Quantification of Desiccated Extracellular Vesicles by Quartz Crystal Microbalance
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cell Culture
2.3. EV Isolation by ExoQuick-TC
2.4. Dot-Blotting
2.5. Nanoparticle Tracking Analysis (NTA)
2.6. Quartz Crystal Microbalance (QCM)
2.7. Transmission Electron Microscopy (TEM)
2.8. Scanning Electron Microscopy (SEM)
2.9. Atomic Force Microscopy (AFM)
3. Results
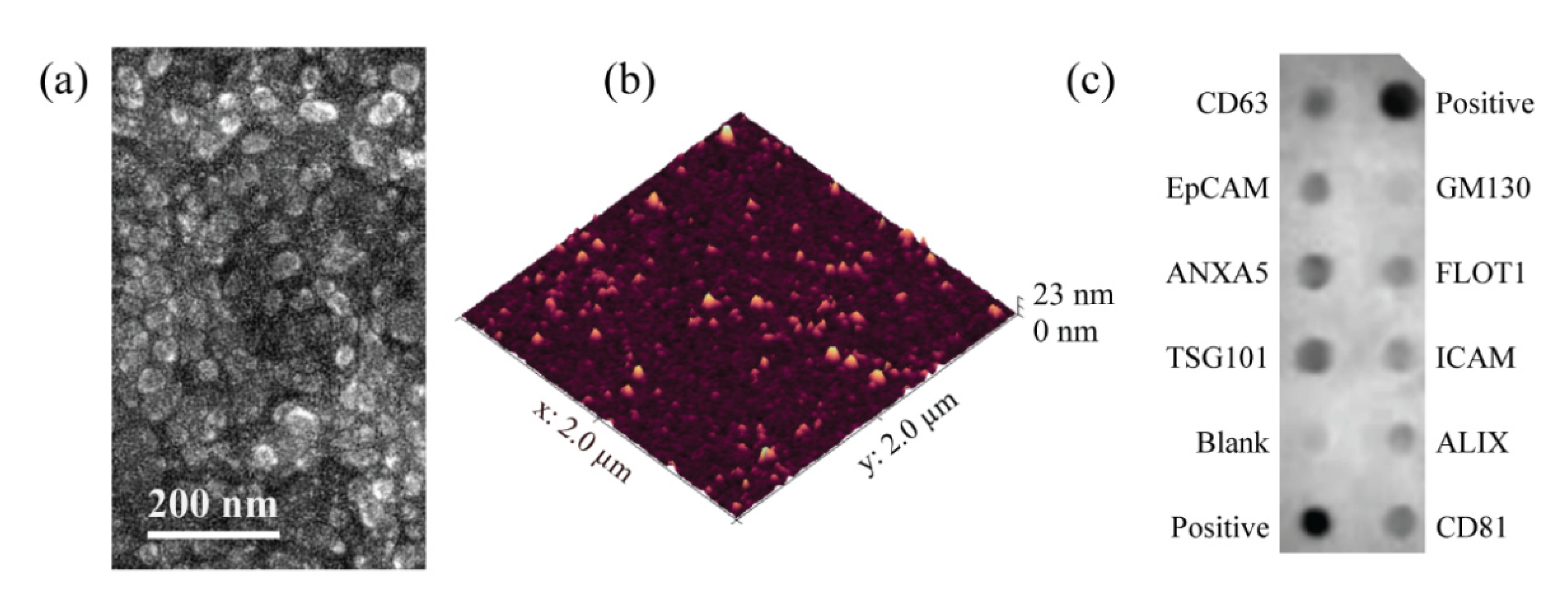
3.1. EV Isolation
3.2. QCM Biosensor Calibration
3.3. Testing the Calibration Curve
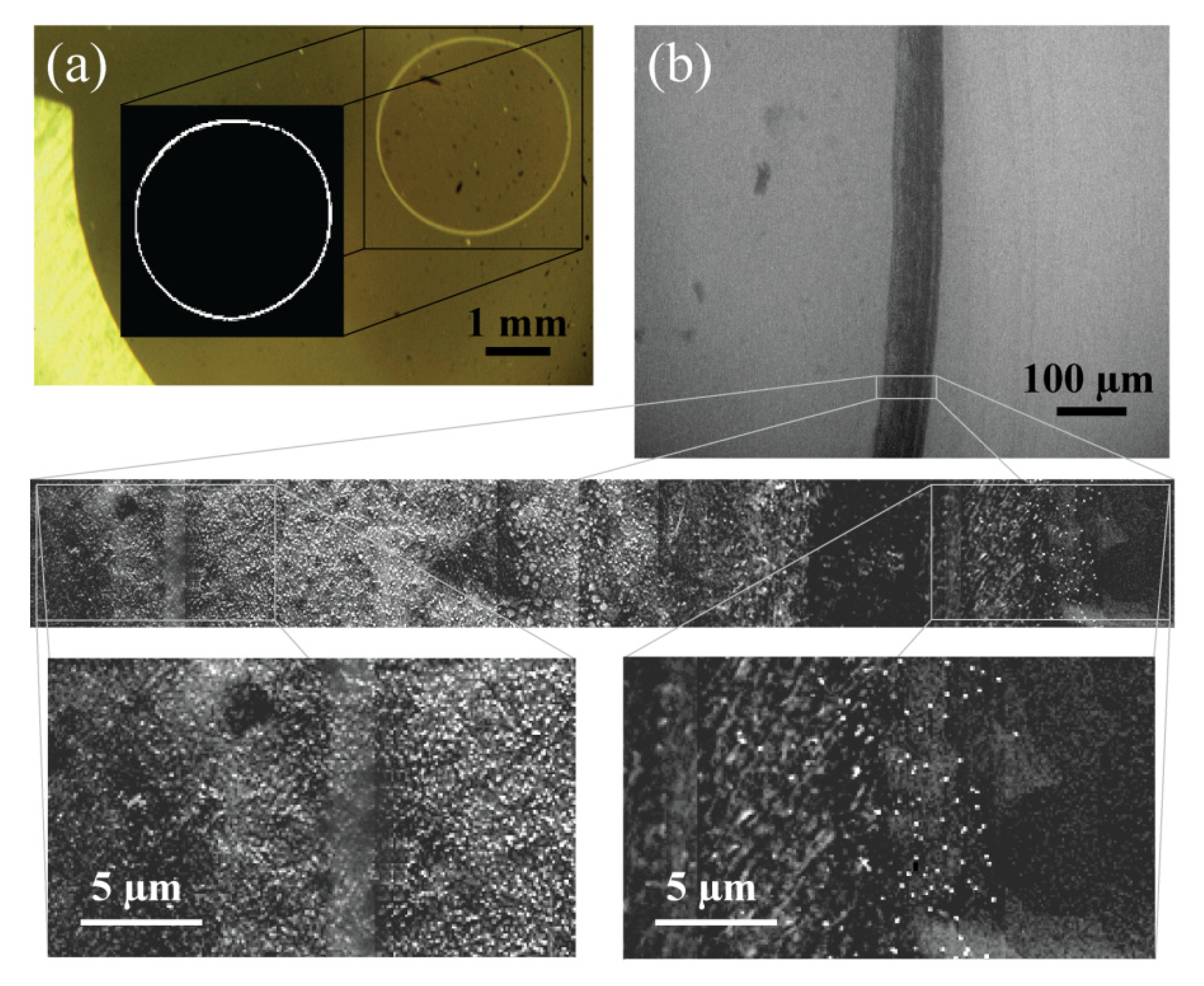
3.4. EV Desiccation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Simons, M.; Raposo, G. Exosomes-vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 2009, 21, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Palma, J.; Yaddanapudi, S.C.; Pigati, L.; Havens, M.A.; Jeong, S.; Weiner, G.A.; Weimer, K.M.E.; Stern, B.; Hastings, M.L.; Duelli, D.M. MicroRNAs are exported from malignant cells in customized particles. Nucleic Acids Res. 2012, 40, 9125–9138. [Google Scholar] [CrossRef] [PubMed]
- Vlassov, A.V.; Magdaleno, S.; Setterquist, R.; Conrad, R. Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim. Biophys. Acta 2012, 1820, 940–948. [Google Scholar] [CrossRef] [PubMed]
- Kharaziha, P.; Ceder, S.; Li, Q.; Panaretakis, T. Tumor cell-derived exosomes: A message in a bottle. Biochim. Biophys. Acta 2012, 1826, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Ge, R.; Tan, E.; Sharghi-Namini, S.; Asada, H.H. Exosomes in Cancer Microenvironment and Beyond: Have we Overlooked these Extracellular Messengers? Cancer Microenviron. 2012, 5, 323–332. [Google Scholar] [CrossRef] [Green Version]
- Pucci, F.; Pittet, M.J. Molecular pathways: Tumor-derived microvesicles and their interactions with immune cells in vivo. Clin. Cancer Res. 2013, 19, 2598–2604. [Google Scholar] [CrossRef] [Green Version]
- King, H.W.; Michael, M.Z.; Gleadle, J.M. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer 2012, 12, 421. [Google Scholar] [CrossRef] [Green Version]
- Rupert, D.L.M.; Claudio, V.; Lässer, C.; Bally, M. Methods for the physical characterization and quantification of extracellular vesicles in biological samples. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 3164–3179. [Google Scholar] [CrossRef]
