Abstract
Large-scale food-borne outbreaks caused by Salmonella are rarely seen nowadays, thanks to the advanced nature of the medical system. However, small, localised outbreaks in certain regions still exist and could possess a huge threat to the public health if eradication measure is not initiated. This review discusses the progress of Salmonella detection approaches covering their basic principles, characteristics, applications, and performances. Conventional Salmonella detection is usually performed using a culture-based method, which is time-consuming, labour intensive, and unsuitable for on-site testing and high-throughput analysis. To date, there are many detection methods with a unique detection system available for Salmonella detection utilising immunological-based techniques, molecular-based techniques, mass spectrometry, spectroscopy, optical phenotyping, and biosensor methods. The electrochemical biosensor has growing interest in Salmonella detection mainly due to its excellent sensitivity, rapidity, and portability. The use of a highly specific bioreceptor, such as aptamers, and the application of nanomaterials are contributing factors to these excellent characteristics. Furthermore, insight on the types of biorecognition elements, the principles of electrochemical transduction elements, and the miniaturisation potential of electrochemical biosensors are discussed.
1. Introduction
Salmonella infections cause various morbidity and mortality globally, especially in developing countries [1]. The prevalence is high in low-resource regions, where poor sanitation and lack of clean water supply are an issue [2]. Enteric fever (typhoid and paratyphoid fever) is the febrile illness caused by Salmonella enterica serotype Typhi and Paratyphi infection, characterised by a long persistent fever and other gastrointestinal complications. Late treatment or misdiagnosis could increase the risk of death and can lead to other problems such as antibiotic resistance [3].
To date, there are various types of Salmonella detection techniques available with unique mechanisms of detection. They are immunological-based assays, such as enzyme-linked immunosorbent assay (ELISA), latex agglutination method, and immunochromatography assay [4,5,6]; molecular-based assays, e.g., polymerase chain reaction (PCR), loop-mediated isothermal amplification (LAMP), nucleic acid sequence-based amplification (NASBA), recombinase polymerase amplification (RPA), DNA microarrays, whole genome-sequencing (WGS) [7,8,9,10,11,12]; mass spectrometry-based methods, e.g., matrix-assisted laser desorption ionisation-time-of-flight mass spectrometry (MALDI-TOF MS), liquid chromatography-mass spectrometry (LC-MS) [13,14]; spectroscopy-based methods, e.g., Raman spectroscopy, near-infrared (NIR) spectroscopy, and hyperspectral imaging (HSI) [15,16,17], optical phenotyping with light diffraction technology [18], and electrochemical biosensors [19]. These methods can identify and discriminate Salmonella down to their serotype level [14,20,21,22,23]. Apart from these detection methods, electrochemical biosensors could potentially be an ideal tool for Salmonella detection, since they are capable of detecting the presence of Salmonella or their cellular components in just a few hours.
Immunological-based assays and molecular-based assays provide excellent detection of Salmonella with high affinity. However, the low stability issue of antibody and high operation costs of PCR were deemed to be the drawback of these methods [23]. These two methods are also prone to cross-reactivity issues among Salmonella serovar [24,25,26]. DNA microarray and whole-genome sequencing methods were able to provide detailed information on Salmonella genotypes [14,27]. Mass spectrometry methods, spectroscopy methods, optical phenotyping are culture-independent methods as these methods utilised the phenotypic characteristics of Salmonella, such as peptides fingerprints, absorbance, and imaging characteristics [23,28].
Electrochemical biosensors were developed to detect a variety of analytes of interest such as glucose, cancer biomarkers, viruses, pathogenic bacteria, and heavy metals, among others. In the case of pathogenic bacteria, current studies have reported extensively on the use of electrochemical biosensors in detecting foodborne pathogens, especially Salmonella. Electrochemical biosensors are known for their rapid, highly sensitive, specific detection response, and can be integrated and miniaturised into a biosensor device, such as in a microfluidic platform, which can provide an advantage in point of care (POC) testing [22,29]. Additionally, the introduction of aptamer technology as a recognition element in electrochemical biosensors has undoubtedly enhanced the specificity, selectivity, and stability of biosensors [30]. Moreover, the integration of nanomaterials, such as graphene derivates, carbon nanotubes, metallic nanoparticles, metal oxide nanoparticles, and silica nanoparticles, as the surface modifier, has tremendously improved the limit of detection (LOD), as compared to the electrochemical biosensors with only antibody and molecular probes [31]. Thus, the selection of an exceptional biorecognition element combined with a favourable surface modifier in electrochemical biosensor development could contribute to rapid and highly sensitive POC diagnostic devices, particularly for Salmonella detection.
2. Salmonella and Their Related Diseases
The Salmonella bacteria was discovered in 1855 by Theobald Smith from pig intestines that been infected with classical swine fever [32]. Upon discovery, Salmonella underwent controversial naming as it was named after Dr. Daniel Elmer Salmon, one of Smith’s co-workers. Later, the Centre for Disease Control and Prevention (CDC) suggested a resolution to the nomenclature issue of Salmonella by following the recommendation by the World Health Organisation (WHO) collaborating centre [33]. This system suggested the classification of Salmonella into two species namely Salmonella enterica and Salmonella bongori, which were classified based on their 16S rRNA sequence analysis. Salmonella enterica can be further grouped into six subspecies, where these subspecies are denoted with a roman numeral symbol from I to VI [34]. Salmonella enterica I can be further classified into typhoidal Salmonella (S. Typhi and S. Paratyphi), which only infect humans, and non-typhoidal Salmonella (S. Typhimurium and S. Enteritidis), which infect both humans and animals [35].
Salmonella is a flagellated Gram-negative bacteria, facultative anaerobe characterised by O, H, and Vi antigens [36]. It is famously known as a foodborne pathogen, as most infections are acquired from food sources. They cause salmonellosis, clinically ranging from common gastroenteritis (e.g., diarrhoea, abdominal cramps, and fever) to enteric fevers (typhoid and paratyphoid), which could be a life-threatening febrile illness if left untreated [2,36,37,38]. However, the degree of severity depends on the serotype involved and human host status. Infants or children under five years old, elderly, and immunocompromised persons are among the high-risk groups who are susceptible to Salmonella infection [32].
Salmonellosis can be acquired from ingestion of food contaminated with Salmonella. The sources of contamination could be from the consumption of undercooked food from infected animals in poultry products or other meats, ready-to-eat foods such as fruits and vegetables that have been contaminated with faeces of infected animals, or water that has been contaminated with faeces of infected people or animals [38]. In urban areas, contaminated water sources contribute significantly to Salmonella outbreaks [3]. Contact infection can also be acquired through direct exposure to an object or environment contaminated with faeces containing Salmonella [38].
Enteric fever is a potentially fatal systemic illness caused by Salmonella enterica subspecies serovars Typhi and Paratyphi A, B and C [3,39]. This illness, which is also known as typhoid or paratyphoid fever, was classified based on the serotype of Salmonella bacterium infecting the host. Fatality in cases involving typhoid fever was reported to be 1.89 times higher than in paratyphoid fever. These infections resulted in bacteraemic febrile illness, with prolonged high fever, malaise, and headache [39]. If left untreated, it would lead to gastrointestinal bleeding, intestinal perforation, ileus, septic shock, altered mental states (termed the typhoid state) and, consequently, death [3,39,40,41].
In 2017, it was estimated that 14.3 million typhoid and paratyphoid cases occurred globally with 136,000 estimated deaths. Children and the elderly were among the groups of highest recorded fatality, especially in lower-income countries. Salmonella enterica serotype Typhi is a major contributor to enteric fever with 76.3% [39]. Typhoid disease is rare in developed countries, but still prevalent in certain areas of developing countries with limited access to clean water and poor sanitation [42,43]. In Malaysia, it was estimated that 0.5 to 0.7 confirmed cases of Salmonella infection per 100,000 population were reported in 2015 [44]. Even though most of the outbreaks are endemic, the cases of both typhoid and paratyphoid fever declined globally by 45% from the year 1990 to 2017. Since there was limited use of typhoid vaccine in typhoid-endemic countries, other factors such as the improvement of water supply and sanitation infrastructure, improved food handling practices, and easy access to antibiotic treatment were likely to be the crucial factors that contributed to this decline [39].
The incidence of antimicrobial resistance among bacterial pathogens is currently at an alarming state which includes concern towards resistance in Salmonella. This issue was relatively not new to Salmonella species as the first chloramphenicol-resistant S. Typhi was discovered two years after it was used to treat patients with enteric fever in 1948 [3]. In the 1980s, multidrug-resistant S. Typhi against chloramphenicol, ampicillin, trimethoprim, and sulfonamides has become prevalent, followed by nalidixic-acid-resistant S. Typhi and S. Paratyphi A in the 1990s [45]. In Asia, the majority of S. Typhi and Paratyphi A strains are nalidixic-acid resistant and completely resistant towards ciprofloxacin and extended-spectrum cephalosporins have emerged in some parts of the world [46]. It was reported that the antimicrobial resistance of Salmonella was linked with the horizontal gene transfer from the virulent multi-drug resistant S. Typhimurium DT 104 through its mobile plasmids to other bacteria [47,48,49]. Thus, early detection of Salmonella could help to prevent the spread of resistance genes and the optimal use of antimicrobial drugs are deemed necessary to combat this issue.
3. Detection of Salmonella
Early and accurate detection of Salmonella infection could be a lifesaver, as necessary and effective treatment can be provided to patients, thereby reducing the possibility of selective pressure that can contribute to the emergence of antimicrobial drug resistance [50]. As the technology progresses, there are many new inventions and innovative approaches for Salmonella detection that provide fast, accurate, and reliable detection. Figure 1 illustrates the currently available methods for Salmonella detection.

Figure 1.
Available methods for Salmonella detection (adapted from ref. [14,20,21,23]).
3.1. Conventional Detection of Salmonella
The clinical diagnosis of typhoid in tropical disease-endemic areas is very difficult and sometimes unreliable. This is due to the difficulties in distinguishing typhoid diagnosis from other co-endemic acute febrile illnesses such as malaria, dengue, leptospirosis, influenza, brucellosis and other systemic infection [50].
The culture method has been the gold standard since the discovery of enteric fever etiological agent in 1880 (as cited in [50]). This traditional isolation method involves enumeration of the targeted bacteria according to their unique morphological and biochemical properties, and it has been standardised by several regulatory agencies [20]. In a clinical diagnostic setting, blood culture is a basic method of detection for Salmonella, or else body fluids such as bone marrow, urine, stool, duodenal aspirates, and rose spot extracts can be used as well [51].
