Procedural Data Processing for Single-Molecule Identification by Nanopore Sensors
Abstract
:1. Introduction
2. Materials and Methods
3. Results and Discussion
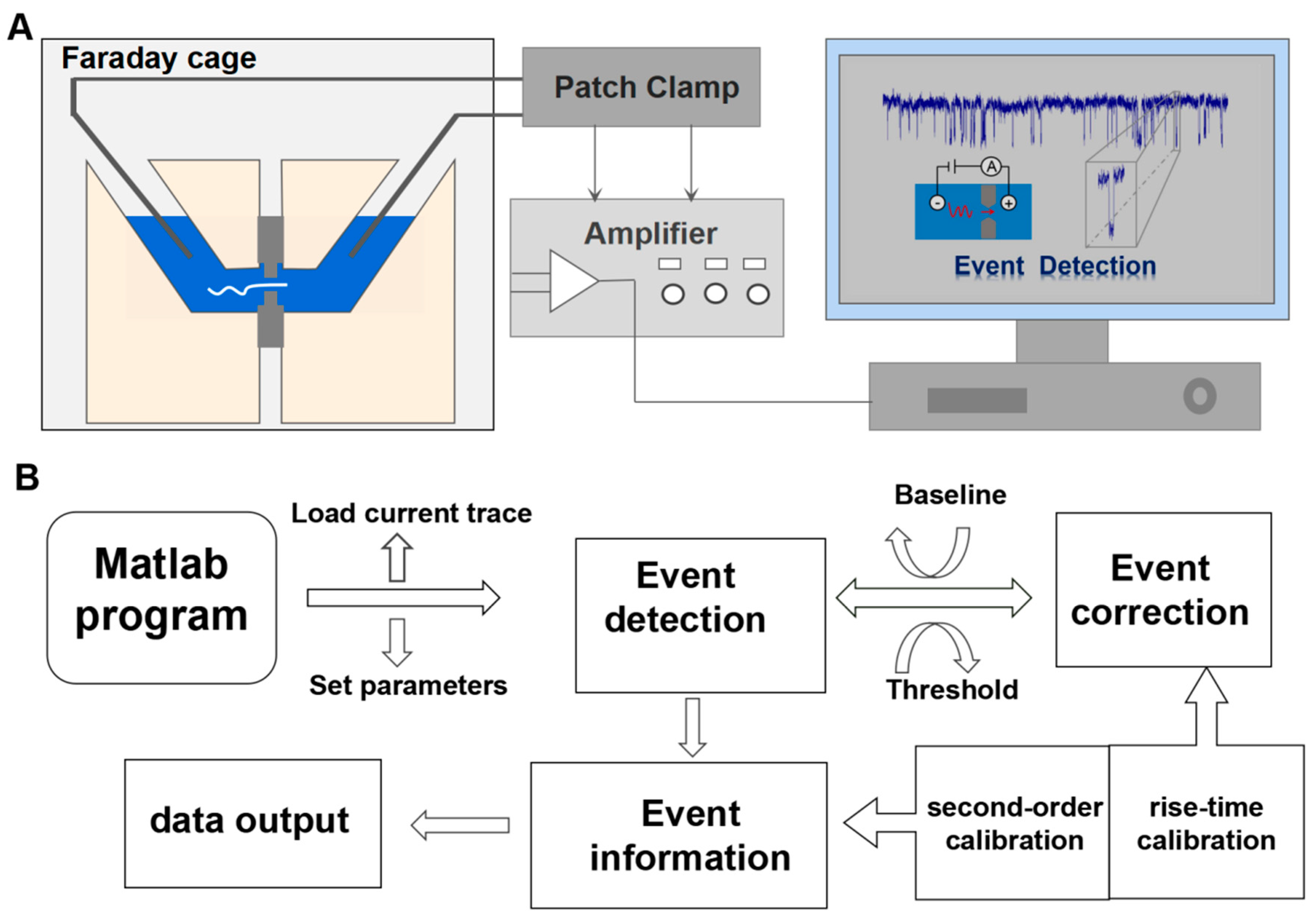
3.1. Nanopore Signal Processing Program
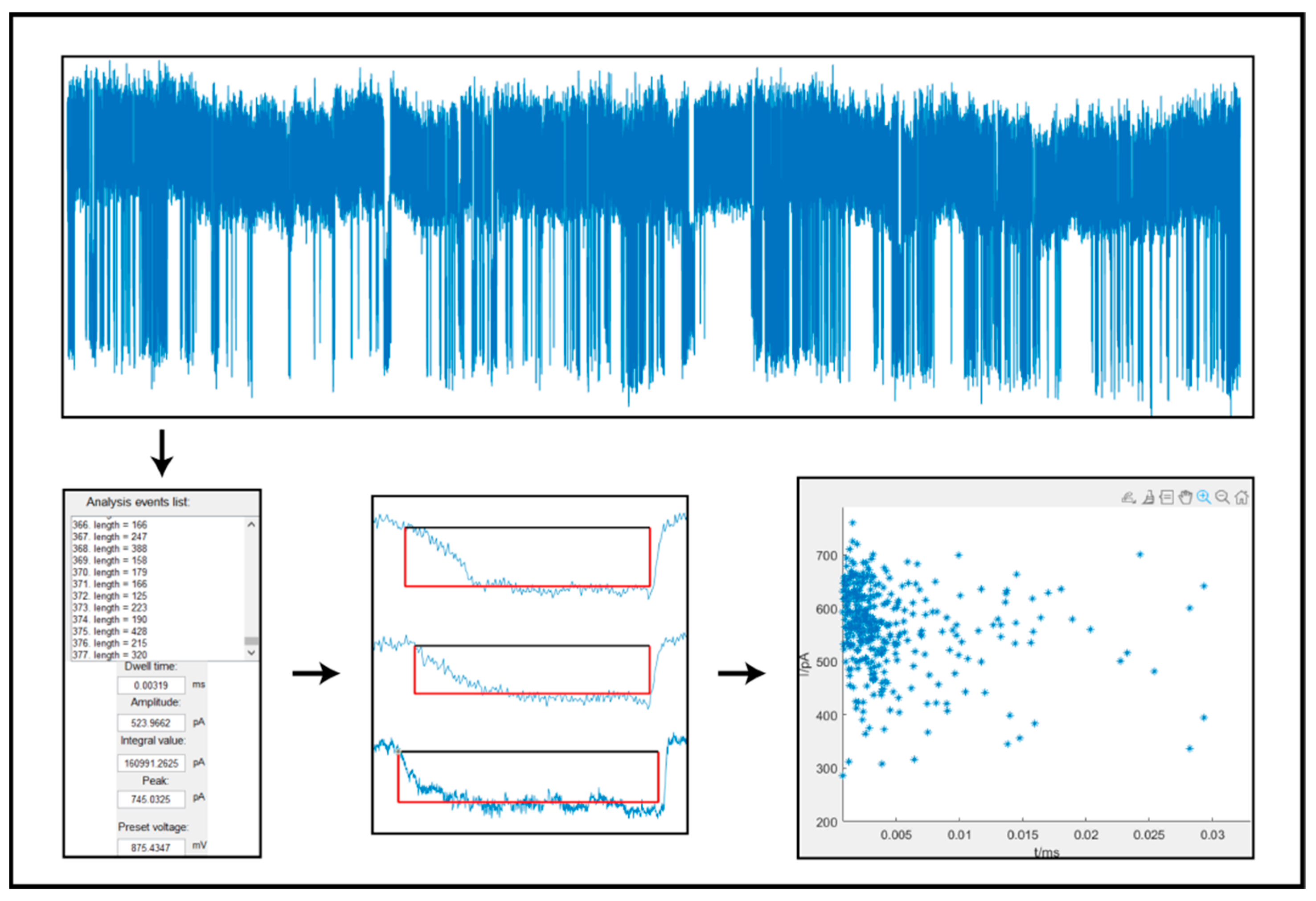
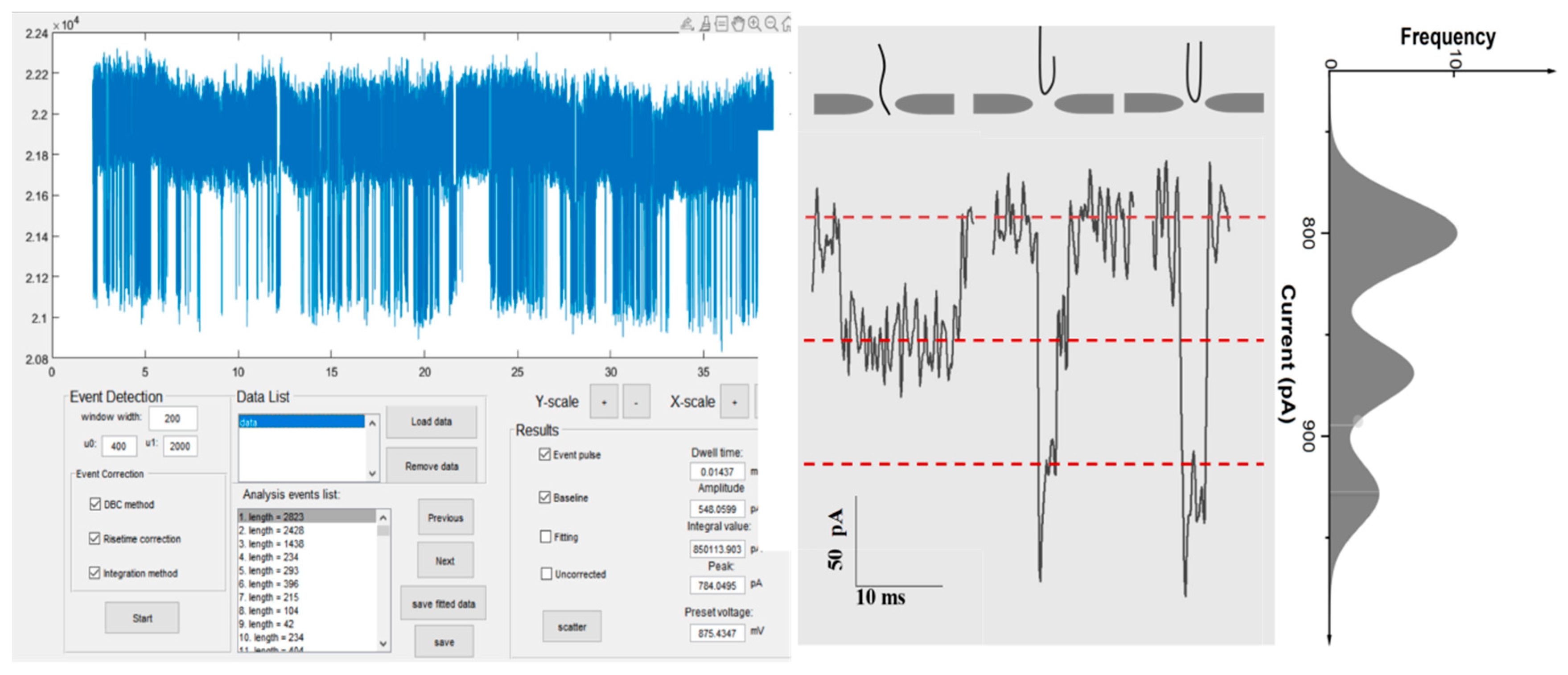
3.2. Determination of Molecular Events
3.3. Correction for Molecular Events
3.4. Electrical Signal Feature Information Extraction
3.5. Applications to DNA Detection
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wan, Y.K.; Hendra, C.; Pratanwanich, P.N.; Göke, J. Beyond Sequencing: Machine Learning Algorithms Extract Biology Hidden in Nanopore Signal Data. Trends Genet. 2022, 38, 246–257. [Google Scholar] [CrossRef] [PubMed]
- Albrecht, T. How to Understand and Interpret Current Flow in Nanopore/Electrode Devices. ACS Nano 2011, 5, 6714–6725. [Google Scholar] [CrossRef] [PubMed]
- Wanunu, M.; Bhattacharya, S.; Xie, Y.; Tor, Y.; Aksimentiev, A.; Drndic, M. Nanopore analysis of individual RNA/antibiotic complexes. ACS Nano 2011, 5, 9345–9353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brinkerhoff, H.; Kang, A.S.W.; Liu, J.; Aksimentiev, A.; Dekker, C. Multiple rereads of single proteins at single-amino acid resolution using nanopores. Science 2021, 374, 1509–1513. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, Y.; Bollas, A.; Wang, Y.; Au, K.F. Nanopore Sequencing Technology, Bioinformatics and Applications. Nat. Biotechnol. 2021, 39, 1348–1365. [Google Scholar] [CrossRef]
- Lastra, L.S.; Bandara, Y.; Nguyen, M.; Farajpour, N.; Freedman, K.J. On the origins of conductive pulse sensing inside a nanopore. Nat. Commun. 2022, 13, 2186. [Google Scholar] [CrossRef]
