A Cascade Signal Amplification Strategy for the Ultrasensitive Fluorescence Detection of Cu2+ via λ-Exonuclease-Assisted Target Recycling with Mismatched Catalytic Hairpin Assembly
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Apparatus
2.3. Procedure for Cu2+ Detection
2.4. Detection of Cu2+ in Real Samples
3. Results and Discussion
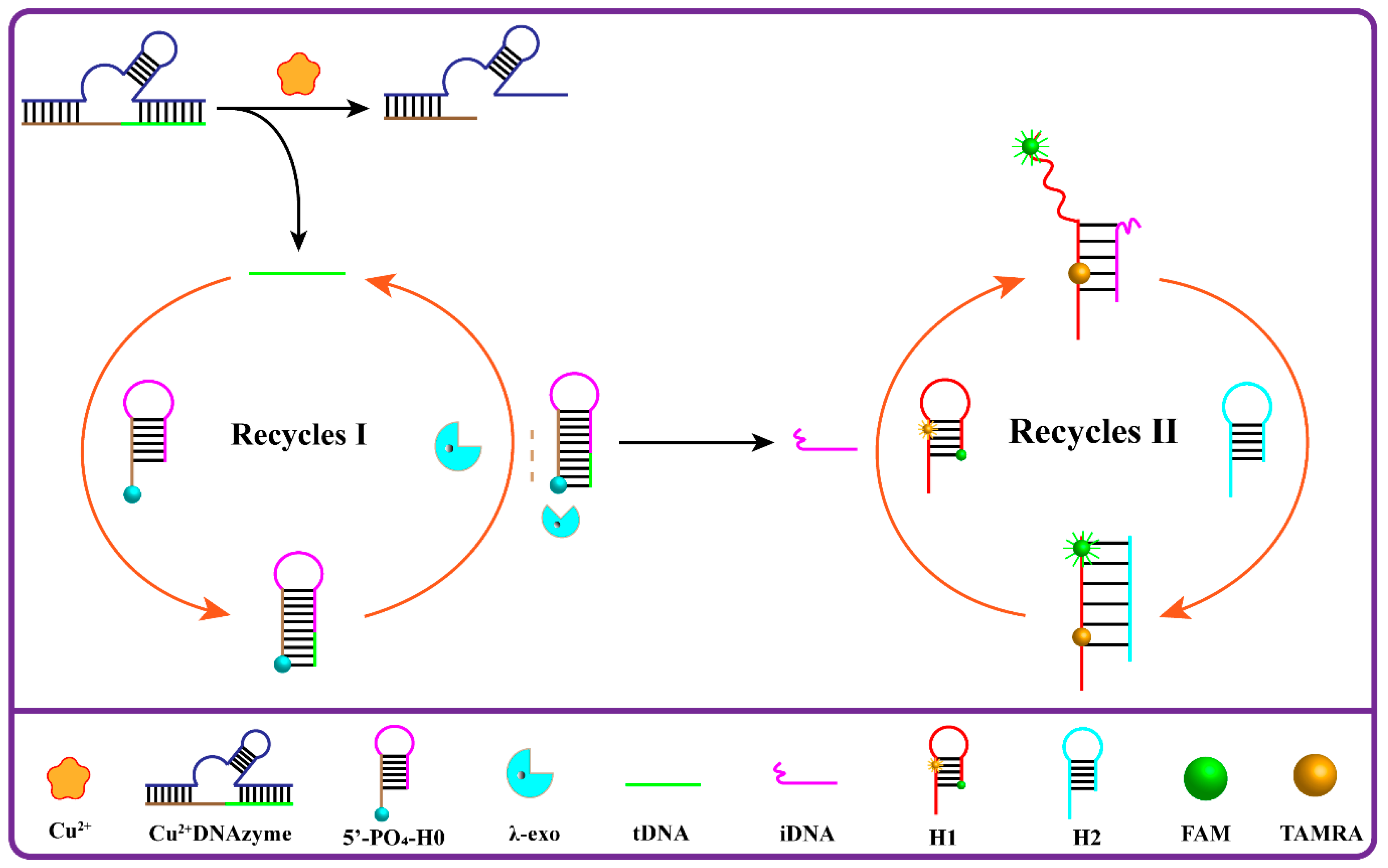
3.1. Principle of the Designed Cu2+ Fluorescence Biosensor
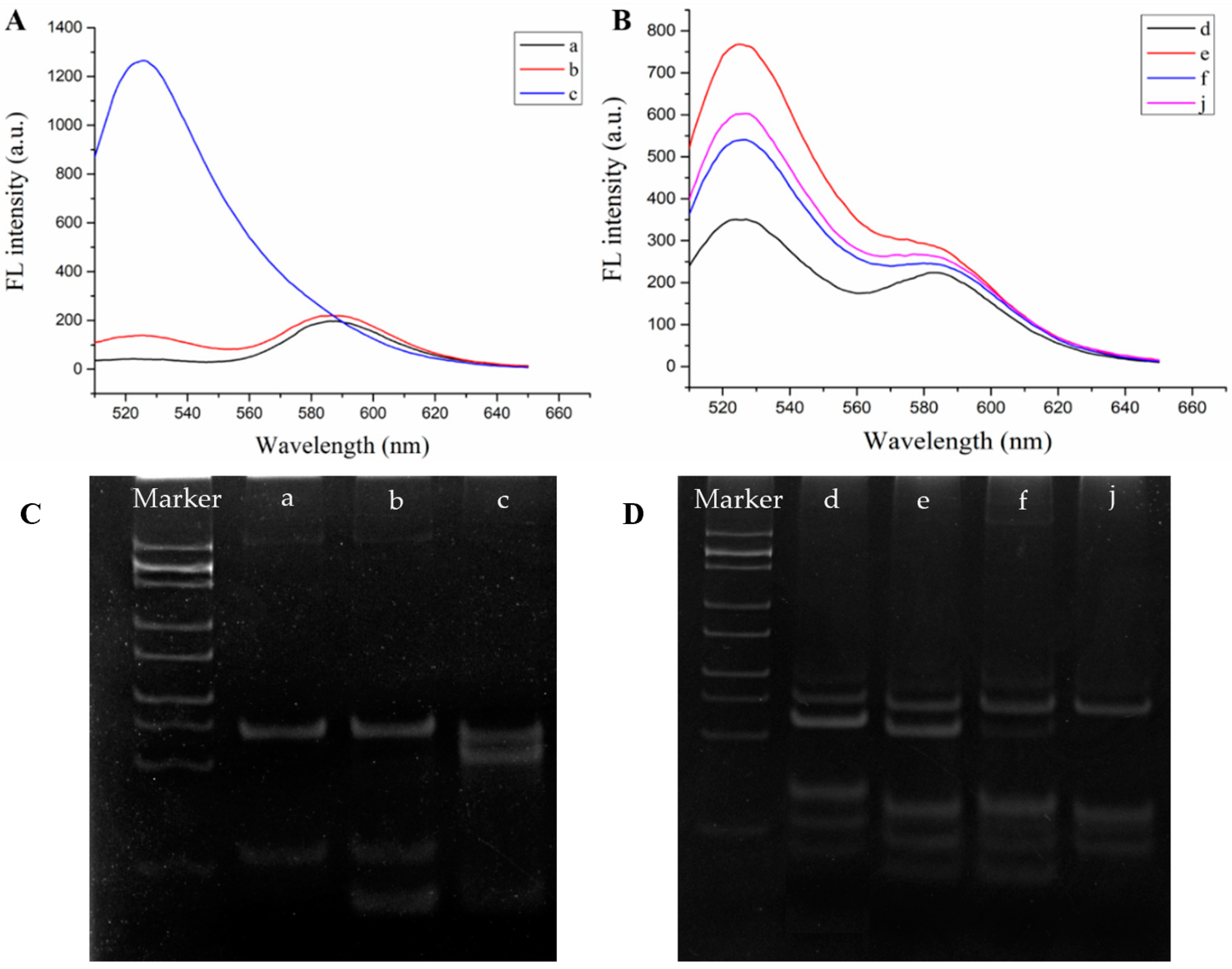
3.2. Feasibility of the Fluorescence Biosensor for Cu2+ Detection
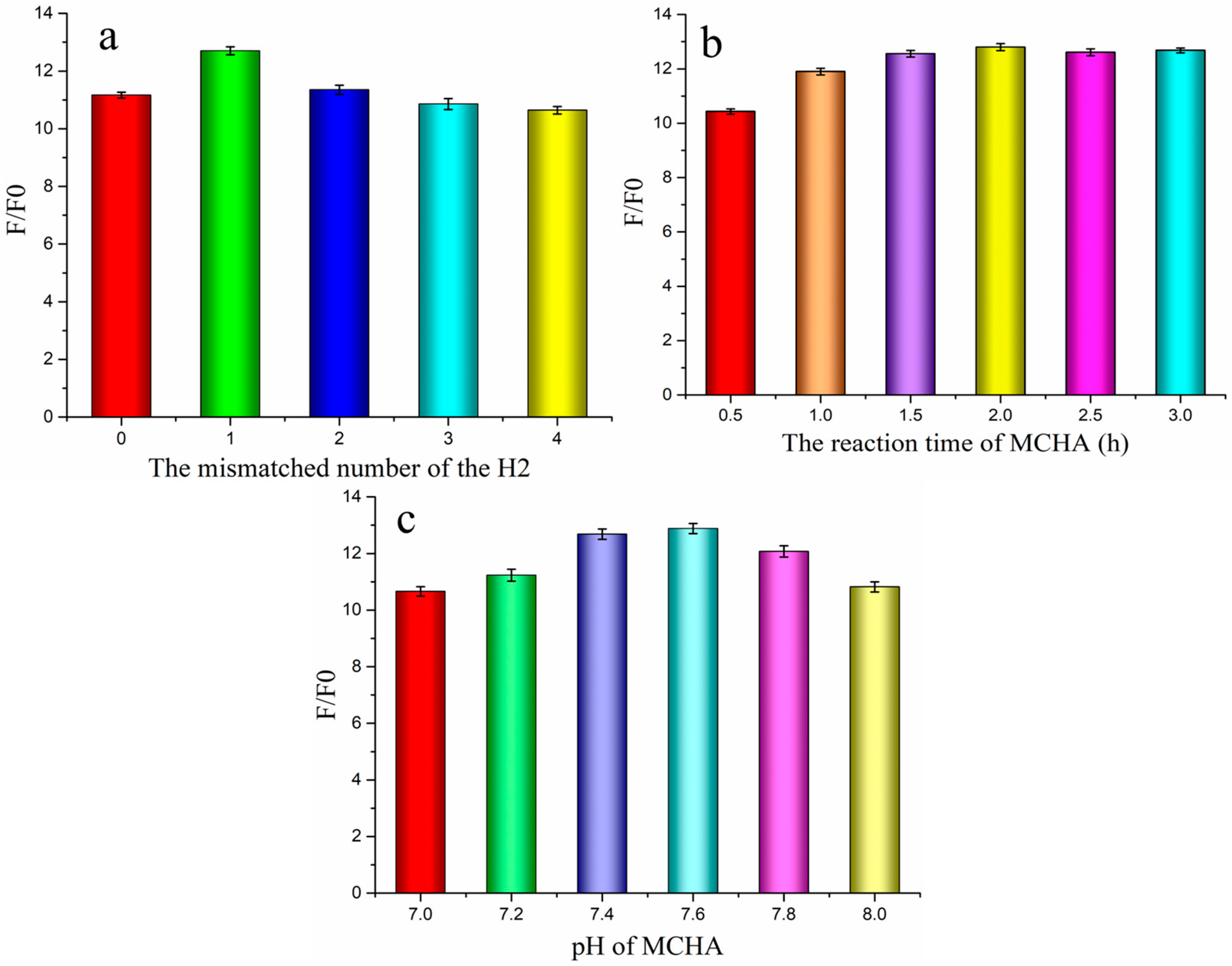
3.3. Optimization of Experimental Conditions
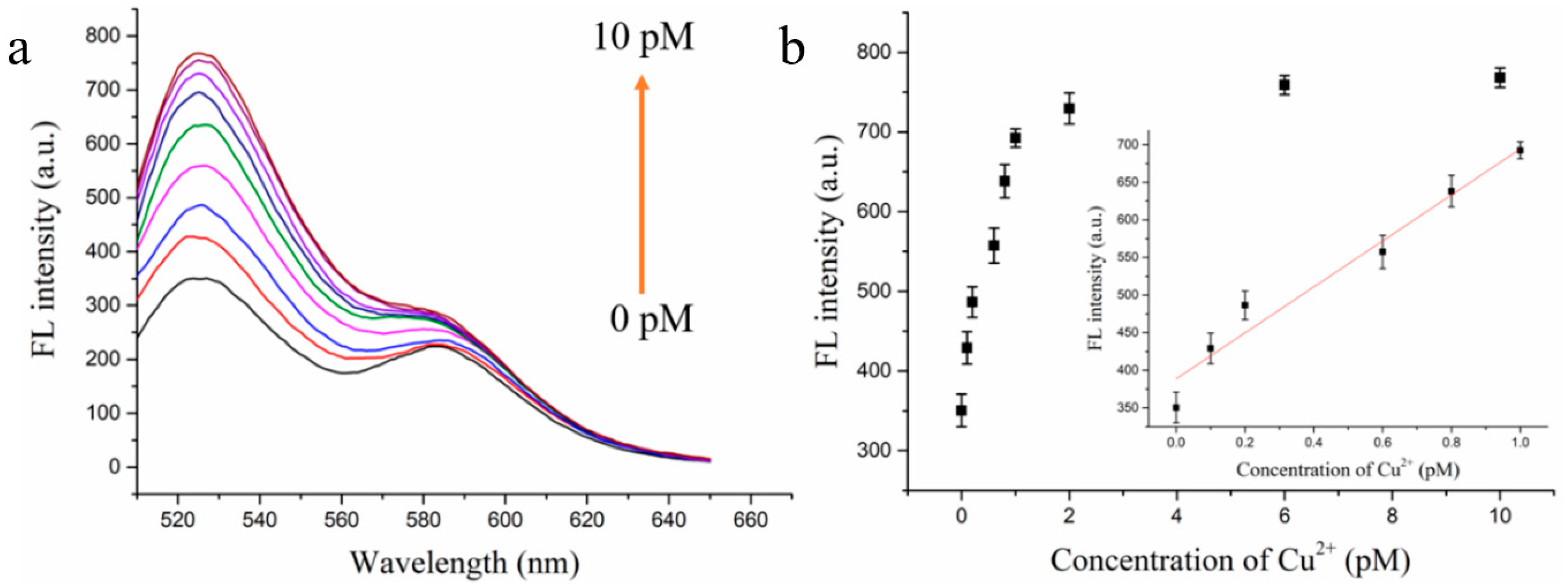
3.4. Detection Performance of the Cu2+ Fluorescence Biosensor
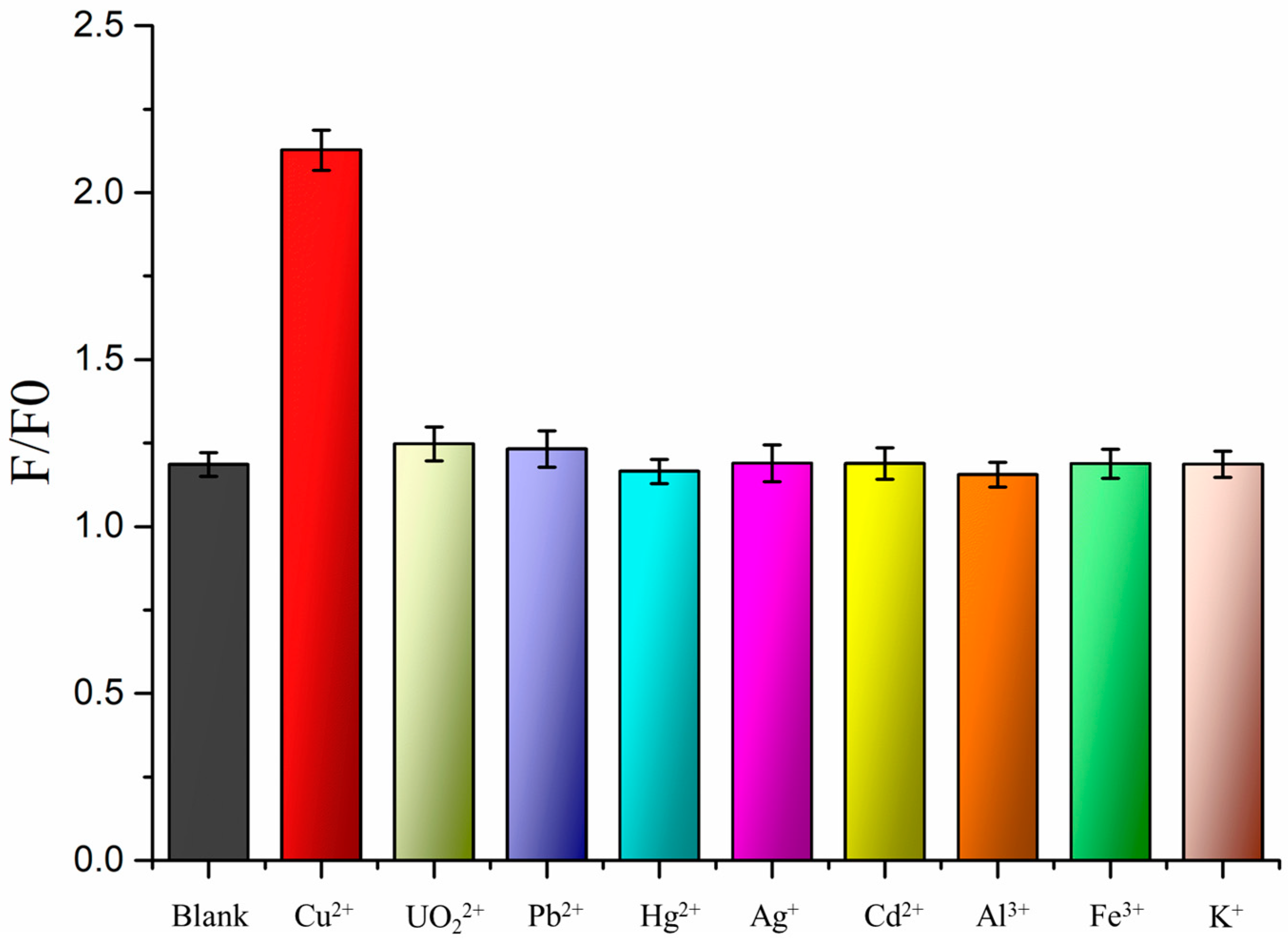
3.5. Specificity of the Cu2+ Fluorescence Biosensor
3.6. Detection of Cu2+ in Real Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, L.Q.; Zhou, H.H.; Chen, X.; Liu, G.J.; Jiang, C.L.; Zheng, L.G. Study of the micromorphology and health risks of arsenic in copper smelting slag tailings for safe resource utilization. Ecotoxicol. Environ. Saf. 2021, 219, 112321. [Google Scholar] [PubMed]
- Paithankar, J.G.; Saini, S.; Dwivedi, S.; Sharma, A.; Chowdhuri, D.K. Heavy metal associated health hazards: An interplay of oxidative stress and signal transduction. Chemosphere 2021, 262, 128350. [Google Scholar]
- Beula, D.; Sureshkumar, M. A review on the toxic E-waste killing health and environment—Today’s global scenario. Mater. Today Proc. 2021, 47, 2168–2174. [Google Scholar]