To date, the guideline has been standardised by the International Organisation of Standards (ISO) (ISO 6579:2002) for Salmonella detection [5]. Through this method, the sample will undergo a pre-enrichment by using buffered peptone water followed by a selective enrichment by using Rappaport Vassialidis soy (RVS) broth and Muller Kauffmann tetrathionate-novobiocin. Finally, the enriched sample will be streaked on a differential medium (e.g., Xylose Lysine Deoxycholate (XLD) and Hoektoen) [5,52]. In general, the proposed guidelines by other regulatory agencies are essentially similar to ISO 6579:2002, which involve four principle stages: non-selective pre-enrichment, selective enrichment, plating on selective isolation agar, and biochemical and serological tests [20,53].
The colonies that appeared on the medium will then proceed for biochemical and serological detection. Until present, a miniaturised form of the biochemical assay was developed to assist in rapid confirmation of isolated organisms from a large number of samples [20]. This system greatly reduces the volume of reagents, media, and apparatus required, as compared to conventional biochemical assay, and produces more results in a short period of time [54,55]. This system comprises a sterilised, disposable microtiter plate, which contains up to 20 specific media or substrates targeting specific microorganisms. Identification of Salmonella to species level will be determined after a 16–24 h incubation period at desired temperature. A biochemical identification test kit API 20E and Biolog automated microbial identification system are some examples of commercial biochemical assays available that are still currently being used [56,57].
Salmonella serogroup can be assessed through an agglutination test with polyvalent antisera for somatic O antigens. Specific serovar can then be determined by slide agglutination tests with monovalent antisera to specific O antigens and tube agglutination tests with antisera to flagella H antigens [53]. However, there is a critical downside of serotyping identification where the same Salmonella serotype could have a different antigenicity. This is due to the loss or modification of surface antigen of the bacterial cells and consequently lowering of the sensitivity of serological tests [58]. If this problem occurs, pulsed-field gel electrophoresis (PFGE) characterisation could be a suitable replacement method [20].
3.2. Rapid Salmonella Detection Methods
There is no doubt that the conventional culture method is preferred in many foods’ safety and health diagnostic laboratories as a routine detection due to its ease of use, high sensitivity, reliability, and reasonable operating cost. However, this method requires a minimum of three days to be completed, starting from pre-enrichment to the identification process depending on the biochemical test used [20]. Furthermore, in a certain condition that requires high throughput screening with high number of samples to be analysed, this method might be of huge disadvantage. Some modifications to the conventional method have been made by researchers to reduce labour, time, cost, and it offers much more rapid detection and identification. For instance, the use of fluorogenic and chromogenic growth media (e.g., Rambach agar, SM-ID agar, and BBL CHROMagar Salmonella), which shows direct detection, enumeration, and identification on isolation media are some of the good ways to reduce the processing time by one day, compared to the conventional method [59,60,61,62]. Yet, this improvement is still considered lacking in response to Salmonella outbreak, bioterrorism, or product recall [20].
To date, several rapid methods with different identification techniques were developed which include immunological-based assays, molecular-based assays, mass spectrometry-based detection, spectroscopy-based detection, optical phenotyping detection, and electrochemical biosensors. From the past two decades, immunological-based, and molecular-based assays are the fastest analytical detection methods developed due to the advancement in molecular sciences. A huge improvement to these methods has been made with the introduction of more specific target antibodies and genes [21]. Vibrational spectroscopy, spectral imaging, and machine vision have gained interest in bacterial detection due to their non-destructive analysis, rapid, and convenient ability to retrieve visual data from specimens and cloud libraries [17]. Another famous method to date is the electrochemical biosensors method, which already received tremendous attention and innovation in research communities in various kinds of applications.
3.2.1. Immunological-Based Assay
Immunological-based assays utilise the specificity of antibodies (monoclonal or polyclonal) for antigen capture that is usually located at the surface of the Salmonella cellular membrane [22]. These assays include enzyme-linked immunosorbent assay (ELISA), the latex agglutination method, and immunochromatography assay. Figure 2 summarises the illustration process of immunological-based assays.

Figure 2.
Illustration of a rapid Salmonella detection method based on immunological methods: (a) schematic diagram of direct enzyme-linked immunosorbent assay (ELISA) that generates a coloured signal with the presence of the target (adapted from ref. [63]); (b) illustration of the latex agglutination method that formed a visible clump in the presence of Salmonella (adapted from ref. [64]); and (c) the basic structure of a lateral flow assay (reproduced from ref. [65] with permission from Elsevier).
Enzyme-linked immunosorbent assay ELISA is the most famous and frequently used labelled immunological-based assay for Salmonella detection, where it is considered the gold standard of all immunoassays [4]. It can be used for the detection and quantification of a variety of samples such as antibodies, antigens, hormones, proteins, and glycoproteins [4]. This assay utilises an immobilised anti-Salmonella antibody in a solid matrix and, once a specific antigen is bound to the antibody and formed antigen–antibody complex, it triggers colour changes due to enzymatic cleavage of a chromogenic substrate (Figure 2a) [66,67].
Improvement on the ELISA method also has been made with a fluorogenic reporter [68], polymerase chain reaction (PCR) [69] and electrochemiluminescence reporters [70] to improve signal generation. The recent major advancement of ELISA was the manipulation of gold nanoparticles, as chromogenic reporters that greatly improve the sensitivity of detection. This assay provides qualitative results by generating a distinct coloured solution with the presence of analyte. The lowest detection limit was recorded at 1 × 10−18 g/mL (attomolar) of prostate-specific antigen (PSA) and HIV-1 capsid antigen p24 in a whole serum sample [71].
The latex agglutination method is one of the simplest Salmonella detection techniques, which utilises latex particles coated with antibodies. The presence of Salmonella will form a visible latex agglutination as the antigens on the Salmonella surface react with the immobilised antibody on latex particles (Figure 2b) [54,72]. Even though this assay is relatively outdated, it can still be used as a confirmatory analysis, since it offers a specific, easy, and reliable technique [73,74,75,76,77].
Immunochromatographic assays also known as lateral flow assays were the basis behind the dipstick assay, lateral flow device (LFD), and lateral flow immunochromatographic assay (LFIA) [6]. This assay consists of four parts namely the sample pad, conjugate pad, reaction membrane and absorbent pad (Figure 2c). The sample pad is the sample receiving region, the conjugate pad is a region where probe hybridisation takes place, the reaction membrane is where the test line and control line for target antigen–antibody or DNA–probe hybridisation interaction is located and absorbent pad served as a waste collection region [65]. These separations of component mixture based on their movement through the reaction membrane are called a chromatographic system [78].
3.2.2. Molecular-Based Assay
Molecular-based assay involves the hybridisation of short oligonucleotide fragment known as a DNA/RNA probe or primer to detect specific targeted DNA/RNA sequences [21]. Specific primer/probe can be isolated from microorganisms or engineered according to their specific target bioreceptor [79]. This method offers high sensitivity and specificity and greatly reduced detection time to only a few hours. They were polymerase chain reaction (PCR), loop-mediated isothermal amplification (LAMP), nucleic acid sequence-based amplification (NASBA), recombinase polymerase amplification (RPA), DNA microarrays, and whole-genome sequencing (WGS). Figure 3 shows the illustration process of a few molecular-based assays based on DNA/RNA amplification.

Figure 3.
Schematic illustration of a molecular detection method based on an amplification technique: (a) the polymerase chain reaction (PCR) amplification process (adapted from ref. [80]); (b) the nucleic acid sequence-based amplification (NASBA) process (reproduced from ref. [81] with permission from Elsevier); (c) the loop-mediated isothermal amplification (LAMP) process (reproduced from ref. [82] with permission from John Wiley and Sons); and (d) Recombinase polymerase amplification (RPA) flow (reproduced from ref. [9] with permission from Elsevier).
The polymerase chain reaction (PCR) is considered the gold standard in bacterial identification, as well as in diagnostic application due to its reliability, high accuracy, and very specific detection outcome. PCR utilises the amplification of nucleic acid and has been extensively utilised for Salmonella detection and identification for quite some time [7]. PCR works by amplifying or selectively amplifying a unique defined DNA region, by using special molecular ingredients at a specific condition, generating thousands to millions of copies of targeted DNA sequences (Figure 3a) [21,83]. A short turnaround time within 16 h from start until detection has made PCR an excellent tool in a diagnostic application [84,85]. To date, PCR is still being used as a mandatory microbial identification confirmation and PCR technology continues to evolve.
Multiplex PCR (mPCR) is one of the PCR methods developed for multiple detections of microorganisms. This method has been used for Salmonella detection and other foodborne pathogens as it allows rapid multiplex detection in various food matrices [82,84,85]. Real-time PCR or known as quantitative PCR (qPCR) is a PCR method that is capable of quantitatively detecting the target sample in real-time [21]. This method can detect the target DNA and bacterial cells as time progresses by using a fluorescent technology known as SYBR green and TaqMan dyes. These dyes will bind on the DNA groove as the double-strand DNA is amplified, which then increases the fluorescent intensity [86,87,88]. qPCR has been widely used for Salmonella detection in various food, poultry, and veterinary products [89,90,91]. In the majority of studies, the invasion gene (invA) and tetrathionate reductase gene (ttr) were the most targeted gene for Salmonella identification using the qPCR method [92]. However, as the research progressed, new target genes such as Salmonella enterotoxin gene (stn) [93], outer membrane porin F gene (ompF) [94], hyperinvasive locus A (hilA) [95], virulence plasmid gene (spvC) [96], and many more have been used, providing high sensitivity and specificity. Multiplex qPCR targeting many genes simultaneously in real-time was also developed to assist in food safety inspection [21].
Even though PCR is considered an excellent choice for reliable and specific microbial identification, there are some limitations and disadvantages to this assay that can contribute to a major problem. In PCR, the presence of walnut components was reported, inhibiting the product amplification [97]. Meanwhile, in qPCR, the presence of fat, black tea and cocoa also inhibit the amplification process [98,99]. PCR is also unable to differentiate between live and dead cells, since it will amplify any targeted sample available regardless of the cellular condition [20]. This lack of discrimination could cause false-positive results, therefore introducing other problems down the line, such as product recall [100]. The PCR process is also laborious, requiring expensive machines (e.g., a thermal cycler, gel electrophoresis, etc.) and trained personnel to conduct and analyse the PCR product [86].
Loop-mediated isothermal amplification (LAMP) is a novel alternative method to amplify DNA aside from PCR. This method can amplify DNA with excellent specificity, rapidity and efficiency under isothermal conditions (constant temperature) [8]. LAMP employs a special DNA polymerase (Bst) and a set of four primers that can recognise six distinct target regions of DNA (Figure 3c) [8,101]. This method has several advantages over the traditional PCR, as it requires only a simple heating block, e.g., a water bath or dry bath, to keep the temperature constant during operation, apart from its lesser sensitivity to the PCR inhibitor and higher yield outcome [101,102,103]. Due to these excellent characteristics of LAMP, it has been widely used for Salmonella detection [104,105,106]. A multiplex LAMP-based system was also developed to detect multiple bacterial targets. Liu N. et al. developed multiplex-LAMP to detect Salmonella spp. and Vibrio parahaemolyticus with 100% specificity [107] and EMA-Rti-LAMP assay ethidium bromide monoazide (EMA) treatment to detect and quantify S. Enteritidis in real-time [108].