- Ghimire, M.L.; Gibbs, D.R.; Mahmoud, R.; Dhakal, S.; Reiner, J.E. Nanopore Analysis as a Tool for Studying Rapid Holliday Junction Dynamics and Analyte Binding. Anal. Chem. 2022, 94, 10027–10034. [Google Scholar] [CrossRef]
- Robertson, J.W.F.; Reiner, J.E. Highlights on the current state of proteomic detection and characterization with nanopore sensors. Proteomics 2022, 22, e2100061. [Google Scholar] [CrossRef]
- Wu, J.; Liang, L.; Zhang, M.; Zhu, R.; Wang, Z.; Yin, Y.; Yin, B.; Weng, T.; Fang, S.; Xie, W.; et al. Single-Molecule Identification of the Conformations of Human C-Reactive Protein and Its Aptamer Complex with Solid-State Nanopores. ACS Appl. Mater. Interfaces 2022, 14, 12077–12088. [Google Scholar] [CrossRef]
- Tu, J.; Meng, H.; Wu, L.; Xi, G.; Fu, J.; Lu, Z. EasyNanopore: A ready-to-use pocessing software for translocation events in nanopore translocation experiments. Langmuir 2021, 37, 10177–10182. [Google Scholar] [CrossRef]
- Raillon, C.; Granjon, P.; Graf, M.; Steinbock, L.J.; Radenovic, A. Fast and Automatic Processing of Multi-Level Events in Nanopore Translocation Experiments. Nanoscale 2012, 4, 4916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forstater, J.H.; Briggs, K.; Robertson, J.W.F.; Ettedgui, J.; Marie-Rose, O.; Vaz, C.; Kasianowicz, J.J.; Tabard-Cossa, V.; Balijepalli, A. MOSAIC: A Modular Single-Molecule Analysis Interface for Decoding Multistate Nanopore Data. Anal. Chem. 2016, 88, 11900–11907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Z.; Liu, X.; Liu, W.; Li, J.; Yang, J.; Qiao, F.; Ma, J.; Sha, J.; Li, J.; Xu, L.-Q. AutoNanopore: An Automated Adaptive and Robust Method to Locate Translocation Events in Solid-State Nanopore Current Traces. ACS Omega 2022, 7, 37103–37111. [Google Scholar] [CrossRef] [PubMed]
