An Optimized Direct Lysis Gene Expression Microplate Assay and Applications for Disease, Differentiation, and Pharmacological Cell-Based Studies
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Differentiations
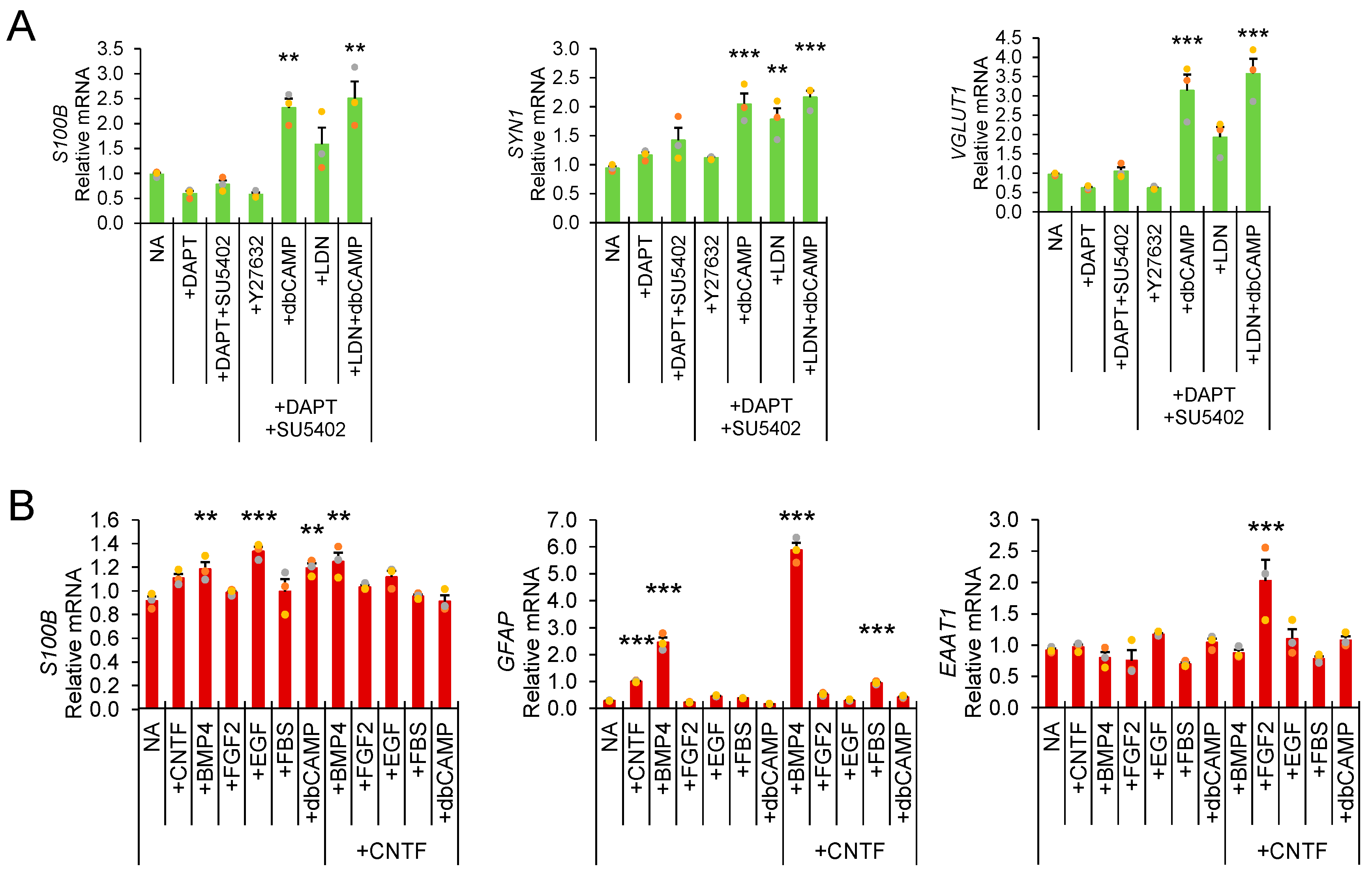
2.3. Inflammatory, ER Stress, and Neural Differentiation Assays
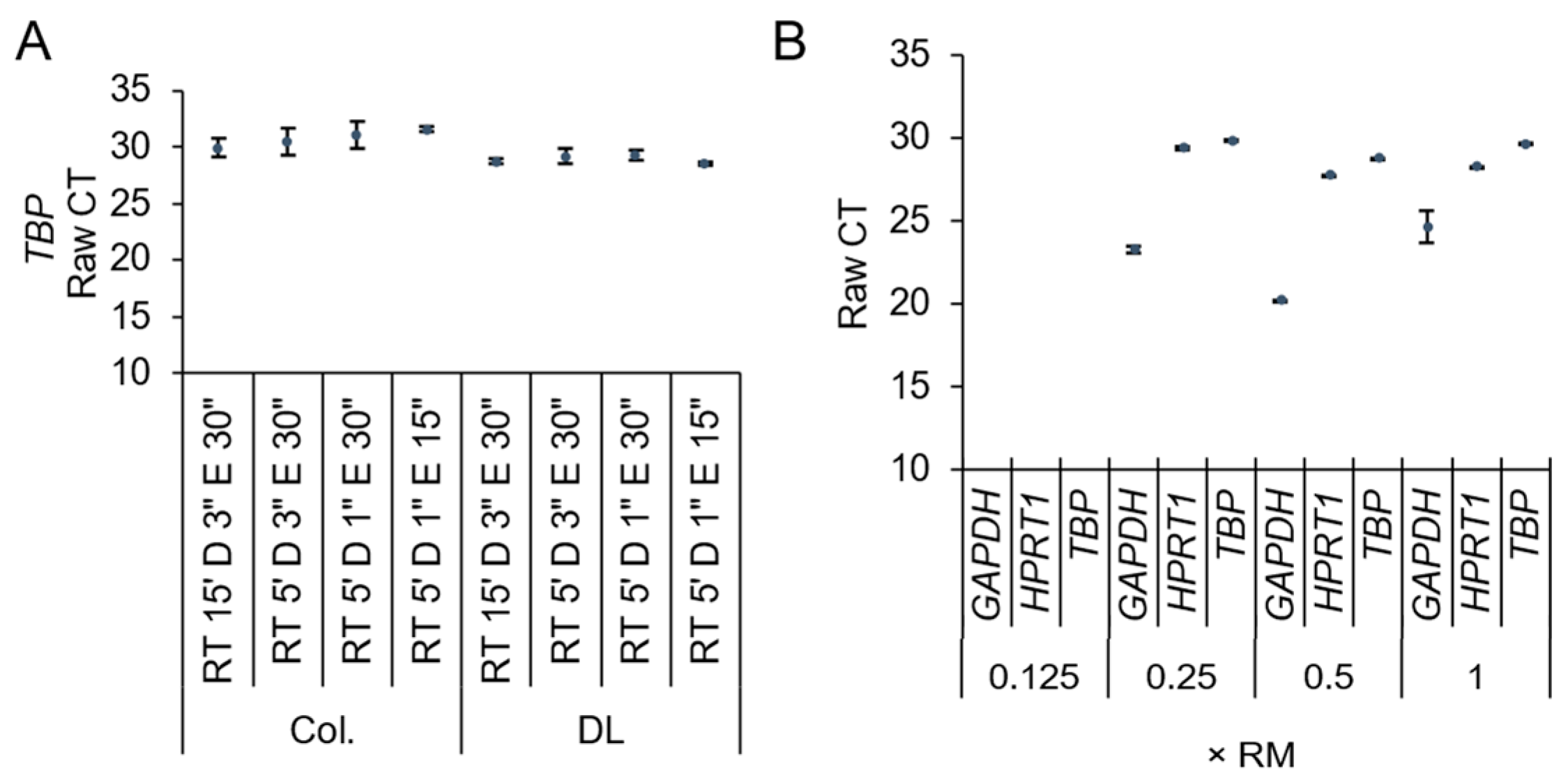
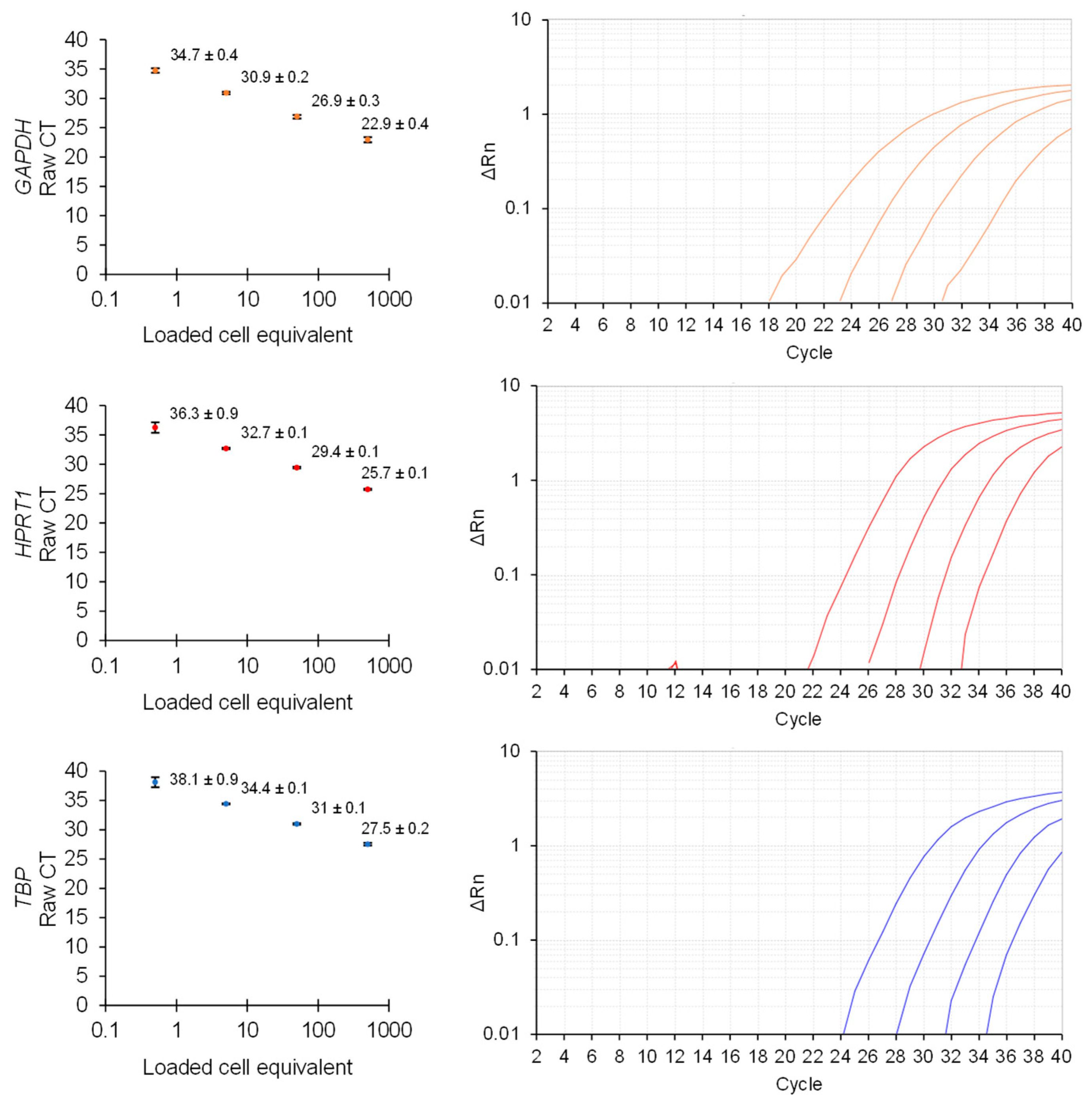
2.4. Direct Lysis RT-qPCR
2.5. RT-qPCR
2.6. Statistical Analysis
3. Results and Discussion
Direct Lysis RT-qPCR Applications
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rio, D.C.; Ares, M.; Hannon, G.J.; Nilsen, T.W. Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb. Protoc. 2010, 2010, 5439. [Google Scholar] [CrossRef] [PubMed]
- Rump, L.V.; Asamoah, B.; Gonzalez-Escalona, N. Comparison of commercial RNA extraction kits for preparation of DNA-free total RNA from Salmonella cells. BMC Res. Notes 2010, 3, 211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le, A.V.