Hou, Y.; Jia, B.; Sheng, P.; Liao, X.; Shi, L.; Fang, L.; Zhou, L.; Kong, W. Aptasensors for mycotoxins in foods: Recent advances and future trends. Compr. Rev. Food Sci. Food Saf. 2022, 21, 2032–2073. [Google Scholar] [CrossRef]
- Xue, Z.; Zhang, Y.; Yu, W.; Zhang, J.; Wang, J.; Wan, F.; Kim, Y.; Liu, Y.; Kou, X. Recent advances in aflatoxin B1 detection based on nanotechnology and nanomaterials–A review. Anal. Chim. Acta 2019, 1069, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Beitollahi, H.; Tajik, S.; Dourandish, Z.; Zhang, K.; Le, Q.V.; Jang, H.W.; Kim, S.Y.; Shokouhimehr, M. Recent advances in the aptamer-based electrochemical biosensors for detecting aflatoxin B1 and its pertinent metabolite aflatoxin M1. Sensors 2020, 20, 3256. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, S.; Kramer, F.R. Molecular beacons: Probes that fluoresce upon hybridization. Nat. Biotechnol. 1996, 14, 303–308. [Google Scholar] [CrossRef]
- Bidar, N.; Amini, M.; Oroojalian, F.; Baradaran, B.; Hosseini, S.S.; Shahbazi, M.-A.; Hashemzaei, M.; Mokhtarzadeh, A.; Hamblin, M.R.; de la Guardia, M. Molecular beacon strategies for sensing purpose. TrAC Trends Anal. Chem. 2021, 134, 116143. [Google Scholar] [CrossRef]
- Moutsiopoulou, A.; Broyles, D.; Dikici, E.; Daunert, S.; Deo, S.K. Molecular aptamer beacons and their applications in sensing, imaging, and diagnostics. Small 2019, 15, 1902248. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Yang, R.; Shi, M.; Wu, C.; Fang, X.; Li, Y.; Li, J.; Tan, W. Rationally designed molecular beacons for bioanalytical and biomedical applications. Chem. Soc. Rev. 2015, 44, 3036–3055. [Google Scholar] [CrossRef] [PubMed]
- Goulko, A.A.; Li, F.; Chris Le, X. Bioanalytical applications of aptamer and molecular-beacon probes in fluorescence-affinity assays. TrAC Trends Anal. Chem. 2009, 28, 878–892. [Google Scholar] [CrossRef]
- Yamamoto, R.; Baba, T.; Kumar, P.K. Molecular beacon aptamer fluoresces in the presence of Tat protein of HIV-1. Genes Cells 2000, 5, 389–396. [Google Scholar] [CrossRef]
- Hamaguchi, N.; Ellington, A.; Stanton, M. Aptamer beacons for the direct detection of proteins. Anal. Biochem. 2001, 294, 126–131. [Google Scholar] [CrossRef]
- Li, J.J.; Fang, X.; Tan, W. Molecular aptamer beacons for real-time protein recognition. Biochem. Biophys. Res. Commun. 2002, 292, 31–40. [Google Scholar] [CrossRef]
- Stojanovic, M.N.; de Prada, P.; Landry, D.W. Aptamer-based folding fluorescent sensor for cocaine. J. Am. Chem. Soc. 2001, 123, 4928–4931. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Mallikaratchy, P.; Yang, R.; Kim, Y.; Zhu, Z.; Wang, H.; Tan, W. Aptamer switch probe based on intramolecular displacement. J. Am. Chem. Soc. 2008, 130, 11268–11269. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Ho, C.M. Aptamer-based optical probes with separated molecular recognition and signal transduction modules. J. Am. Chem. Soc. 2008, 130, 2380–2381. [Google Scholar] [CrossRef]