- Wen, C.; Dematties, D.; Zhang, S.L. A Guide to Signal Processing Algorithms for Nanopore Sensors. ACS Sens. 2021, 6, 3536–3555. [Google Scholar] [CrossRef] [PubMed]
- Fragasso, A.; Schmid, S.; Dekker, C. Comparing Current Noise in Biological and Solid-State Nanopores. ACS Nano 2020, 14, 1338–1349. [Google Scholar] [CrossRef] [Green Version]
- Balan, A.; Machielse, B.; Niedzwiecki, D.; Lin, J.; Ong, P.; Engelke, R.; Shepard, K.L.; Drndic, M. Improving signal-to-noise performance for DNA translocation in solid-state nanopores at MHz bandwidths. Nano Lett. 2014, 14, 7215–7220. [Google Scholar] [CrossRef]
- Smeets, R.M.; Keyser, U.F.; Dekker, N.H.; Dekker, C. Noise in solid-state nanopores. Proc. Natl. Acad. Sci. USA 2008, 105, 417–7221. [Google Scholar] [CrossRef] [Green Version]
- O’Donnell, C.R.; Wiberg, D.M.; Dunbar, W.B. A Kalman Filter for Estimating Nanopore Channel Conductance in Voltage-Varying Experiments. In Proceedings of the IEEE 51st Conference on Decision and Control (IEEE CDC), Maui, HI, USA, 10−13 December 2012; pp. 2304–2309. [Google Scholar]
- Pedone, D.; Firnkes, M.; Rant, U. Data Analysis of Translocation Events in Nanopore Experiments. Anal. Chem. 2009, 81, 9689–9694. [Google Scholar] [CrossRef]
- Shekar, S.; Chien, C.-C.; Hartel, A.; Ong, P.; Clarke, O.B.; Marks, A.; Drndic, M.; Shepard, K.L. Wavelet Denoising of High-Bandwidth Nanopore and Ion-Channel Signals. Nano Lett. 2019, 19, 1090–1097. [Google Scholar] [CrossRef]
- Plesa, C.; Dekker, C. Data analysis methods for solid-state nanopores. Nanotechnology 2015, 26, 084003. [Google Scholar] [CrossRef]