Nucleic acid sequence-based amplification (NASBA) is another isothermal amplification method, which is based on a molecular transcription system targeting exclusively RNA (Figure 3b). [81,109]. This method can selectively detect viable microorganisms in the sample and successfully eliminate the problems that arise from PCR techniques [10]. NASBA was found to be effective and sensitive for Salmonella detection [110,111,112].
Recombinase polymerase amplification (RPA) is another new, novel isothermal amplification technique that is capable of amplifying a minimum of 1–10 DNA target copies in less than 20 min [9]. This selective and highly sensitive technique is operated at 37–42 °C, requires minimal sample preparation without prior DNA extraction and purification (Figure 3d). It has been used to amplify ssDNA, dsDNA, RNA, miRNA from diverse samples and organisms [9,113]. Integration of RPA with other detection strategies such as lateral flow [114], real-time fluorescent detection [113], and microfluidic [115] was found to be a success. As soon as their inception, RPA-based methods have been applied in the detection of Salmonella in food samples [116,117,118]. Since the success of the isothermal amplification method for Salmonella detection, the RPA system is likely the most convenient choice for point of care application due to its short turnaround time, simple implementation, and minimum sample preparation [119].
DNA microarray is an advanced molecular technology that performs a parallel hybridisation of hundreds to thousands of specific and selective DNA probes to their respective target DNA in a single assay [11,27]. This method was initially used to study gene expression analysis [120], but its application had gained widespread expansion for use in comparative genomics, sequence analysis and diagnostics [27]. Reports on microarray-based S. enterica detection are available [121,122,123]. Guo D. et al. successfully developed a microarray system that can detect and identify 40 Salmonella O serogroups from a simulated food samples [124]. This method is highly specific, as it correctly identifies 98% (n = 288) of Salmonella strains in a mixed sample with other bacterial species.
Whole-genome sequencing (WGS) or next-generation sequencing (NGS) is another advanced molecular method referring to highly automated and parallelised genome sequencers that are used to sequence the entire genomes of bacterial pathogens [14]. Unlike a DNA microarray that targets certain genes, WGS works by sequencing the entire fragments of bacterial DNA, aligning them into a complete genomic sequence and subsequently compare them in genome sequence databases [23,125]. There are numbers of established genome sequence databases available such as NCBI genome database (https://www.ncbi.nlm.nih.gov/genome/) (accessed on 8 September 2021), CFSAN-FDA (https://github.com/FDA/open.fda.gov) (accessed on 8 September 2021) and other public domains such as GenomTrakr (https://www.fda.gov/food/science-research-food/whole-genome-sequencing-wgs-program) (accessed on 8 September 2021) [14].
With the aid of these databases, WGS can provide detailed information on identified bacterial species including their antimicrobial resistance genes, any mutations to the sequence and even their source-level, such as source of isolation, geographic regions, etc. [14]. Ibrahim G. M. and Morin P. M. in their study successfully predicted Salmonella according to their serotypes with a success rate of 89% (n = 899) by using WGS and SEQSERO programme as a data analysis tool [12].
3.2.3. Mass Spectrometry-Based Method
The matrix-assisted laser desorption ionisation-time-of-flight mass spectrometry (MALDI-TOF MS) is a recent mass spectrometry technology used for rapid and reliable microorganism identification [126]. This method utilises a protein spectrum profile of a microorganism as a basis of the identification system, which is called a peptide mass fingerprint (PMF) [23,126]. The obtained PMF is compared in an open PMF database, using a scoring algorithm to match the analysed PMF spectrum with reference spectra and consequently identify the microbial species with a high degree of certainty [23].
There are many reports on the use of MALDI-TOF MS for Salmonella detection. Mangmee S. et al. develop a MALDI-TOF MS-based method for simultaneous identification of non-typhoidal Salmonella (NTS) in the Thai broiler industry. They successfully identified NTS with high accuracy up to species and subspecies level and the method could support faster large-scale screening with efficient cost [13]. Yang S. M. et al. uses MALDI-TOF MS to identify and discriminate three different Salmonella serovars, Enteritidis, Typhimurium, and Thompson, that are epidemiologically important in Korea [127].
Liquid chromatography-mass spectrometry (LC-MS) is an alternative mass spectrometry method to MALDI-TOF MS. This method showed promising results in the identification of a closely related bacterial species and has been used for serovar level identification of Salmonella ([128] as cited in [14]). This method separates the intact bacterial protein lysates through chromatographic separation and detected with MS [14]. Mc Farland M. A. et al., uses LC-MS coupled with electrospray ionisation (ESI) to identify Salmonella serovars (S. Typhimurium vs. S. Heidelberg) based on their single nucleotide polymorphisms (SNPs) [129]. However, the LC-MS method is slower compared to MALDI-TOF MS analysis [14].
3.2.4. Spectroscopy Method
Raman spectroscopy is a spectroscopy technique based on inelastic scattering of monochromatic light (usually from a laser source). Upon the interaction with the sample, the frequency of photons in monochromatic light changes as they are absorbed by the sample and then reemitted. The shifting frequency of the reemitted photons from the sample is then compared with the original monochromatic frequency to form a Raman effect [23,130]. Surface-enhanced Raman spectroscopy (SERS) is the enhanced version of Raman spectroscopy with a better amplification of electromagnetic fields created by the excitation of localised surface plasmon. SERS can detect samples in a low concentration of analytes with high sensitivity [23]. This technique has been used to rapidly detect foodborne pathogens [15]. Duan N. et al. utilised a SERS-based aptasensor to detect S. Typhimurium in a real food sample where they successfully detected the pathogen with a detection limit of 15 CFU/mL [131].
Near-infrared spectroscopy (NIR) is a technique that utilises a specific spectral region called near-infrared (780–2526 nm), where the occurrence of the overtones and combination of vibration response is likely related to the changes in chemical bonds such as O-H, C-H, N-H, C-O, and other organic molecules [23,132]. Since the membrane structure of bacteria consists of combinations of macromolecules that are unique to their respective species, thus the NIR spectrum of each bacterium has a highly specific absorption signature [133]. Pereira J. M. et al. reported fast detection of S. Typhimurium in a milk sample by using NIR spectroscopy and evaluated using the chemometric method of partial least squares with discriminant analysis [16]. Their study was able to discriminate S. Typhimurium with a good prediction and all samples were correctly classified. Gao X. et al. also reported a compilation studies of foodborne bacteria detection and identification using NIR spectroscopy [133]. This technique provides a non-destructive, fast, and accurate measurement of the target in complex sample matrices.
Hyperspectral imaging (HSI) is an emerging rapid method for bacterial identification. This method utilises the integration of conventional imaging and spectroscopy to provide both spatial and spectral information of a sample [23,134]. Thus, HSI will provide more specific detection and identification compared to a single source of modality. This non-destructive method is usually being used in foodborne pathogen detection and the development of a complete hyperspectral graph of common foodborne pathogens is still under development [135]. Seo Y. N. et al. reported on the development of an HSI method for automated screening of S. Enteritidis and S. Typhimurium in poultry carcass rinses [136]. They use five different machine learning algorithms to analyse and train the systems and the best performance was achieved by quadratic discriminant analysis (QDA) with a prediction accuracy of 99%.
3.2.5. Optical Phenotyping Using Light Diffraction Technology Method
Microbial identification through light diffraction or a forward light scattering phenomenon is an interesting new way to identify bacteria. This method provides a non-contact and non-destructive measurement as it directly observes the microorganism colonies grown on a culture plate to produce acquisition of scattering images and consequently compare it with reference scatter image libraries of known bacteria [28,137]. BARDOT (Bacterial Rapid Detection using Optical Scattering Technology) and BISLD (Bacteria Identification System by Light Diffraction) are two available methods based on this technique [137,138]. Abdelhaseib M. U. et al. utilised BARDOT the system in their study coupled with a multi-pathogen selective medium to identify S. enterica, E. coli and L. monocytogenes in a single assay [18]. Singh A. K. et al. utilised BARDOT and a laser optical sensor to detect 36 different Salmonella serovars in a food sample [139]. Real-time detection in food samples showed 84% detection accuracy in 24 h comparable to those of the USDA Food Safety Inspection Service method, which require ~72 h.
3.2.6. Electrochemical Biosensors
Electrochemical biosensor is an integrated receptor–transducer device that captures a biological signal, processes it through electrochemical means, and translates it into a detectable electrical signal [140]. To date, electrochemical biosensors have received tremendous attention in Salmonella detection due to their rapid detection, high specificity and sensitivity and possible on-site testing [23,141,142,143]. A study by Jia F. et al. showed a very sensitive detection of S. Typhimurium ATCC 50761 with a limit of detection of 25 CFU/mL in one hour [144]. A similar result was obtained when tested in a spiked chicken sample, which represents good reproducibility. Further details on electrochemical biosensors will be discussed in Section 4.
3.2.7. Advantages and Disadvantages of Salmonella Detection Methods
Table 1 summarises the advantages and disadvantages of each discussed Salmonella detection method and their examples. The electrochemical biosensor method was found to be the most promising method, as it offers attractive advantages such as low cost per test, user-friendliness, and possible on-site testing. Despite rapid, sensitive, and specific detection, the electrochemical biosensor method struggles with drawbacks such as high early instrument cost (potentiostat), non-standardised sample preparation due to its dependence on the bioreceptor used, and lack of multiplex detection [23,145]. However, recent creation of a low-cost smartphone-controlled potentiostat could solve the expensive potentiostat issue in the near future [146].

Table 1.
Overview of Salmonella identification method used in the literature including their advantages, disadvantages, and examples.
Detection sensitivity is one of the important aspects in Salmonella detection and it is dependent on the minimum amount of analyte that can be detected. The detection limit represents the lowest concentration of analyte required to give a positive result [154]. The electrochemical biosensor showed the highest sensitivity with the lowest detection limit, and it can detect an analyte down to the femto-molar scale or a small number of cells [155,156]. Meanwhile, in the molecular-based method, the detection limit is expressed in the form of minimum genomic copies for the amplification process. RPA can amplify a minimum of 1–10 DNA targets and PCR requires 32 genome copies in 1 µL of the DNA sample [9,157]. Meanwhile, for other methods, they normally require the initial sample in high concentration. Thus, a pre-enrichment process is one crucial step to increase the analyte concentration.