-P.; Huang, D.; Blick, T.; Thompson, E.W.; Dobrovic, A. An optimised direct lysis method for gene expression studies on low cell numbers. Sci. Rep. 2015, 5, 12859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Svec, D.; Andersson, D.; Pekny, M.; Sjöback, R.; Kubista, M.; Ståhlberg, A. Direct cell lysis for single-cell gene expression profiling. Front. Oncol. 2013, 3, 274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shatzkes, K.; Teferedegne, B.; Murata, H. A simple, inexpensive method for preparing cell lysates suitable for downstream reverse transcription quantitative PCR. Sci. Rep. 2014, 4, 4659. [Google Scholar] [CrossRef] [PubMed]
- Ng, N.; Cabral-da-Silva, M.C.; Maksour, S.; Berg, T.; Engel, M.; Silva, D.M.; Do-Ha, D.; Lum, J.S.; Muñoz, S.S.; Suarez-Bosche, N.; et al. Identification of repurposable cytoprotective drugs in vanishing white matter disease patient-derived cells. Transl. Med. Commun. 2020, 5, 18. [Google Scholar] [CrossRef]
- Hulme, A.J.; McArthur, J.R.; Maksour, S.; Miellet, S.; Ooi, L.; Adams, D.J.; Finol-Urdaneta, R.K.; Dottori, M. Molecular and functional characterization of Neurogenin-2 induced human sensory neurons. Front. Cell. Neurosci. 2020, 14, 425. [Google Scholar] [CrossRef]