- Gu, Z.; Ying, Y.L.; Cao, C.; He, P.; Long, Y.T. Accurate data process for nanopore analysis. Anal. Chem. 2015, 87, 907–913. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, X.; Ying, Y.L.; Gu, Z.; Meng, F.N.; Long, Y.T. High-bandwidth nanopore data analysis by using a modified hidden Markov model. Nanoscale 2017, 9, 3458–3465. [Google Scholar] [CrossRef]
- Wei, Z.X.; Ying, Y.L.; Li, M.Y.; Yang, J.; Zhou, J.L.; Wang, H.F.; Yan, B.Y.; Long, Y.T. Learning Shapelets for Improving Single-Molecule Nanopore Sensing. Anal. Chem. 2019, 91, 10033–10039. [Google Scholar] [CrossRef] [PubMed]
- Saharia, J.; Bandara, Y.; Goyal, G.; Lee, J.S.; Karawdeniya, B.I.; Kim, M.J. Molecular-Level Profiling of Human Serum Transferrin Protein through Assessment of Nanopore-Based Electrical and Chemical Responsiveness. ACS Nano 2019, 13, 4246–4254. [Google Scholar] [CrossRef] [PubMed]
- Bandara, Y.M.; Nuwan, D.Y.; Saharia, J.; Karawdeniya, B.I.; Kluth, P.; Kim, M.J. Nanopore data analysis: Baseline construction and abrupt change-based multi-level fitting. Anal. Chem. 2021, 93, 11710–11718. [Google Scholar] [CrossRef]
- Dematties, D.; Wen, C.; Pérez, M.D.; Zhou, D.; Zhang, S.L. Deep Learning of Nanopore Sensing Signals Using a Bi-Path Network. ACS Nano 2021, 15, 14419–14429. [Google Scholar] [CrossRef] [PubMed]
- Diaz Carral, A.; Ostertag, M.; Fyta, M. Deep learning for nanopore ionic current blockades. J. Chem. Phys. 2021, 154, 044111. [Google Scholar] [CrossRef]
- Tsutsui, M.; Takaai, T.; Yokota, K.; Kawai, T.; Washio, T. Deep Learning-Enhanced Nanopore Sensing of Single-Nanoparticle Translocation Dynamics. Small Methods 2021, 5, e2100191. [Google Scholar] [CrossRef]
- Arima, A.; Harlisa, I.H.; Yoshida, T.; Tsutsui, M.; Tanaka, M.; Yokota, K.; Tonomura, W.; Yasuda, J.; Taniguchi, M.; Washio, T.; et al. Identifying Single Viruses Using Biorecognition Solid-State Nanopores. J. Am. Chem. Soc. 2018, 140, 16834–16841. [Google Scholar] [CrossRef]