Cross-reactivity among Salmonella serotypes is a common selectivity problem in Salmonella identification. This problem not only occurs in immunological-based assay but also in molecular-based identification, which might lead to a false positive result [24,25,26]. This problem can be easily avoided in DNA microarray and whole-genome sequencing (WGS) as a database library of specific serogroups is available [12,124]. Other methods such as MALDI-TOF MS, LC-MS, Raman spectroscopy, NIR spectroscopy, HSI imaging and optical phenotyping usually developed their own specific spectral data, mass fingerprint data or scattering images data that are specific enough to discriminate Salmonella serovars [16,127,128,136,139]. In the electrochemical biosensor method, the use of aptamer-based bioreceptor is the best strategy to avoid cross-reactivity problems. In a study conducted by Hyeon J. Y. et al., they successfully developed an RNA aptamer for S. Enteritidis with no cross-reactivity to other Salmonella serovars [158]. Thus, the use of a highly specific aptamer biorecognition element could prevent cross-reactivity problems. Few studies that utilise aptamer as a biorecognition element showed a good selectivity detection between Salmonella serovar [153,159,160].
On the other hand, sample preparation is also a crucial part of the Salmonella detection process as the sample might be isolated in variety of forms (solid/liquid). Most of the Salmonella detection methods discussed require an input sample in a liquid form except for MALDI-TOF MS, NIR spectroscopy, HSI spectroscopy, and optical phenotyping (BARDOT) as they require bacterial colonies grown on an agar plate [18,127,132,135]. Most of the immunological and molecular-based methods also require sample pre-treatment or pre-enrichment, while other methods such as mass spectrometry, spectroscopy and electrochemical biosensor can be perform in a mixed sample with minimal pre-treatment. These pre-treatment and pre-enrichment processes will increase the overall detection time of sample.
4. Biosensors Developed for Salmonella Detection
The discovery of a glucose sensor by Clark and Lyons in 1962 was considered the starting point for the development of biosensors in the biomedical field [161]. This technology was then commercialised by the Yellow Springs Instrument Company, by introducing the world first commercial glucose sensor (Model 23A YSI analyser) for the direct measurement of glucose in 1975 [162]. Later, biosensor technology has bloomed with the development of many kinds of biosensor such as cancer biomarker biosensors [163], protein biomarker biosensors [164], and many more.
4.1. Biosensors
A biosensor is defined as an analytical device that converts a biological signal or response into a quantifiable and processible signal [165,166]. Generally, a biosensor consists of three main components, namely a biorecognition element, transducer components and the electronic system, e.g., a signal amplifier, processor and electronic display as shown in Figure 4 [167]. The biosensor can detect biological samples in a very specific, fast, and reliable manner utilising a sensing method such as optical, physical changes such as strain or piezo effect due to difference in sample mass or volume, electronic or field-effect transistor (FET) and electrochemical techniques [31]. The generated signal will then be translated into an electrical signal, proportional to the number of analyte–bioreceptor interaction events [31].

Figure 4.
Schematic diagram of three main components of a biosensor and examples (adapted from ref. [167]): (A) biorecognition elements that interact with the analytes such as cells, tissues, antigens, nucleic acids and many more; (B) transducer elements that convert the analyte–bioreceptor interaction into a quantifiable signal; and (C) electronic systems amplify and process the signal from the transducer and display the output as digital data.
Some researchers have also further classified the biorecognition element into analyte and bioreceptor and the electronic system into electronics and display [154]. The analyte is a component of interest or a solution containing a component of interest to be detected such as nucleic acid, proteins, enzymes, antibodies, microorganisms, cellular components, tissues and also drugs [20]. A bioreceptor is a biorecognition element used to detect the presence of the analyte. It could be an enzyme, antibodies, nucleic acid, aptamers, viruses, and cells. A recognition signal will be generated once the target analyte binds to the biorecognition element and the signal can be generated in a variety of ways such as current, charge transfer resistance, potential difference, mass change, absorbance, reflectance, and many more [73,154]. The transducer is an element that converts the biorecognition signals into measurable signals. Electronics is a part that processes the transduced signal through complex electronic circuitry, which then amplifies or transforms the signals into a digital form. Finally, display components will quantify the processed signal generated in a form of a specific graph, numerical data or any format depending on the requirements of the end-user [154].
Biosensors have been broadly classified based on their biorecognition element and transducer [31,168]. These two components are a crucial first step in developing a reliable, sensitive, and robust biosensor combined with good microelectronics systems for data processing and interpretation. The bioreceptor must bind with high affinity to the targeted analyte to ensure high selectivity and sensitivity, and the transducer must be highly sensitive and accurately transform the signals generated for processing [154].
4.2. Electrochemical Biosensors
Each type of sensing mechanism has its advantages and disadvantages. Among many kinds of biosensors, electrochemical biosensors have sparked a lot of interest in pathogen detection including Salmonella [169]. Electrochemical biosensors offer significant advantages over other biosensors due to their high sensitivity, low cost, versatile detection strategy, automation, miniaturisation potential, sustainability (i.e., low sample volume and minimal solvent use in its development and application) and possible real-time quantification [22,170]. The electrochemical biosensor is also capable of sensing a sample with high turbidity and not being affected by quenching or interference from absorbance and fluorescent compounds as optical-based techniques struggle with [171]. Moreover, the electrochemical biosensor set-up requires relatively simple instrumentation that is low-power compared to surface plasmon resonance techniques that need a light source and receiver, which adds more complexity to the system [171]. This sensing system also can detect a sample in a large linear detection range and a wide variety of solvents and electrolytes compared to a field-effect transistor (FET), which only can detect the target at a certain range of concentration due to a Debye length effect [171,172,173]. Those excellent characteristics are the advantages of electrochemical systems, which make them a reliable sensing system for pathogen detection. A discussion on other transducing elements, such as optical and piezoelectric ones, can be found elsewhere. Figure 5 shows the schematic of typical electrochemical biosensors, including their types of receptors and materials used for electrode surface modification, which will be further discussed in the next section.
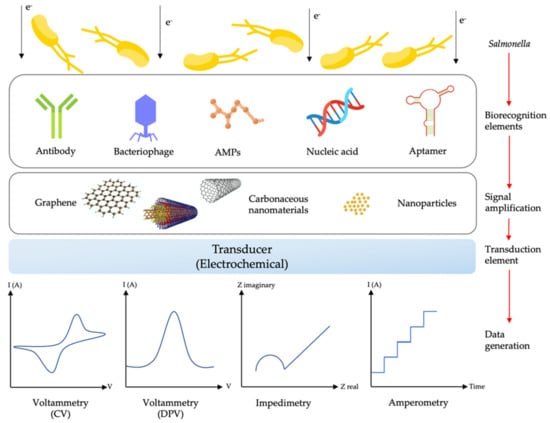
Figure 5.
Schematic illustration of a typical electrochemical biosensor with different types of biorecognition elements, materials for signal amplification and electrochemical transducing techniques for Salmonella detection (adapted from ref. [22] and the graphene scheme from ref. [174]).
Theoretically, the electrochemical biosensor uses electrical means to analyse or examine biochemical reaction (the charge transfer process) that occurs on the surface of the electrode (electrode–solution interface) [166,170]. The electrode itself is one of the crucial components in an electrochemical reaction as its materials, surface modification and dimensions can greatly influence the performance of electrochemical reactions. The working electrode serves as the main region where an electrochemical transduction process takes place, and the counter electrode will complete the circuit by establishing a connection to the working electrode through the analyte as the current is applied. The reference electrode, usually positioned at a distance from the reaction site, serves to maintain a known stable potential [166]. As the reaction takes place, the electrochemical reaction (redox potential changes) can be detected and measured in terms of current, voltage, impedance and capacitance [175,176]. Table 2 summarises the types of electrochemical biosensors based on their electrochemical transduction principle namely potentiometric, amperometric, impedimetric, and voltammetric biosensors.

Table 2.
Summary of electrochemical biosensor types according to their transduction principle, their performances and examples of studies conducted.
4.3. Bioreceptors Used in Salmonella Biosensors
Bioreceptors or the biorecognition elements are one of the crucial parts in developing Salmonella biosensors. The interaction of the bioreceptor and the Salmonella will determine the specificity, selectivity, and sensitivity of the developed biosensor [167]. Each type of biorecognition element available has its advantages and disadvantages. Table 3 presents the summary of the pros and cons of each biorecognition element, which are an important consideration for biosensor development.

Table 3.
Summary of biorecognition elements used in biosensors.
4.3.1. Antibody–Antigen or Immunosensors
Immunosensors, also known as an antibody–antigen biosensors are a widely utilised analytical tool for Salmonella detection especially in dairy and food processing settings [191]. This biosensor works by immobilising a specific anti-Salmonella antibody on a surface of a transducer, and the coupling of an antigen to the antibody will trigger an immunochemical reaction, which will be used as a detection signal [192]. Recently, there were quite extensive studies reported on Salmonella detection using an immunosensor.
Melo et al. reported on the detection of S. Typhimurium in contaminated milk samples via the chronoamperometry method [143]. The polyclonal anti-Salmonella antibodies were immobilised on the gold working electrode using carboxymethylated cashew gum (CMCG) film through electrodeposition and the limit of detection (LOD) was observed at 10 CFU mL−1 with a detection time of 125 min. Alexandre et al. also reported similar results as Melo in their study for S. Typhimurium detection in milk samples with LOD of 10 CFU mL−1 and 125 min of detection time [193]. The only difference in this study was that the polyclonal antibodies were immobilised using the cysteine–thiol self-assembly monolayer (SAM) method on a gold screen-printed electrode. A study by Sannigrahi et al. used magnetosome (biogenic nanoparticles synthesised in a magnetotactic bacteria Magnetospirillum sp. through biomineralisation) functionalised with anti-Salmonella antibody to detect lipopolysaccharide (somatic “O” antigen) of S. Typhimurium in food and water samples [194]. Electrochemical impedance spectroscopy (EIS) confirmed the detection of lipopolysaccharide at 0.001–0.1 μg/mL and the LOD of bacteria at 1 × 101 CFU/mL in water and milk samples. A labelled immunoassay was also reported by Bu et al., where they used ferrocene (Fc)-functionalised nanocomposites to amplify the electrochemical signals [195].
4.3.2. Bacteriophage-Based or Phagosensors
A phagosensor is a type of biosensor that uses bacteriophages as their detection vector. Bacteriophages, also known as phages, are viruses that strictly infect and replicate inside bacterial cells [196]. The nature of bacteriophages that only infect a very specific, single-cell host species, and sometimes even specific strains within the species, is an excellent characteristic to employ as a bioreceptor [196]. This strategy is similar to antigen–antibody binding and DNA hybridisation, which provide an exclusive, specific binding of the target [19].