- Al-Sulami, A.I.; Basha, M.T.; Althagafy, H.S.; Al-Zaydi, K.M.; Davaasuren, B.; Al-Kaff, N.S.; Said, M.A. Azo-based multifunctional molecules and their copper (II) complexes as potential inhibitors against Alzheimer’s disease: XRD/Hirshfeld analysis/DFT/molecular docking/cytotoxicity. Inorg. Chem. Commun. 2022, 142, 109535. [Google Scholar]
- Xia, Y.D.; Sun, Y.Q.; Cheng, Y.; Xia, Y.; Yin, X.B. Rational design of dual-ligand Eu-MOF for ratiometric fluorescence sensing Cu2+ ions in human serum to diagnose Wilson’s disease. Anal. Chim. Acta 2022, 1204, 339731. [Google Scholar]
- Zhang, D.; Wang, Z.M.; Yang, J.; Yi, L.; Liao, L.F.; Xiao, X.L. Development of a method for the detection of Cu2+ in the environment and live cells using a synthesized spider web-like fluorescent probe. Biosens. Bioelectron. 2021, 182, 113174. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.Z.; Lin, X.C.; Wu, X.P.; Xie, Z.H. Solid phase extraction of lead (II), copper (II), cadmium (II) and nickel (II) using gallic acid-modified silica gel prior to determination by flame atomic absorption spectrometry. Talanta 2008, 74, 836–843. [Google Scholar] [CrossRef]
- Shen, M.; Chen, L.Y.; Han, W.L.; Ma, A. Methods for the determination of heavy metals in indocalamus leaves after different preservation treatment using inductively-coupled plasma mass spectrometry. Microchem. J. 2018, 139, 295–300. [Google Scholar]
- Liu, Y.; Wu, Y.P.; Guo, X.Y.; Wen, Y.; Yang, H.F. Rapid and selective detection of trace Cu2+ by accumulation-reaction-based Raman spectroscopy. Sens. Actuators B 2019, 283, 278–283. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, H.; Xu, G.L.; Guan, M.D.; Zhang, Q.Y.; Wang, Z.Y.; Dong, Z.Z.; Chen, W.H.; Yang, X.X.; Qiao, A.B. A multiplexed electrochemical quantitative polymerase chain reaction platform for single-base mutation analysis. Biosens. Bioelectron. 2022, 214, 114496. [Google Scholar] [CrossRef]