- Balijepalli, A.; Ettedgui, J.; Cornio, A.T.; Robertson, J.W.; Cheung, K.P.; Kasianowicz, J.J.; Vaz, C. Quantifying short-lived events in multistate ionic current measurements. ACS Nano 2014, 8, 1547–1553. [Google Scholar] [CrossRef]
- Fologea, D.; Brandin, E.; Uplinger, J.; Branton, D.; Li, J. DNA conformation and base number simultaneously determined in a nanopore. Electrophoresis 2007, 28, 3186–3192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fologea, D.; Gershow, M.; Ledden, B.; McNabb, D.S.; Golovchenko, J.A.; Li, J. Detecting single stranded DNA with a solid state nanopore. Nano Lett. 2005, 5, 1905–1909. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Yuan, J.; Deng, H.; Zhang, Z.; Ma, Q.D.Y.; Wu, L.; Weng, L. Procedural Data Processing for Single-Molecule Identification by Nanopore Sensors. Biosensors 2022, 12, 1152. https://doi.org/10.3390/bios12121152
Wang Y, Yuan J, Deng H, Zhang Z, Ma QDY, Wu L, Weng L. Procedural Data Processing for Single-Molecule Identification by Nanopore Sensors. Biosensors. 2022; 12(12):1152. https://doi.org/10.3390/bios12121152
Chicago/Turabian StyleWang, Yupeng, Jianxuan Yuan, Haofeng Deng, Ziang Zhang, Qianli D. Y. Ma, Lingzhi Wu, and Lixing Weng. 2022. "Procedural Data Processing for Single-Molecule Identification by Nanopore Sensors" Biosensors 12, no. 12: 1152. https://doi.org/10.3390/bios12121152
APA StyleWang, Y., Yuan, J., Deng, H., Zhang, Z., Ma, Q. D. Y., Wu, L., & Weng, L. (2022). Procedural Data Processing for Single-Molecule Identification by Nanopore Sensors. Biosensors, 12(12), 1152. https://doi.org/10.3390/bios12121152