Reports on electrochemical phagosensors for Salmonella detection are not widely available as compared to immunosensor and DNA sensors as they are opting for different detection methods. A study by Vinay M. et al. successfully developed a prototype of a phage-based fluorescent biosensor to detect enteric bacteria such as E. coli and S. Typhimurium in water samples [197]. Their limit of detection was 10 bacteria per mL of solution without concentrating or enriching the sample. Moreover, their prototype is also robust as it can detect target bacteria in seawater sample. Another rapid, sensitive, and direct detection of S. Typhimurium based on a wireless magnetoelastic (ME) biosensor has been reported and tested on eggshell samples [198]. This biosensor utilises the E2 phage as the biomolecular recognition element that selectively binds with S. Typhimurium and their limit of detection was found at 1.6 × 102 CFU/cm2 in a humidity-controlled chamber.
4.3.3. Antimicrobial Peptide-Based Biosensor (AMPs)
Antimicrobial peptides (AMPs) is a short fragment of peptides consisting of around 12 to 15 amino acid residues [199]. AMPs can be found as an innate immune system of living organisms, which provide defence mechanisms against invading pathogens [200]. Generally, AMPs are amphipathic and cationic and they can bind to the bacterial cell membrane via electrostatic and hydrophobic interaction [201]. Their excellent properties such as being intrinsically stable in harsh conditions, their possibility of being produced synthetically in a large quantity, and ease of modification with low production costs, make them suitable candidates as a bioreceptor [168,202].
The number of recent reports for AMP biosensors for Salmonella detection is also limited. For example, novel AMPs known as nisin were used in an electrochemical impedance biosensor to detect pathogenic and non-pathogenic Salmonella spp. in milk samples. The lowest limit of detection (LOD) was recorded at 1.5 × 101 CFU/mL [142]. Another study by Mannor et al. reported that immobilisation of semi selective AMPs called magainine I on micro capacitive electrode arrays showed a good recognition capability towards E. coli and Salmonella [203].
4.3.4. Nucleic Acid-Based Sensors or Genosensors
Genosensors also known as DNA biosensors utilise genetic materials such as DNA and RNA sequences as the biorecognition element. Similar to immunosensors and phagosensor, this approach utilises the complementary hybridisation signal of the DNA probe with the target nucleic acid of pathogens, which then translates into a specific transduction signal [204]. In this method, the majority of the target DNA/RNA isolated or extracted from microorganisms undergoes denaturation and is then exposed to the DNA/RNA probe. The hybridisation of probe and target occurs at the sensor surface, which then triggers signal generation [205].
A study by Das Ritu et al. on a Salmonella genosensor showed promising results. Biosensor detection response was found to be linear with the target sequence concentrations ranging from 1.0 × 10−11 to 0.5 × 10−8 M and the lowest detection limit found at 50 (±2.1) pM. This study employed the Salmonella Vi gene as a molecular marker. The DNA probe was immobilised on a gold nanoparticle (AuNP)-modified SPE and the signal was monitored using the differential pulse voltammetry (DPV) method [206].
4.3.5. Aptamers as a New Bioreceptor for Salmonella Biosensors
Aptamer-based biosensors or aptasensors can be considered as a new technology in biosensors. The aptamer is a single-stranded nucleic acid (ssDNA) or RNA known as oligonucleotides, firstly discovered and reported by Gold and Szostak in the early 1990s [207,208]. The first aptasensor was reported by Kenneth A. Davis and colleagues in 1996 with the development of an optical biosensor based on a fluoresceinated DNA ligand for detection of human neutrophil elastase (HNE) antigen [209]. The aptamer can be obtained through a process called Systematic Evolution of Ligands by Exponential Enrichment (SELEX). Through this process, a pool of nucleotides (nucleotides library) will be incubated with the targeted molecules, and a specific random sequence will form, binding with the target. This process will be repeated for many cycles until a sequence with strong binding to the target molecule is found (Figure 6a) [210]. Aptamers can bind with high affinity and specificity to a broad spectrum of molecules such as nucleotides [211], proteins [212], peptides [213], toxins [214], antibiotics [215], and small molecules [216]. The whole cell SELEX is one of the SELEX methods that received much attention for aptamer development for Salmonella [217,218,219]. Through this method, an aptamer specific to the characteristics of the target cell, e.g., specific surface protein, that can be developed and enriched in further steps (Figure 6b) [167].

Figure 6.
(a) Schematic diagram of the SELEX method. (b) Whole-cell SELEX method for aptamer selection against live bacterial cells. (Reprinted with permission from ref. [167]).
Aptasensors have also been widely utilised for Salmonella detection. For instance, a study by Dinshaw et al. utilised an aptamer specific for the outer membrane of S. Typhimurium for detection in artificially spiked raw chicken samples. The aptasensor exhibited a low limit of detection of 101 CFU mL−1 [220]. Ma et al. also reported a Salmonella biosensor using a Salmonella-specific recognition aptamer with a low detection limit of 3 CFU mL−1 [221]. Other studies reported a duplex detection of pathogenic microorganisms using an evanescent wave dual-colour fluorescence aptasensor and a fibre nanoprobe. Two fluorescence labelled aptasensors, namely, Cy3-apt-E and Cy5.5-apt-S, for the detection of E. coli O157:H7 and S. Typhimurium, respectively, were able to perform detection in less than 35 min. The limits of detection were recorded at 340 CFU/mL for E. coli O157:H7 and 180 CFU/mL for S. Typhimurium [222]. Bagheryan Z. et al. developed a label-free impedimetric aptasensor by using diazonium-modified screen-printed carbon electrodes for the detection of S. Typhimurium in food samples [141]. They successfully detect the Salmonella with a limit of detection of 6 CFU mL−1. Figure 7 illustrates the overview of their aptasensor preparation method and the EIS results obtained. To date, the aptamer biosensor is in the limelight due to its prominent advantages compared to other bioreceptors. Table 4 demonstrates the number of available publications regarding electrochemical aptasensors targeting Salmonella sp. from 2010 to 2021.

Figure 7.
(a) The overview of the Salmonella aptasensor preparation utilising a screen-printed electrode (SPE); and (b) EIS results of aptasensor-based charge transfer resistance (Rct) when incubated with different concentrations of S. Typhimurium. This figure has been reproduced from ref. [141] with permission from Elsevier.

Table 4.
Research published on electrochemical aptasensors for the detection of Salmonella.
5. Miniaturisation of Electrochemical Biosensors
Sensor miniaturisation could promise an excellent return in terms of possible large-scale production, low production cost, low sample volume analysis, low power analysis, reduced weight, and on-site testing and monitoring on a real-world sample, which is hardly to be achieved with bulky conventional electrodes [226]. Screen-printed electrodes (SPEs) are one of the miniaturised electrode technologies widely utilised in the development of commercial sensors nowadays. They have been used for the detection of heavy metals [227], toxins [228], antibiotics [229] as well as microorganisms [230]. Aside from portability, SPEs are also excellent in avoiding some of the common problems faced by the traditional solid electrode such as memory effects and tedious cleaning process [231]. Figure 8 shows the differences between the conventional electrode platform compared to SPE. The conventional electrodes (e.g., glassy carbon, graphite, etc.) require the presence of external counter electrode (CE) and reference (RE), rigorous polishing and cleaning process prior to use and higher sample volume. Meanwhile, SPE is ready to use without a polishing and cleaning process, smaller in size, disposable and requires much lower sample volume [232]. There are several reports available of SPE-based electrochemical biosensors for the detection of Salmonella in various samples (Table 5). However, the reports on the use of SPEs with an aptamer as the biorecognition elements are scarce.

Figure 8.
Schematic overview of a classical electrochemical sensing process using a conventional electrode (a) and SPE (b) adapted from ref. [232].

Table 5.
Salmonella electrochemical biosensors utilised in the use of SPE.
SPEs are usually configured based on three-electrode systems, which include a working electrode, counter electrode, and reference electrode similar to those conventional electrodes. They are printed on an inert, non-conductive substrate such as plastics, ceramic, or a printed circuit board (PCB). The nature of SPE development through layer by layer deposition of a conductive ink using a screen or mesh system on a solid substrate provides wide opportunity and flexibility of using different materials of choice based on the application of interest [226]. Moreover, the introduction of binders, mediators or any conductive components that modify the electrode surface such as polymers or electroactive metals could enhance the sensor efficiency by having better electron transfer kinetics compared to the unmodified electrode [233,234].
However, the miniaturisation process of the sensor does not always translate into good results. Some problems related to the miniaturisation of the sensor have been addressed by Andreas B. Dahlin in his finding [239]. These include the complicated fabrication process as they require more sophisticated and expensive machines to reach precision up to a micro or nano-scale level, are typically more hassle to handle due to their miniaturised size, and produce reduction in measurement precision, which lead to a poor detection limit as well as noise and stability issues [239]. Nevertheless, plasmonic nanoparticle sensors could be one important exception as they are relatively simple to produce and measure on [240]. The development of an open-source format in the form of printed electrodes, circuit boards, electric circuits, components, microcontrollers, software, etc. has made this miniaturised sensor system accessible to the ground as they offer lower production costs [232].
5.1. Nanomaterials as the Surface Modifier of Electrochemical Biosensors
In the construction of a sensor, the types of material used, their formula and compositions directly affect the capability and performance of the sensor. The number of particles loaded to build the electrode itself strongly influence the electron transfer process, which translates into a better performance [226]. However, the maximum threshold of the sensor performance is restricted to the surface area of the working electrode itself. This is the area where the miniaturised biosensor is lacking, as the surface area for the biomolecule immobilisation is limited.
To date, the utilisation of nanomaterials as surface modification materials has become eminent. These nanomaterials not only provide a large surface area for biomolecule attachment but also have excellent electrical conductivity, electron transfer and biocompatibility with capture biomolecules [241,242]. Aside from immobilisation, various strategies can also be implemented by using these nanoparticles as an electrochemical label [185]. In general, these nanomaterials can be classified into two groups, which are carbon-based nanomaterials and non-carbon nanomaterials [242].
5.1.1. Carbon-Based Nanomaterials
Graphene and carbon nanotubes are examples of carbon-based nanomaterials. These types of nanomaterials are famous in the research setting and widely applied in the industrial field [243]. Graphene is a two-dimensional (2D) hexagonal pattern of carbon atoms with one single atomic layer, which is densely organised in a regular sp2 orbital hybridisation and serves as a basic structure of other sp2 carbon-bonded materials such as fullerenes and carbon nanotubes [244,245]. Since its discovery in 2004 [246], graphene-based research has been tremendously popular in a wide variety of research fields with substantial publication beginning in 2013, including the biosensor field [245,247].