- Zhang, Y.; Pak, C.; Han, Y.; Ahlenius, H.; Zhang, Z.; Chanda, S.; Marro, S.; Patzke, C.; Acuna, C.; Covy, J.; et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 2013, 78, 785–798. [Google Scholar] [CrossRef] [Green Version]
- Newbery, M.; Maksour, S.; Hulme, A.; Ng, N.S.; Dottori, M.; Ooi, L. Efficient Third Generation Lentiviral Particle Production. Protocols-io. 2021. Available online: https://doi.org/10.17504/protocols.io.bugxntxn (accessed on 11 January 2022).
- Hulme, A.J.; Maksour, S.; Glover, M.S.-C.; Miellet, S.; Dottori, M. Making neurons, made easy: The use of Neurogenin-2 in neuronal differentiation. Stem Cell Rep. 2021, 17, 14–34. [Google Scholar] [CrossRef]
- Canals, I.; Ginisty, A.; Quist, E.; Timmerman, R.; Fritze, J.; Miskinyte, G.; Monni, E.; Hansen, M.G.; Hidalgo, I.; Bryder, D.; et al. Rapid and efficient induction of functional astrocytes from human pluripotent stem cells. Nat. Meth. 2018, 15, 693–696. [Google Scholar] [CrossRef]
- Careaga, M.; Taylor, S.L.; Chang, C.; Chiang, A.; Ku, K.M.; Berman, R.F.; van de Water, J.A.; Bauman, M.D. Variability in PolyIC induced immune response: Implications for preclinical maternal immune activation models. J. Neuroimmunol. 