- Szatanek, R.; Baj-Krzyworzeka, M.; Zimoch, J.; Lekka, M.; Siedlar, M.; Baran, J. The Methods of Choice for Extracellular Vesicles (EVs) Characterization. Int. J. Mol. Sci. 2017, 18, 1153. [Google Scholar] [CrossRef]
- Pospichalova, V.; Svoboda, J.; Dave, Z.; Kotrbova, A.; Kaiser, K.; Klemova, D.; Ilkovics, L.; Hampl, A.; Crha, I.; Jandakova, E.; et al. Simplified protocol for flow cytometry analysis of fluorescently labeled exosomes and microvesicles using dedicated flow cytometer. J. Extracell. Vesicles 2015, 4, 25530. [Google Scholar] [CrossRef]
- O’Sullivan, C.K.; Guilbault, G.G. Commercial quartz crystal microbalances–Theory and applications. Biosens. Bioelectron. 1999, 14, 663–670. [Google Scholar] [CrossRef]
- Sauerbrey, G. The use of quartz oscillators for weighing thin layers and for microweighing. Z. Phys. 1959, 155, 206–222. [Google Scholar] [CrossRef]
- King, W.H. Piezoelectric Sorption Detector. Anal. Chem. 1964, 36, 1735–1739. [Google Scholar] [CrossRef]
- Shons, A.; Dorman, F.; Najarian, J. The piezoelectric quartz immunosensor. J. Biomed. Mater. Res. 1972, 6, 565–570. [Google Scholar] [CrossRef]
- Fawcett, N.C.; Evans, J.A.; Chen, L.C.; Drozda, K.A.; Flowers, N. A quartz crystal detector for DNA. Anal. Lett. 1988, 21, 1099–1110. [Google Scholar] [CrossRef]
- Guleryuz, H.; Kaus, I.; Buron, C.C.; Filiâtre, C.; Hedin, N.; Bergström, L.; Einarsrud, M.A. Deposition of silica nanoparticles onto alumina measured by optical reflectometry and quartz crystal microbalance with dissipation techniques. Colloids Surf. A Physicochem. Eng. Asp. 2014, 443, 384–390. [Google Scholar] [CrossRef] [Green Version]
- Latif, U.; Qian, J.; Can, S.; Dickert, F.L. Biomimetic receptors for bioanalyte detection by quartz crystal microbalances—From molecules to cells. Sensors 2014, 14, 23419–23438. [Google Scholar] [CrossRef]
- Schulz, W.W.; King, W.H., Jr. A Universal Mass Detector for Liquid Chromatography. J. Chromatogr. Sci. 1973, 11, 343–348. [Google Scholar] [CrossRef]
- Reipa, V.; Purdum, G.; Choi, J. Measurement of nanoparticle concentration using quartz crystal microgravimetry. J. Phys. Chem. B 2010, 114, 16112–16117. [Google Scholar] [CrossRef]