Graphene in a 2D hexagonal lattice form has received tremendous attention in sensor research [248]. Its excellent physicochemical characteristics such as large surface area, exceptional electron transfer, high thermal conductivity, good mechanical stability, flexibility and biocompatibility have made graphene a preferable choice in biosensor development compared to other carbon materials [249]. However, the use of the hydrophilic solution as the analyte target of the biosensor (e.g., buffers and fluid samples) has exposed the downside of graphene as graphene itself is hydrophobic. However, this problem can easily encountered by the functionalisation of graphene with hydrophilic functional groups such as a carboxyl group (-COOH) or hydroxyl group (-OH) to obtain a graphitic structure called graphene oxide (GO) [242]. This process can be performed by oxidative stripping of graphite [250,251]. To remove the oxygen-rich functional group on GO, a simple heat or chemical treatment can be conducted to obtain another derivative of graphene called reduced graphene oxide (rGO) [251]. Both GO and rGO retained the basic structure of graphene with modification only applied on the surface and at the edge structure of graphene, but their excellent physicochemical characteristics remain unchanged [245]. To date, there are many biosensor studies reporting on the use of graphene-based nanomaterials for Salmonella detection (Table 6). However, since graphene is still considered a new material of interest in the biosensor field, the reliability and reproducibility for high-performance analysis and real-world performance are still under development.

Table 6.
Integration of nanomaterials in biosensor application for Salmonella detection.
Carbon nanotubes (CNTs) are a carbon structure with an electron orbital hybridisation type sp2 between adjacent carbon atoms such as graphene. Unlike sheet-structured graphene, the CNTs’ structure comprises a distinct tube-like hollow cylindrical carbon nanostructure, which is made up of rolled-up graphene sheets [244,252]. This structure grants CNTs a strong mechanical structure, large surface area and excellent electrical conductivity [248]. Most of the carbon nanotubes’ physical properties are derived from graphene [244]. There are two types of CNTs, namely single-walled carbon nanotubes (SWCNTs) and multi-walled carbon nanotubes (MWCNTs).
As the name suggests, SWCNTs are made up of a single cylindrical shape of carbon nanostructure while MWCNTs are composed of multilayer concentric single-walled graphene cylinders supported by Van der Waals forces [242]. MWCNTs have a few advantages over SWCNTs such as high purity, easy bulk synthesis, catalyst-free synthesis, being less prone to defects, have more accumulation in the CNTs’ body (provides more surface area) and having a rigid structure. Meanwhile, SWCNTs offer less purity, are difficult to synthesis in bulk, require a catalyst for synthesis, are prone to defects during functionalisation, have less accumulation of the CNTs’ body and can easily bend and be twisted [244]. Both CNTs have been used in biosensor technology to enhance the electrical properties. Their huge surface area could provide a better immobilisation base for the bioreceptor, improved electrical conductivity and translated into a better signal response [253].
5.1.2. Non-Carbon Nanomaterials
Non-carbon nanomaterials are nanomaterials that originated from sources other than carbon such as metals, silica, polymers and many more. They have been employed as alternative supporting materials of the electrode to improve the electrochemical performance of biosensors. Examples of non-carbon nanomaterials are metallic nanoparticles, silica nanoparticles, nanowire, indium tin oxide (ITO) as well as organic polymers and they have become more popular options in biosensor research [242]. Table 6 shows some of the Salmonella biosensors that utilise non-carbon nanoparticles in their systems.
Gold nanoparticles (AuNPs) are the most famous metallic nanoparticles studied in the biosensor field [269]. They comprise thousands of atoms, which can be electrochemically oxidised or reduced and essentially serve as an electrochemical mediator to improve electron transfer [270]. They offer a large surface to volume ratio due to that nanoscale size, unique electron transfer ability, and great biocompatibility [271]. AuNPs also can be handily conjugated with many biomolecules without affecting their respective biochemical activities [272]. Thiolation of respective biomolecules, e.g., oligonucleotides (DNA-AuNPs) is probably the most simple and best way to attach biomolecules to AgNPs through a self-assembled monolayer (SAM) [273]. In the electrochemical DNA sensor field, AuNPs can be utilised through many kinds of strategies such as a modified DNA probe with AuNPs for signal enhancement, DNA hybridisation detection through AuNP labels, surface modification of electrode with AuNPs to increase DNA probe adsorption, and silver deposition on AuNPs [270]. There are other metallic nanoparticles such as silver, copper, and platinum nanoparticles, but the research on their function as Salmonella biosensors are quite scarce. Iron oxide (Fe3O4) magnetic nanoparticles are also an excellent nanoparticle in biosensor study. It has been used in various research fields such as biotechnology, pharmacology, drug delivery and cell separation. This is due to their excellent superparamagnetic property, biocompatibility, fast electron transfer, low toxicity, and good catalytic action [274,275].
Mesoporous silica nanoparticles (MSN) have received interest in biosensors due to their high load capacity, simple preparation and easy manipulation of morphology, size and pore diameter [276]. An excellent electron transfer of MSN could be achieved through proper manipulation, design and tailoring of the nanostructure [277]. In biosensors, MSN are usually used as a capture agent or controlled release process of biomolecules or redox probes by external stimuli [278].
Indium tin oxide (ITO) is usually employed as an electrode due to its great electrical conductivity, cost-effective nature and special optoelectronic characteristics and transmittance [279,280]. ITO contains hydroxyl groups (-OH) on its surface, which can be further functionalised with a wide variety of chemical compounds to capture biomolecules [242]. ITO can be deposited as a thin film on the electrode and its glass-like properties provide an effective immobilisation of biomolecules through surface modification. However, ITO has a slow electron transfer rate compared to a metallic and carbon-based electrodes [242,248].
Nanowires are a cylindrical nanostructure quite similar to those of carbon nanotubes, but with a lower length to diameter ratio. Usually, the aspect ratios of nanowires are in the range of thousands while nanotubes are far beyond that [281]. Nanowires have an excellent surface to volume ratio, electron transfer properties and improved charge carrier motions compared to bulky wires [281,282]. They can be synthesised from metals (e.g., Cu, Ni, Pt, Au, etc.), metal oxides (e.g., Fe2O3, ZnO, SnO2, etc.) and semiconductors (e.g., Si, InP, GaN, etc.), where each material will directly influence their electrical conductivity [248]. For instance, silver nanowires were found to have extraordinary electrical properties such as rapid response, excellent electrocatalytic behaviour and reproducibility, which then was used as a component in electrochemical immunosensors [277].
The conductive polymer is a class of organic polymer, which has characteristics similar to some inorganic semiconductors and metals (e.g., good electrical conductor) while retaining polymer properties such as flexibility, ease of synthesis and processing, low cost and possible miniaturisation [283]. It can be printed on diverse solid supports and has been used in transistor and electrode fabrication [242]. For example, the conductive organic polymer has been used in glucose oxidase sensors by incorporating organic thin-film transistors (OTFT) into the sensor [284]. To date, conductive organic polymers are usually used as a synergy combination nanocomposite with metal nanoparticles or graphene derivatives. This combination has received considerable attention due to the excellence in electrochemical properties, as well as improved catalytic stability [285].
5.2. Lab-on-Chip Platforms for Rapid Detection of Salmonella
A lab-on-chip (LoC) is a system that brings all the analytical assays conducted on a laboratory scale into a single, miniaturised, and autonomous device. This system works by integrating microfluidic technology coupled with biochemical or chemical processes that serially work to conduct the analysis [286]. Some diagnostic applications that successfully utilise the LoC concept are glucose and HIV detection systems [287,288].
LoC systems have also been implemented for pathogen detection including Salmonella. For example, Tsougeni et al. developed a LoC platform based on an oxygen plasma nanotextured polymeric chip containing bacteria immunoaffinity capturing chamber, chemical lysis, DNA isothermal amplification and a label-free Surface Acoustic Wave (SAW) biosensor. This system successfully detects the bacteria in less than 4.5 h [289]. Another LoC system integrated with loop-mediated isothermal amplification (LAMP) showed a promising detection result within 1.5 h with a low detection limit of 14 CFU mL−1 [290]. The VerebeefTM Detection Kit is one of the commercial detection kits that incorporate multiplex PCR and microarray technology on the LoC system for the detection of multiple pathogenic microorganisms including Salmonella species. This kit has been extensively tested on raw beef trim samples, and it was able to successfully detect each pathogen without any false-positive or false-negative detection [291].
Even though there are only a few numbers of LoC systems have been reported for Salmonella detection, the likelihood of finding the LoC system integrated with an electrochemical system, especially the electrochemical aptasensor system, is scarce. This might be due to the aptamer technology still being at its early stage in miniaturised biosensor technology and probably progressing well in the near future. The integration of LoC systems with a miniaturised electrochemical aptasensor could be a huge leap on Salmonella detection, as this system can provide a very specific, rapid, sensitive, and sustainable microorganism detection.
6. Conclusions and Future Impact of Salmonella Biosensors
The research on pathogen detection, especially on the Salmonella enterica species, is still progressing from year to year, emphasising the importance of a rapid and precise detection to prevent any casualty and fatality to humans. Even though the world statistics of Salmonella infection are gradually declining, provident preparation must be made to prevent any potential outbreak. In this review, we have presented the progression of Salmonella detection technology from the conventional culture-based method to an advanced electrochemical biosensing system. The conventional culture method is still being used nowadays, especially in clinical settings where the target sample (e.g., blood, plasma, stool, etc.) is sometimes difficult to process with the latest sensing technology or the samples might need to be clinically diagnosed in detail, such as for their antimicrobial resistance properties, etc. Rapid detection methods based on immunological assay, molecular assay, mass spectrometry, spectroscopy, optical phenotyping are also reliable and they have their specific niche in the Salmonella detection system. Biosensors are undoubtedly one of the best Salmonella detection systems available, offering versatile applications in various fields, such as medical diagnostic, food safety, environmental monitoring, drug delivery, and many more. The bioreceptor is one of the important aspects of the biosensor to ensure specific and selective detection of the target. Amongst the discussed bioreceptors (i.e., antibody–antigen, bacteriophage, AMPs, nucleic acids, and aptamer), the aptamer was found to be the most reliable bioreceptor due to its outstanding advantages compared to that of the others. The integration of nanomaterials to the already sensitive electrochemical sensing technology can further enhance the Salmonella detection limit down to the femtomolar concentration, as well as the detection of individual Salmonella cells. Theoretically, these combinations could be a plausible move for the development of Salmonella point of care detection devices (in a form of a lab-on-chip device) that have rapid, highly specific, selective, and sensitive detection, are small enough for on-site application (miniaturised form), are potentially easy and cheap to build, and are user friendly. Furthermore, the development of big data analytics and artificial intelligence (AI) technology could further enhance Salmonella biosensor technology to reach the next milestone.