- Wang, C.; Sun, L.; Zhao, Q. A simple aptamer molecular beacon assay for rapid detection of aflatoxin B1. Chinese Chem. Lett. 2019, 30, 1017–1020. [Google Scholar] [CrossRef]
- Chen, L.; Wen, F.; Li, M.; Guo, X.; Li, S.; Zheng, N.; Wang, J. A simple aptamer-based fluorescent assay for the detection of aflatoxin B1 in infant rice cereal. Food Chemistry 2017, 215, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Wu, F.; Liu, P.; Zhou, G.; Yu, B.; Lou, X.; Xia, F. A label-free fluorescent aptasensor for the detection of aflatoxin B1 in food samples using AIEgens and graphene oxide. Talanta 2019, 198, 71–77. [Google Scholar] [CrossRef]
- Sabet, F.S.; Hosseini, M.; Khabbaz, H.; Dadmehr, M.; Ganjali, M.R. FRET-based aptamer biosensor for selective and sensitive detection of aflatoxin B1 in peanut and rice. Food Chemistry 2017, 220, 527–532. [Google Scholar] [CrossRef]
- Xia, X.; Wang, Y.; Yang, H.; Dong, Y.; Zhang, K.; Lu, Y.; Deng, R.; He, Q. Enzyme-free amplified and ultrafast detection of aflatoxin B1 using dual-terminal proximity aptamer probes. Food Chemistry 2019, 283, 32–38. [Google Scholar] [CrossRef]
- Lu, Z.; Chen, X.; Wang, Y.; Zheng, X.; Li, C.M. Aptamer based fluorescence recovery assay for aflatoxin B1 using a quencher system composed of quantum dots and graphene oxide. Microchimica Acta 2014, 182, 571–578. [Google Scholar] [CrossRef]
- Zhao, Z.; Yang, H.; Deng, S.; Dong, Y.; Yan, B.; Zhang, K.; Deng, R.; He, Q. Intrinsic conformation response-leveraged aptamer probe based on aggregation-induced emission dyes for aflatoxin B1 detection. Dye. Pigment. 2019, 171, 107767. [Google Scholar] [CrossRef]
- Mousivand, M.; Javan-Nikkhah, M.; Anfossi, L.; Di Nardo, F.; Salina, M.; Bagherzadeh, K. High performance aptasensing platform development through in silico aptamer engineering for aflatoxin B1 monitoring. Food Control 2023, 145, 109418. [Google Scholar] [CrossRef]



| Name | Base Sequence (5′ to 3′) | Labels |
|---|---|---|
| MB | CGTGTTGTCTCTCTGTGT CTCG | 5′-BHQ1, 3′-FAM |
| FDNA | CGTGTTGTCTCTCTGTGT CTCG | 3′-FAM |
| BDNA | CGTGTTGTCTCTCTGTGT CTCG | 5′-BHQ1 |
| cDNA | GAGACAACACG | no |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.; Zhu, K.; Yu, J.; Shi, P. Complementary DNA Significantly Enhancing Signal Response and Sensitivity of a Molecular Beacon Probe to Aflatoxin B1. Biosensors 2023, 13, 195. https://doi.org/10.3390/bios13020195
Wang C, Zhu K, Yu J, Shi P. Complementary DNA Significantly Enhancing Signal Response and Sensitivity of a Molecular Beacon Probe to Aflatoxin B1. Biosensors. 2023; 13(2):195. https://doi.org/10.3390/bios13020195
Chicago/Turabian StyleWang, Chao, Kexiao Zhu, Jie Yu, and Pengfei Shi. 2023. "Complementary DNA Significantly Enhancing Signal Response and Sensitivity of a Molecular Beacon Probe to Aflatoxin B1" Biosensors 13, no. 2: 195. https://doi.org/10.3390/bios13020195
APA StyleWang, C., Zhu, K., Yu, J., & Shi, P. (2023). Complementary DNA Significantly Enhancing Signal Response and Sensitivity of a Molecular Beacon Probe to Aflatoxin B1. Biosensors, 13(2), 195. https://doi.org/10.3390/bios13020195