- Suthar, J.; Prieto-Simon, B.; Williams, G.R.; Guldin, S. Dual-Mode and Label-Free Detection of Exosomes from Plasma Using an Electrochemical Quartz Crystal Microbalance with Dissipation Monitoring. Anal. Chem. 2022, 94, 2465–2475. [Google Scholar] [CrossRef]
- Liangsupree, T.; Multia, E.; Forssén, P.; Fornstedt, T.; Riekkola, M.-L. Kinetics and interaction studies of anti-tetraspanin antibodies and ICAM-1 with extracellular vesicle subpopulations using continuous flow quartz crystal microbalance biosensor. Biosens. Bioelectron. 2022, 206, 114151. [Google Scholar] [CrossRef] [PubMed]
- Suthar, J.; Parsons, E.S.; Hoogenboom, B.W.; Williams, G.R.; Guldin, S. Acoustic Immunosensing of Exosomes Using a Quartz Crystal Microbalance with Dissipation Monitoring. Anal. Chem. 2020, 92, 4082–4093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Priglinger, E.; Strasser, J.; Buchroithner, B.; Weber, F.; Wolbank, S.; Auer, D.; Grasmann, E.; Arzt, C.; Sivun, D.; Grillari, J.; et al. Label-free characterization of an extracellular vesicle-based therapeutic. J. Extracell. Vesicles 2021, 10, e12156. [Google Scholar] [CrossRef] [PubMed]
- Stratton, D.; Lange, S.; Kholia, S.; Jorfi, S.; Antwi-Baffour, S.; Inal, J. Label-free real-time acoustic sensing of microvesicle release from prostate cancer (PC3) cells using a Quartz Crystal Microbalance. Biochem. Biophys. Res. Commun. 2014, 453, 619–624. [Google Scholar] [CrossRef]
- Zhao, L.; Yu, J.; Wang, J.; Li, H.; Che, J.; Cao, B. Isolation and Identification of miRNAs in exosomes derived from serum of colon cancer patients. J. Cancer 2017, 8, 1145–1152. [Google Scholar] [CrossRef] [Green Version]
- Zlotogorski-Hurvitz, A.; Dayan, D.; Chaushu, G.; Korvala, J.; Salo, T.; Sormunen, R.; Vered, M. Human Saliva-Derived Exosomes: Comparing Methods of Isolation. J. Histochem. Cytochem. 2014, 63, 181–189. [Google Scholar] [CrossRef] [Green Version]
- Gooch, A.; Skliar, M.; Zhang, P.; Hu, Z.; Westenfelder, C. Administration of exosomes from Mesenchymal Stem Cells provides effective survival benefits and functional rescue from severe progressive ischemia-reperfusion injury-induced AKI in rats. J. Am. Soc. Nephrol. 2020, 31, 101. [Google Scholar]
- Skliar, M.; Chernyshev, V.S. Imaging of extracellular vesicles by atomic force microscopy. J. Vis. Exp. 2019, 151, e59254. [Google Scholar] [CrossRef]