Author Contributions
Conceptualisation, A.A.M. and H.H.H.; formal analysis, M.S.A.; investigation, M.S.A.; resources, M.S.A., H.H.H., N.S.Z. and M.A.N.; data curation, A.A.M., Y.B. and H.H.H.; writing—original draft preparation, M.S.A.; writing—review and editing, A.A.M., Y.B., H.H.H., N.S.Z., M.A.N., M.F.K. and I.A.; visualisation, M.S.A., Y.B. and H.H.H.; supervision, A.A.M., Y.B., H.H.H. and I.A.; project administration, A.A.M., M.F.K. and I.A.; funding acquisition, A.A.M. and I.A. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the Collaborative Research in Engineering, Science, and Technology (CREST) Grant (304/CIPPM/6150181/C121) and the Higher Institution Centre of Excellence (HICoE) Grant (311/CIPPM/4401005) from the Ministry of Higher Education, Malaysia.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
Authors would like to thank Collaborative Research in Engineering, Science & Technology Center (CREST) for their continuous support in this research.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kurtz, J.R.; Goggins, J.A.; McLachlan, J.B. Salmonella infection: Interplay between the bacteria and host immune system. Immunol. Lett. 2017, 190, 42–50. [Google Scholar] [CrossRef]
- Waddington, C.S.; Darton, T.C.; Pollard, A.J. The challenge of enteric fever. J. Infect. 2014, 68, S38–S50. [Google Scholar] [CrossRef] [PubMed]
- Harris, J.B.; Brooks, W.A. Typhoid and paratyphoid (enteric) fever. In Hunter’s Tropical Medicine and Emerging Infectious Diseases; Elsevier: Amsterdam, The Netherlands, 2020; pp. 608–616. [Google Scholar]
- Alhajj, M.; Farhana, A. Enzyme Linked Immunosorbent Assay [Updated 6 February 2021]. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2020. Available online: https://www.ncbi.nlm.nih.gov/books/NBK555922/ (accessed on 8 September 2021).
- International Organization for Standardization (ISO). ISO 6579: 2002 Microbiology of Food and Animal Feeding Stuffs–Horizontal Method for the Detection of Salmonella spp., 2002; International Organization for Standardization IOS: Geneva, Switzerland, 2002; p. 27. [Google Scholar]
- Dilek, Ç.A.M. Lateral Flow Assay for Salmonella Detection and Potential Reagents. In New Insight into Brucella Infection and Foodborne Diseases; IntechOpen: London, UK, 2019. [Google Scholar]
- Fenollar, F.; Raoult, D. Molecular genetic methods for the diagnosis of fastidious microorganisms. APMIS 2004, 112, 785–807. [Google Scholar] [CrossRef] [PubMed]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, E63. [Google Scholar] [CrossRef]
- Lobato, I.M.; O’Sullivan, C.K. Recombinase polymerase amplification: Basics, applications and recent advances. TrAC Trends Anal. Chem. 2018, 98, 19–35. [Google Scholar] [CrossRef] [PubMed]
- Fakruddin, M.D.; Mazumdar, R.M.; Chowdhury, A.; Mannan, K.S.B. Nucleic acid sequence based amplification (NASBA)-prospects and applications. Int. J. Life Sci. Pharma Res. 2012, 2, 106–121. [Google Scholar]
- Ehrenreich, A. DNA microarray technology for the microbiologist: An overview. Appl. Microbiol. Biotechnol. 2006, 73, 255–273. [Google Scholar] [CrossRef]
- Ibrahim, G.M.; Morin, P.M. Salmonella Serotyping Using Whole Genome Sequencing. Front. Microbiol. 2018, 9, 2993. [Google Scholar] [CrossRef]
- Mangmee, S.; Reamtong, O.; Kalambaheti, T.; Roytrakul, S.; Sonthayanon, P. MALDI-TOF mass spectrometry typing for predominant serovars of non-typhoidal Salmonella in a Thai broiler industry. Food Control 2020, 113, 107188. [Google Scholar] [CrossRef]
- Bell, R.L.; Jarvis, K.G.; Ottesen, A.R.; McFarland, M.A.; Brown, E.W. Recent and emerging innovations in Salmonella detection: A food and environmental perspective. Microb. Biotechnol. 2016, 9, 279–292. [Google Scholar] [CrossRef]
- Zhao, X.; Li, M.; Xu, Z. Detection of Foodborne Pathogens by Surface Enhanced Raman Spectroscopy. Front. Microbiol. 2018, 9, 1236. [Google Scholar] [CrossRef]
- Pereira, J.M.; Leme, L.M.; Perdoncini, M.R.F.G.; Valderrama, P.; Março, P.H. Fast Discrimination of Milk Contaminated with Salmonella sp. Via Near-Infrared Spectroscopy. Food Anal. Methods 2018, 11, 1878–1885. [Google Scholar] [CrossRef]
- Bonah, E.; Huang, X.; Aheto, J.H.; Osae, R. Application of hyperspectral imaging as a nondestructive technique for foodborne pathogen detection and characterization. Foodborne Pathog. Dis. 2019, 16, 712–722. [Google Scholar] [CrossRef]
- Abdelhaseib, M.U.; Singh, A.K.; Bhunia, A.K. Simultaneous detection of Salmonella enterica, Escherichia coli and Listeria monocytogenes in food using a light scattering sensor. J. Appl. Microbiol. 2019, 126, 1496–1507. [Google Scholar] [CrossRef] [PubMed]
- Liébana, S.; Brandão, D.; Alegret, S.; Pividori, M.I. Electrochemical immunosensors, genosensors and phagosensors for Salmonella detection. Anal. Methods 2014, 6, 8858–8873. [Google Scholar] [CrossRef]
- Lee, K.-M.; Runyon, M.; Herrman, T.J.; Phillips, R.; Hsieh, J. Review of Salmonella detection and identification methods: Aspects of rapid emergency response and food safety. Food Control 2015, 47, 264–276. [Google Scholar] [CrossRef]
- Lin, L.; Zheng, Q.; Lin, J.; Yuk, H.-G.; Guo, L. Immuno-and nucleic acid-based current technique for Salmonella detection in food. Eur. Food Res. Technol. 2020, 246, 373–395. [Google Scholar] [CrossRef]
- Silva, N.F.D.; Magalhães, J.M.C.S.; Freire, C.; Delerue-Matos, C. Electrochemical biosensors for Salmonella: State of the art and challenges in food safety assessment. Biosens. Bioelectron. 2018, 99, 667–682. [Google Scholar] [CrossRef] [PubMed]
- Ferone, M.; Gowen, A.; Fanning, S.; Scannell, A.G.M. Microbial detection and identification methods: Bench top assays to omics approaches. Compr. Rev. Food Sci. Food Saf. 2020, 19, 3106–3129. [Google Scholar] [CrossRef]
- Tiwari, H.; Kamat, R.S. Cross-reactions in cell-mediated immunity to Salmonella causing enteric fever. J. Med. Microbiol. 1986, 21, 233–237. [Google Scholar] [CrossRef] [PubMed]
- Dehghani, B.; Rasooli, I.; Gargari, S.L.M.; Nadooshan, M.R.J.; Owlia, P.; Nazarian, S. Immunogenicity of Salmonella enterica serovar Enteritidis virulence protein, InvH, and cross-reactivity of its antisera with Salmonella strains. Microbiol. Res. 2013, 168, 84–90. [Google Scholar] [CrossRef]
- Nga, T.V.T.; Karkey, A.; Dongol, S.; Thuy, H.N.; Dunstan, S.; Holt, K.; Tu, L.T.P.; Campbell, J.I.; Chau, T.T.; Chau, N.V.V.; et al. The sensitivity of real-time PCR amplification targeting invasive Salmonella serovars in biological specimens. BMC Infect. Dis. 2010, 10, 125. [Google Scholar] [CrossRef]
- Kostić, T.; Sessitsch, A. Microbial diagnostic microarrays for the detection and typing of food-and water-borne (bacterial) pathogens. Microarrays 2012, 1, 3–24. [Google Scholar] [CrossRef] [PubMed]
- Minoni, U.; Signoroni, A.; Nassini, G. On the application of optical forward-scattering to bacterial identification in an automated clinical analysis perspective. Biosens. Bioelectron. 2015, 68, 536–543. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, D.; Wang, J.; Wu, L.; Huang, Y.; Zhang, Y.; Zhu, M.; Wang, Y.; Zhu, Z.; Yang, C. Trends in miniaturized biosensors for point-of-care testing. TrAC Trends Anal. Chem. 2020, 122, 115701. [Google Scholar] [CrossRef]
- Ansari, N.; Yazdian-Robati, R.; Shahdordizadeh, M.; Wang, Z.; Ghazvini, K. Aptasensors for quantitative detection of Salmonella Typhimurium. Anal. Biochem. 2017, 533, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Naresh, V.; Lee, N. A Review on Biosensors and Recent Development of Nanostructured Materials-Enabled Biosensors. Sensors 2021, 21, 1109. [Google Scholar] [CrossRef] [PubMed]
- Eng, S.-K.; Pusparajah, P.; Ab Mutalib, N.-S.; Ser, H.-L.; Chan, K.-G.; Lee, L.-H. Salmonella: A review on pathogenesis, epidemiology and antibiotic resistance. Front. Life Sci. 2015, 8, 284–293. [Google Scholar] [CrossRef]
- Popoff, M.Y.; Bockemühl, J.; Gheesling, L.L. Supplement 2001 (no. 45) to the Kauffmann–White scheme. Res. Microbiol. 2003, 154, 173–174. [Google Scholar] [CrossRef]
- Reeves, M.W.; Evins, G.M.; Heiba, A.A.; Plikaytis, B.D.; Farmer, J.J. Clonal nature of Salmonella typhi and its genetic relatedness to other salmonellae as shown by multilocus enzyme electrophoresis, and proposal of Salmonella bongori comb. nov. J. Clin. Microbiol. 1989, 27, 313–320. [Google Scholar] [CrossRef]
- Hurley, D.; McCusker, M.P.; Fanning, S.; Martins, M. Salmonella–Host Interactions—Modulation of the Host Innate Immune System. Front. Immunol. 2014, 5, 481. [Google Scholar] [CrossRef] [PubMed]
- Giannella, R.A. Medical Microbiology. In Medical Microbiology, 4th ed.; University of Texas Medical Branch at Galveston: Texas, TX, USA, 1996. [Google Scholar]