- Liu, C.; Li, Y.J.; Liu, J.Q.; Liao, L.F.; Zhou, R.L.; Yu, W.Z.; Li, Q.; He, L.Q.; Li, Q.X.; Xiao, X.L. Recent advances in the construction of functional nucleic acids with isothermal amplification for heavy metal ions sensor. Microchem. J. 2021, 175, 107077. [Google Scholar]
- Xia, X.H.; Yang, H.; Cao, J.J.; Zhang, J.Q.; He, Q.; Deng, R.J. Isothermal nucleic acid amplification for food safety analysis. TrAC Trends Anal. Chem. 2022, 153, 116641. [Google Scholar]
- Zhong, M.; Yang, S.Y.; Chen, L.; Liu, C.; Shi, J.; Liang, H.; Xiao, X.L.; Li, L.; Liu, J.Q. Highly sensitive and efficient fluorescent sensing for Hg2+ detection based on triple-helix molecular switch and exonuclease III-assisted amplification. Anal. Chim. Acta 2022, 1205, 339751. [Google Scholar]
- Li, X.Y.; Du, Z.H.; Lin, S.H.; Tian, J.J.; Tian, H.T.; Xu, W.T. ExoIII and TdT dependent isothermal amplification (ETDA) colorimetric biosensor for ultra-sensitive detection of Hg2+. Food Chem. 2020, 316, 126303. [Google Scholar] [CrossRef]
- Li, H.K.; Cai, Q.Q.; Wu, D.; Jie, G.F.; Zhou, H. Fluorescence energy transfer biosensing platform based on hyperbranched rolling circle amplification and multi-site strand displacement for ultrasensitive detection of miRNA. Anal. Chim. Acta 2022, 1222, 340190. [Google Scholar]