- Chernyshev, V.S.; Rachamadugu, R.; Tseng, Y.H.; Belnap, D.M.; Jia, Y.; Branch, K.J.; Butterfield, A.E.; Pease, L.F.; Bernard, P.S.; Skliar, M. Size and Shape Characterization of Hydrated and Desiccated Exosomes. Anal. Bioanal. Chem. 2015, 407, 3285–3301. [Google Scholar] [CrossRef]
- Conde-Vancells, J.; Rodriguez-Suarez, E.; Embade, N.; Gil, D.; Matthiesen, R.; Valle, M.; Elortza, F.; Lu, S.C.; Mato, J.M.; Falcon-Perez, J.M. Characterization and comprehensive proteome profiling of exosomes secreted by hepatocytes. J. Proteome Res. 2008, 7, 5157–5166. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Xu, H.; Xu, W.; Wang, B.; Wu, H.; Tao, Y.; Zhang, B.; Wang, M.; Mao, F.; Yan, Y.; et al. Exosomes released by human umbilical cord mesenchymal stem cells protect against cisplatin-induced renal oxidative stress and apoptosis in vivo and in vitro. Stem Cell Res. Ther. 2013, 4, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coleman, B.M.; Hanssen, E.; Lawson, V.A.; Hill, A.F. Prion-infected cells regulate the release of exosomes with distinct ultrastructural features. FASEB J. 2012, 26, 4160–4173. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Das, K.; Woo, J.; Gimzewski, J.K. Nanofilaments on glioblastoma exosomes revealed by peak force microscopy. J. R. Soc. Interface 2014, 11, 20131150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hardij, J.; Cecchet, F.; Berquand, A.; Gheldof, D.; Chatelain, C.; Mullier, F.; Chatelain, B.; Dogné, J.-M. Characterisation of tissue factor-bearing extracellular vesicles with AFM: Comparison of air-tapping-mode AFM and liquid Peak Force AFM. J. Extracell. Vesicles 2013, 2, 21045. [Google Scholar] [CrossRef] [PubMed]
- Deville, S.; Berckmans, P.; Van Hoof, R.; Lambrichts, I.; Salvati, A.; Nelissen, I. Comparison of extracellular vesicle isolation and storage methods using high-sensitivity flow cytometry. PLoS ONE 2021, 16, e0245835. [Google Scholar] [CrossRef]
- Brennan, K.; Martin, K.; FitzGerald, S.P.; O’Sullivan, J.; Wu, Y.; Blanco, A.; Richardson, C.; Mc Gee, M.M. A comparison of methods for the isolation and separation of extracellular vesicles from protein and lipid particles in human serum. Sci. Rep. 2020, 10, 1039. [Google Scholar] [CrossRef] [Green Version]