2018, 323, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Wittwer, C.T.; Garling, D.J.J.B. Rapid cycle DNA amplification: Time and temperature optimization. Biotechniques 1991, 10, 76–83. [Google Scholar]
- Bustin, S.A. How to speed up the polymerase chain reaction. Biomol. Detect. Quantif. 2017, 12, 10–14. [Google Scholar] [CrossRef] [PubMed]
- Piovesan, A.; Caracausi, M.; Antonaros, F.; Pelleri, M.C.; Vitale, L. GeneBase 1.1: A tool to summarize data from NCBI gene datasets and its application to an update of human gene statistics. Database 2016, 2016, baw153. [Google Scholar] [CrossRef]
- Ng, N.S.; Gately, R.; Ooi, L. Automated liquid handling for microplate assays: A simplified user interface for the Hamilton Microlab STAR. J. Appl. Bioanal. 2021, 7, 11–18. [Google Scholar] [CrossRef]
- Perach, M.; Hizi, A. Catalytic features of the recombinant reverse transcriptase of bovine leukemia virus expressed in bacteria. Virology 1999, 259, 176–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coffin, J.M.; Hughes, H.S.; Varmus, H.E. Biochemistry of Reverse Transcription. 1997. Available online: https://www.ncbi.nlm.nih.gov/books/NBK19435/ (accessed on 1 January 2021).