- Chen, M.; He, Q.D.; Tong, Y.L.; Chen, Z.G. A universal fluorescent sensing system for pathogen determination based on loop-mediated isothermal amplification triggering dual-primer rolling circle extension. Sens. Actuators B 2021, 331, 129436. [Google Scholar] [CrossRef]
- Luo, Z.W.; Li, Y.X.; Zhang, P.; He, L.; Feng, Y.T.; Feng, Y.Q.; Qian, C.; Tian, Y.H.; Duan, Y.X. Catalytic hairpin assembly as cascade nucleic acid circuits for fluorescent biosensor: Design, evolution and application. TrAC Trends Anal. Chem. 2022, 151, 116582. [Google Scholar]
- Zhang, M.R.; Ding, Q.; Zhu, M.H.; Yuan, R.; Yuan, Y.L. An ultrasensitive electrochemical biosensor with amplification of highly efficient triple catalytic hairpin assembly and tetris hybridization chain reaction. Sens. Actuators B 2022, 361, 131683. [Google Scholar] [CrossRef]
- Xia, Y.K.; Huang, Z.N.; Chen, T.T.; Xu, L.L.; Zhu, G.Z.; Chen, W.Q.; Chen, G.Y.; Wu, S.X.; Lan, J.M.; Lin, X. Sensitive fluorescent detection of exosomal microRNA based on enzymes-assisted dual-signal amplification. Biosens. Bioelectron. 2022, 209, 114259. [Google Scholar]
- Pan, J.F.; Li, Q.; Zhou, D.H.; Chen, J.H. Label-free and highly sensitive fluorescence detection of lead (ii) based on DNAzyme and exonuclease III-assisted cascade signal amplification. New J. Chem. 2019, 43, 5857–5862. [Google Scholar] [CrossRef]
- Zhang, Y.Y.; Wang, J.L.; Chen, S.H.; Yuan, R. A novel magnetic beads-assisted highly-ordered enzyme-free localized DNA cascade reaction for the fluorescence detection of Pb2+. Sens. Actuators B 2021, 342, 130040. [Google Scholar] [CrossRef]