- Skliar, M.; Chernyshev, V.S.; Belnap, D.M.; Sergey, G.V.; Al-Hakami, S.M.; Bernard, P.S.; Stijleman, I.J.; Rachamadugu, R. Membrane proteins significantly restrict exosome mobility. Biochem. Biophys. Res. Commun. 2018, 501, 1055–1059. [Google Scholar] [CrossRef]
- Curie, J.; Curie, P. An oscillating quartz crystal mass detector. Rendu 1880, 91, 294–297. [Google Scholar]
- Alassi, A.; Benammar, M.; Brett, D. Quartz crystal microbalance electronic interfacing systems: A review. Sensors 2017, 17, 2799. [Google Scholar] [CrossRef] [Green Version]
- Ludwig, N.; Yerneni, S.S.; Menshikova, E.V.; Gillespie, D.G.; Jackson, E.K.; Whiteside, T.L. Simultaneous Inhibition of Glycolysis and Oxidative Phosphorylation Triggers a Multi-Fold Increase in Secretion of Exosomes: Possible Role of 2′3′-cAMP. Sci. Rep. 2020, 10, 6948. [Google Scholar] [CrossRef] [Green Version]
- Fukuta, T.; Nishikawa, A.; Kogure, K. Low level electricity increases the secretion of extracellular vesicles from cultured cells. Biochem. Biophys. Rep. 2020, 21, 100713. [Google Scholar] [CrossRef] [PubMed]
- Gardiner, C.; Ferreira, Y.J.; Dragovic, R.A.; Redman, C.W.G.; Sargent, I.L. Extracellular vesicle sizing and enumeration by nanoparticle tracking analysis. J. Extracell. Vesicles 2013, 2, 19671. [Google Scholar] [CrossRef] [PubMed]
- Bosch, S.; De Beaurepaire, L.; Allard, M.; Mosser, M.; Heichette, C.; Chrétien, D.; Jegou, D.; Bach, J.M. Trehalose prevents aggregation of exosomes and cryodamage. Sci. Rep. 2016, 6, 36162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chernyshev, V.S.; Chuprov-Netochin, R.N.; Tsydenzhapova, E.; Van Devener, B.; Leonov, S.; Gorin, D.; Skliar, M. Dynamic surface tension probe for measuring the concentration of extracellular vesicles. Biochem. Biophys. Res. Commun. 2022, 609, 189–194. [Google Scholar] [CrossRef]
- Voinova, M.V.; Jonson, M.; Kasemo, B. “Missing mass” effect in biosensor’s QCM applications. Biosens. Bioelectron. 2002, 17, 835–841. [Google Scholar] [CrossRef]
- Johannsmann, D.; Reviakine, I.; Richter, R.P. Dissipation in films of adsorbed nanospheres studied by quartz crystal microbalance (QCM). Anal. Chem. 2009, 81, 8167–8176. [Google Scholar] [CrossRef]
- Deegan, R.D.; Bakajin, O.; Dupont, T.F.; Huber, G.; Nagel, S.R.; Witten, T.A. Capillary flow as the cause of ring stains from dried liquid drops. Nature 1997, 389, 827–829. [Google Scholar] [CrossRef]