- Wan, Q.-H.; Fang, S.-G. Application of species-specific polymerase chain reaction in the forensic identification of tiger species. Forensic Sci. Int. 2003, 131, 75–78. [Google Scholar] [CrossRef]
- Crawford, T.Q.; Roelink, H. The Notch response inhibitor DAPT enhances neuronal differentiation in embryonic stem cell-derived embryoid bodies independently of sonic hedgehog signaling. Dev. Dyn. 2007, 236, 886–892. [Google Scholar] [CrossRef]
- Serio, A.; Bilican, B.; Barmada, S.J.; Ando, D.M.; Zhao, C.; Siller, R.; Karen, B.; Ghazal, H.; David, S.; Lumi, N.A.; et al. Astrocyte pathology and the absence of non-cell autonomy in an induced pluripotent stem cell model of TDP-43 proteinopathy. Proc. Natl. Acad. Sci. USA 2013, 110, 4697–4702. [Google Scholar] [CrossRef] [Green Version]
- Savchenko, E.; Teku, G.N.; Boza-Serrano, A.; Russ, K.; Berns, M.; Deierborg, T.; Lamas, N.J.; Wichterle, H.; Rothstein, J.; Henderson, C.E.; et al. FGF family members differentially regulate maturation and proliferation of stem cell-derived astrocytes. Sci. Rep. 2019, 9, 9610. [Google Scholar] [CrossRef] [Green Version]
- West, D.M.; Sawyer, J. Freezing complete polymerase chain reaction master mix reagents for routine molecular diagnostics. J. Vet. Diagn. Investig. 2006, 18, 580–582. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ng, N.S.; Maksour, S.; Lum, J.S.; Newbery, M.; Shephard, V.; Ooi, L. An Optimized Direct Lysis Gene Expression Microplate Assay and Applications for Disease, Differentiation, and Pharmacological Cell-Based Studies. Biosensors 2022, 12, 364. https://doi.org/10.3390/bios12060364
Ng NS, Maksour S, Lum JS, Newbery M, Shephard V, Ooi L. An Optimized Direct Lysis Gene Expression Microplate Assay and Applications for Disease, Differentiation, and Pharmacological Cell-Based Studies. Biosensors. 2022; 12(6):364. https://doi.org/10.3390/bios12060364
Chicago/Turabian StyleNg, Neville S., Simon Maksour, Jeremy S. Lum, Michelle Newbery, Victoria Shephard, and Lezanne Ooi. 2022. "An Optimized Direct Lysis Gene Expression Microplate Assay and Applications for Disease, Differentiation, and Pharmacological Cell-Based Studies" Biosensors 12, no. 6: 364. https://doi.org/10.3390/bios12060364
APA StyleNg, N. S., Maksour, S., Lum, J. S., Newbery, M., Shephard, V., & Ooi, L. (2022). An Optimized Direct Lysis Gene Expression Microplate Assay and Applications for Disease, Differentiation, and Pharmacological Cell-Based Studies. Biosensors, 12(6), 364. https://doi.org/10.3390/bios12060364