- Yun, W.; Wu, H.; Yang, Z.H.; Wang, R.Q.; Wang, C.J.; Yang, L.Z.; Tang, Y.J. A dynamic, ultra-sensitive and “turn-on” strategy for fluorescent detection of uranyl based on DNAzyme and entropy-driven amplification initiated circular cleavage amplification. Anal. Chim. Acta 2019, 1068, 104–110. [Google Scholar] [CrossRef]
- Tian, J.J.; Zhang, Y.; Zhu, L.J.; Tian, H.T.; Li, K.; Huang, K.L.; Luo, Y.B.; Zhang, X.J.; Xu, W.T. dsDNA/ssDNA-switchable isothermal colorimetric biosensor based on a universal primer and λ exonuclease. Sens. Actuators B 2020, 323, 128674. [Google Scholar] [CrossRef]
- Yu, Y.J.; Ma, L.; Li, L.D.; Deng, Y.N.; Xu, L.D.; Liu, H.; Xiao, L.H.; Su, X. Digestion of dynamic substrate by exonuclease reveals high single-mismatch selectivity. Anal. Chem. 2018, 90, 13655–13662. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.B.; Li, C.J.; Hu, Y.Q.; Ding, Y.W.; Wu, T.B. A structure change-induced fluorescent biosensor for uracil-DNA glycosylase activity detection based on the substrate preference of Lambda exonuclease. Talanta 2022, 243, 123350. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.Y.; Ma, C.B.; Li, Z.K.; Wu, K.F. An exonuclease-assisted fluorescence sensor for assaying alkaline phosphatase based on SYBR Green I. Mol. Cell. Probes 2019, 45, 26–30. [Google Scholar] [CrossRef]
- Xu, J.Y.; Guo, J.J.; Golob-Schwarzl, N.; Haybaeck, J.; Qiu, X.; Hildebrandt, N. Single-Measurement Multiplexed Quantification of MicroRNAs from Human Tissue Using Catalytic Hairpin Assembly and Forster Resonance Energy Transfer. ACS Sens. 2020, 5, 1768–1776. [Google Scholar] [CrossRef]
- Huang, Q.; Ma, P.Q.; Li, H.D.; Yin, B.C.; Ye, B.C. Catalytic-hairpin-assembly-assisted DNA tetrahedron nanoprobe for intracellular microRNA imaging. ACS Appl. Bio Mater. 2020, 3, 2861–2866. [Google Scholar] [CrossRef]