- Sliz, R.; Czajkowski, J.; Fabritius, T. Taming the Coffee Ring Effect: Enhanced Thermal Control as a Method for Thin-Film Nanopatterning. Langmuir 2020, 36, 9562–9570. [Google Scholar] [CrossRef]
- Yi, J.; Jeong, H.; Park, J. Modulation of nanoparticle separation by initial contact angle in coffee ring effect. Micro Nano Syst. Lett. 2018, 6, 1–7. [Google Scholar] [CrossRef]
- Parsa, M.; Harmand, S.; Sefiane, K. Mechanisms of pattern formation from dried sessile drops. Adv. Colloid Interface Sci. 2018, 254, 22–47. [Google Scholar] [CrossRef] [Green Version]

| MCF7 | MDA-MB-231 | MCF10A | 22Rv1 | LNCaP | PC3 | |
|---|---|---|---|---|---|---|
| Mode (nm) | 117 | 123 | 115 | 106 | 135 | 113 |
| Mean (nm) | 183 | 147 | 205 | 177 | 201 | 220 |
| StDev (nm) | 84 | 74 | 116 | 63 | 141 | 119 |
| MCF7 | MDA-MB-231 | LNCaP | PC3 | |
|---|---|---|---|---|
| QCM (#) × 106 | 4.03 ± 0.70 | 1.14 ± 0.39 | 19.40 ± 1.05 | 16.72 ± 1.24 |
| NTA (#) × 106 | 4.16 ± 0.83 | 1.05 ± 0.27 | 21.21 ± 2.04 | 15.60 ± 0.73 |
| MCF7 | MDA-MB-231 | MCF10A | 22Rv1 | LNCaP | PC3 | |
|---|---|---|---|---|---|---|
| Ring area (mm2) | 0.25 ± 0.02 | 0.15 ± 0.03 | 0.26 ± 0.04 | 0.24 ± 0.01 | 0.29 ± 0.05 | 0.18 ± 0.02 |
| Mass (fg) | 0.16 ± 0.01 | 0.13 ± 0.03 | 0.18 ± 0.04 | 0.16 ± 0.01 | 0.17 ± 0.04 | 0.13 ± 0.01 |
| Diameter (nm) | 52 ± 1 | 49 ± 9 | 54 ± 2 | 51 ± 1 | 53 ± 3 | 48 ± 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chernyshev, V.S.; Skliar, M. Quantification of Desiccated Extracellular Vesicles by Quartz Crystal Microbalance. Biosensors 2022, 12, 371. https://doi.org/10.3390/bios12060371
Chernyshev VS, Skliar M. Quantification of Desiccated Extracellular Vesicles by Quartz Crystal Microbalance. Biosensors. 2022; 12(6):371. https://doi.org/10.3390/bios12060371
Chicago/Turabian StyleChernyshev, Vasiliy S., and Mikhail Skliar. 2022. "Quantification of Desiccated Extracellular Vesicles by Quartz Crystal Microbalance" Biosensors 12, no. 6: 371. https://doi.org/10.3390/bios12060371
APA StyleChernyshev, V. S., & Skliar, M. (2022). Quantification of Desiccated Extracellular Vesicles by Quartz Crystal Microbalance. Biosensors, 12(6), 371. https://doi.org/10.3390/bios12060371