- Zhang, Z.; Xiang, X.; Hu, Y.Q.; Deng, Y.H.; Li, L.J.; Zhao, W.B.; Wu, T.B. A sensitive biomolecules detection device with catalytic hairpin assembly and cationic conjugated polymer-assisted dual signal amplification strategy. Talanta 2021, 223, 121716. [Google Scholar] [CrossRef]
- Yin, P.; Choi, H.M.T.; Calvert, C.R.; Pierce, N.A. Programming biomolecular self-assembly pathways. Nature 2008, 451, 318–322. [Google Scholar] [CrossRef]
- Jiang, Y.S.; Bhadra, S.; Li, B.L.; Ellington, A.D. Mismatches improve the performance of strand-displacement nucleic acid circuits. Angew. Chem. 2014, 126, 1876–1879. [Google Scholar] [CrossRef]
- Zhang, L.L.; Zhang, Y.Y.; Wei, M.J.; Yi, Y.H.; Li, H.T.; Yao, S.Z. A label-free fluorescent molecular switch for Cu2+ based on metal ion-triggered DNA-cleaving DNAzyme and DNA intercalator. New J. Chem. 2013, 37, 1252–1257. [Google Scholar] [CrossRef]
- Wang, D.; Ge, C.C.; Wang, L.; Xing, X.R.; Zeng, L.W. A simple lateral flow biosensor for the rapid detection of copper (II) ions based on click chemistry. RSC Adv. 2015, 5, 75722–75727. [Google Scholar] [CrossRef]
- Ge, C.; Chen, J.; Wu, W.; Fang, Z.; Chen, L.; Liu, Q.; Wang, L.; Xing, X.; Zeng, L. An enzyme-free and label-free assay for copper (II) ion detection based on self-assembled DNA concatamers and Sybr Green I. Analyst 2013, 138, 4737–4740. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, L.; Ou, Y.D.; Wang, Z.H.; Fu, F.F.; Guo, L.Q. DNAzyme-based biosensor for Cu2+ ion by combining hybridization chain reaction with fluorescence resonance energy transfer technique. Talanta 2016, 155, 245–249. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Lee, C.Y.; Park, K.S.; Park, H.G. Enzyme-Free Colorimetric Detection of Cu2+ by Utilizing Target-Triggered DNAzymes and Toehold-Mediated DNA Strand Displacement Events. Chem.-Eur. J. 2017, 23, 17379–17383. [Google Scholar] [CrossRef]
- Shen, R.D.; Zou, L.; Wu, S.X.; Li, T.T.; Wang, J.; Liu, J.M.; Ling, L. A novel label-free fluorescent detection of histidine based upon Cu2+-specific DNAzyme and hybridization chain reaction. Spectrochim. Acta A 2019, 213, 42–47. [Google Scholar] [CrossRef]
- Ming, J.J.; Fan, W.B.; Jiang, T.F.; Wang, Y.H.; Lv, Z.H. Portable and sensitive detection of copper (II) ion based on personal glucose meters and a ligation DNAzyme releasing strategy. Sens. Actuators B-Chem. 2017, 240, 1091–1098. [Google Scholar] [CrossRef]
- Qin, Y.F.; Li, M.; Yang, Y.Y.; Gao, Z.Y.; Zhang, H.S.; Zhao, J.J. A unimolecular DNA fluorescent probe for determination of copper ions based on click chemistry. RSC Adv. 2020, 10, 6017–6021. [Google Scholar] [CrossRef]
- Yan, W.C.; Zhong, Z.S.; Ma, J.; Rujiralai, T. Highly sensitive colorimetric sensing of copper (ii) ions based on “CLICK-17” DNAzyme-catalyzed azide modified gold nanoparticles and alkyne capped dsDNA cycloaddition. RSC Adv. 2021, 11, 24196–24205. [Google Scholar] [CrossRef]
- Szerlauth, A.; Szalma, L.; Muráth, S.; Sáringer, S.; Varga, G.; Li, L.; Szilágyi, I. Nanoclay-based sensor composites for the facile detection of molecular antioxidants. Analyst 2022, 147, 1367–1374. [Google Scholar] [CrossRef] [PubMed]

| Xiangjiang River Water | Added (fM) | Found Mean ± SD * | Recovery (%) |
|---|---|---|---|
| Sample 1 | 400 | 425.55 ± 0.6 | 106.38 |
| Sample 2 | 600 | 587.59 ± 0.4 | 97.93 |
| Sample 3 | 800 | 808.20 ± 0.2 | 101.03 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Liu, C.; He, L.; Liu, J.; Li, L.; Yang, S.; Tan, Y.; Liu, X.; Xiao, X. A Cascade Signal Amplification Strategy for the Ultrasensitive Fluorescence Detection of Cu2+ via λ-Exonuclease-Assisted Target Recycling with Mismatched Catalytic Hairpin Assembly. Biosensors 2023, 13, 918. https://doi.org/10.3390/bios13100918
Liu Z, Liu C, He L, Liu J, Li L, Yang S, Tan Y, Liu X, Xiao X. A Cascade Signal Amplification Strategy for the Ultrasensitive Fluorescence Detection of Cu2+ via λ-Exonuclease-Assisted Target Recycling with Mismatched Catalytic Hairpin Assembly. Biosensors. 2023; 13(10):918. https://doi.org/10.3390/bios13100918
Chicago/Turabian StyleLiu, Zhen, Chen Liu, Liqiong He, Jinquan Liu, Le Li, Shengyuan Yang, Yan Tan, Xing Liu, and Xilin Xiao. 2023. "A Cascade Signal Amplification Strategy for the Ultrasensitive Fluorescence Detection of Cu2+ via λ-Exonuclease-Assisted Target Recycling with Mismatched Catalytic Hairpin Assembly" Biosensors 13, no. 10: 918. https://doi.org/10.3390/bios13100918
APA StyleLiu, Z., Liu, C., He, L., Liu, J., Li, L., Yang, S., Tan, Y., Liu, X., & Xiao, X. (2023). A Cascade Signal Amplification Strategy for the Ultrasensitive Fluorescence Detection of Cu2+ via λ-Exonuclease-Assisted Target Recycling with Mismatched Catalytic Hairpin Assembly. Biosensors, 13(10), 918. https://doi.org/10.3390/bios13100918