- Luo, Y.; Yi, W.; Yao, Y.; Zhu, N.; Qin, P. Characteristic diversity and antimicrobial resistance of Salmonella from gastroenteritis. J. Infect. Chemother. 2018, 24, 251–255. [Google Scholar] [CrossRef] [PubMed]
- Jessica, M.H.; Beau, B.B. Chapter 4: Travel-related infectious diseases, Salmonellosis (Nontyphoidal). In CDC Yellow Book, Centers for Disease Control and Preventions (CDC); Oxford University Press: New York, NY, USA, 2019. [Google Scholar]
- Stanaway, J.D.; Reiner, R.C.; Blacker, B.F.; Goldberg, E.M.; Khalil, I.A.; Troeger, C.E.; Andrews, J.R.; Bhutta, Z.A.; Crump, J.A.; Im, J.; et al. The global burden of typhoid and paratyphoid fevers: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Infect. Dis. 2019, 19, 369–381. [Google Scholar] [CrossRef]
- Vergese, A. The “typhoid state” revisited. Am. J. Med. 1985, 79, 370–372. [Google Scholar] [CrossRef]
- Bhutta, Z.A. Impact of age and drug resistance on mortality in typhoid fever. Arch. Dis. Child. 1996, 75, 214–217. [Google Scholar] [CrossRef]
- Qamar, F.N.; Azmatullah, A.; Bhutta, Z.A. Challenges in measuring complications and death due to invasive Salmonella infections. Vaccine 2015, 33, C16–C20. [Google Scholar] [CrossRef]
- Andoh, L.A.; Ahmed, S.; Olsen, J.E.; Obiri-Danso, K.; Newman, M.J.; Opintan, J.A.; Barco, L.; Dalsgaard, A. Prevalence and characterization of Salmonella among humans in Ghana. Trop. Med. Health 2017, 45, 1–11. [Google Scholar] [CrossRef]
- Muhammad, E.N.; Abdul Mutalip, M.H.; Hasim, M.H.; Paiwai, F.; Pan, S.; Mahmud, M.A.F.; Yeop, N.; Tee, G.H.; Senin, A.A.; Aris, T. The burden of typhoid fever in Klang Valley, Malaysia, 2011–2015. BMC Infect. Dis. 2020, 20, 843. [Google Scholar] [CrossRef]
- Chau, T.T.; Campbell, J.I.; Galindo, C.M.; Hoang, N.V.M.; Diep, T.S.; Nga, T.T.T.; Chau, N.V.V.; Tuan, P.Q.; Page, A.L.; Ochiai, R.L. Antimicrobial drug resistance of Salmonella enterica serovar Typhi in Asia and molecular mechanism of reduced susceptibility to the fluoroquinolones. Antimicrob. Agents Chemother. 2007, 51, 4315–4323. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Rizvi, M.; Berry, N. Rising prevalence of enteric fever due to multidrug-resistant Salmonella: An epidemiological study. J. Med. Microbiol. 2008, 57, 1247–1250. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, X.; Biswas, S.; Paudyal, N.; Pan, H.; Li, X.; Fang, W.; Yue, M. Antibiotic resistance in Salmonella Typhimurium isolates recovered from the food chain through national antimicrobial resistance monitoring system between 1996 and 2016. Front. Microbiol. 2019, 10, 985. [Google Scholar] [CrossRef]
- Mather, A.E.; Reid, S.W.J.; Maskell, D.J.; Parkhill, J.; Fookes, M.C.; Harris, S.R.; Brown, D.J.; Coia, J.E.; Mulvey, M.R.; Gilmour, M.W. Distinguishable epidemics of multidrug-resistant Salmonella Typhimurium DT104 in different hosts. Science 2013, 341, 1514–1517. [Google Scholar] [CrossRef]
- Helms, M.; Ethelberg, S.; Mølbak, K.; Group, D.S. International Salmonella typhimurium DT104 infections, 1992–2001. Emerg. Infect. Dis. 2005, 11, 859. [Google Scholar] [CrossRef]
- Andrews, J.R.; Ryan, E.T. Diagnostics for invasive Salmonella infections: Current challenges and future directions. Vaccine 2015, 33, C8–C15. [Google Scholar] [CrossRef] [PubMed]
- Mogasale, V.; Ramani, E.; Mogasale, V.V.; Park, J. What proportion of Salmonella Typhi cases are detected by blood culture? A systematic literature review. Ann. Clin. Microbiol. Antimicrob. 2016, 15, 32. [Google Scholar] [CrossRef]
- Siala, M.; Barbana, A.; Smaoui, S.; Hachicha, S.; Marouane, C.; Kammoun, S.; Gdoura, R.; Messadi-Akrout, F. Screening and Detecting Salmonella in Different Food Matrices in Southern Tunisia Using a Combined Enrichment/Real-Time PCR Method: Correlation with Conventional Culture Method. Front. Microbiol. 2017, 8, 2416. [Google Scholar] [CrossRef] [PubMed]
- Gast, R.K.; Porter, R.E., Jr. Chapter 16: Salmonella Infections. In Diseases of Poultry, 14th ed.; John Wiley & Son Inc.: Hoboken, NJ, USA, 2020; pp. 717–753. [Google Scholar]
- Fung, D.Y.C. Rapid methods and automation in microbiology. In Proceedings of the Workshop MRAMA, Barcelona, Spain, 20–24 November 2002. [Google Scholar]
- Ge, B.; Meng, J. Advanced technologies for pathogen and toxin detection in foods: Current applications and future directions. JALA J. Assoc. Lab. Autom. 2009, 14, 235–241. [Google Scholar] [CrossRef]
- Klingler, J.M.; Stowe, R.P.; Obenhuber, D.C.; Groves, T.O.; Mishra, S.K.; Pierson, D.L. Evaluation of the Biolog automated microbial identification system. Appl. Environ. Microbiol. 1992, 58, 2089–2092. [Google Scholar] [CrossRef] [PubMed]
- El-Liethy, M.A.; Hemdan, B.A.; El-Taweel, G.E. Prevalence of E. coli, Salmonella, and Listeria spp. as potential pathogens: A comparative study for biofilm of sink drain environment. J. Food Saf. 2020, 40, e12816. [Google Scholar] [CrossRef]
- Sørensen, L.L.; Alban, L.; Nielsen, B.; Dahl, J. The correlation between Salmonella serology and isolation of Salmonella in Danish pigs at slaughter. Vet. Microbiol. 2004, 101, 131–141. [Google Scholar] [CrossRef]
- Alakomi, H.; Saarela, M. Salmonella importance and current status of detection and surveillance methods. Qual. Assur. Saf. Crop. Foods 2009, 1, 142–152. [Google Scholar] [CrossRef]
- Maciorowski, K.G.; Herrera, P.; Jones, F.T.; Pillai, S.D.; Ricke, S.C. Cultural and immunological detection methods for Salmonella spp. in animal feeds—A review. Vet. Res. Commun. 2006, 30, 127–137. [Google Scholar] [CrossRef]
- Manafi, M. New developments in chromogenic and fluorogenic culture media. Int. J. Food Microbiol. 2000, 60, 205–218. [Google Scholar] [CrossRef]
- Perry, J.D.; Freydière, A.M. The application of chromogenic media in clinical microbiology. J. Appl. Microbiol. 2007, 103, 2046–2055. [Google Scholar] [CrossRef] [PubMed]
- Lin, A. V Direct ELISA. In ELISA; Springer: Berlin/Heidelberg, Germany, 2015; pp. 61–67. [Google Scholar]
- Alhabbab, R.Y. Precipitation and Agglutination Reactions. In Basic Serological Testing; Springer: Berlin/Heidelberg, Germany, 2018; pp. 23–30. [Google Scholar]
- Bahadır, E.B.; Sezgintürk, M.K. Lateral flow assays: Principles, designs and labels. TrAC Trends Anal. Chem. 2016, 82, 286–306. [Google Scholar] [CrossRef]
- Blivet, D.; Soumet, C.; Ermel, G.; Colin, P. Rapid detection methods for pathogens. In Proceedings of the 3rd Karlsruhe Nutrition Symposium European Research towards Safer and Better Food, Review and Transfer Congress Centre, Karlsruhe, Germany, 18–20 October 1998; p. 3. [Google Scholar]
- Tietjen, M.; Fung, D.Y.C. Salmonellae and Food Safety. Crit. Rev. Microbiol. 1995, 21, 53–83. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Olson, N.A.; You, W.; Haugland, R.P. A sensitive competitive ELISA for 2, 4-dinitrophenol using 3, 6-fluorescein diphosphate as a fluorogenic substrate. J. Immunol. Methods 1992, 149, 261–266. [Google Scholar] [CrossRef]
- Sue, M.J.; Yeap, S.K.; Omar, A.R.; Tan, S.W. Application of PCR-ELISA in Molecular Diagnosis. Biomed Res. Int. 2014, 2014, 653014. [Google Scholar] [CrossRef]
- Peng, H.; Huang, Z.; Wu, W.; Liu, M.; Huang, K.; Yang, Y.; Deng, H.; Xia, X.; Chen, W. Versatile High-Performance Electrochemiluminescence ELISA Platform Based on a Gold Nanocluster Probe. ACS Appl. Mater. Interfaces 2019, 11, 24812–24819. [Google Scholar] [CrossRef]
- De La Rica, R.; Stevens, M.M. Plasmonic ELISA for the ultrasensitive detection of disease biomarkers with the naked eye. Nat. Nanotechnol. 2012, 7, 821–824. [Google Scholar] [CrossRef]
- Thorns, C.J.; McLaren, I.M.; Sojka, M.G. The use of latex particle agglutination to specifically detect Salmonella enteritidis. Int. J. Food Microbiol. 1994, 21, 47–53. [Google Scholar] [CrossRef]
- Eijkelkamp, J.M.; Aarts, H.J.M.; Van der Fels-Klerx, H.J. Suitability of rapid detection methods for Salmonella in poultry slaughterhouses. Food Anal. Methods 2009, 2, 1–13. [Google Scholar] [CrossRef]
- Love, D.C.; Sobsey, M.D. Simple and rapid F+ coliphage culture, latex agglutination, and typing assay to detect and source track fecal contamination. Appl. Environ. Microbiol. 2007, 73, 4110–4118. [Google Scholar] [CrossRef]
- Magwood, S.E.; Annau, E. The adsorption of somatic antigens of salmonella by polystyrene latex particles. Can. J. Comp. Med. Vet. Sci. 1961, 25, 69. [Google Scholar] [PubMed]
- Cheng-yu, Y. Using latex Agglutination Text Detecting Salmonella in Commercial Feed. Chin. Qinghai J. Anim. Vet. Sci. 2006, 2. [Google Scholar]
- Thompson, W.; Crabtree, D.; Bastin, B.; Koch, K.; Hahs, M.; Sohier, D. AOAC Validation Study of a Real-Time PCR Workflow for Salmonella Detection in Large Test Portions of Cocoa and Chocolate Products. IAFP 2021. Available online: https://assets.thermofisher.cn/TFS-Assets/MBD/posters/Poster-IAFP-2021-QLabs-AOAC-SureTect-Salm-LT2659A.pdf (accessed on 8 September 2021).
- Mao, X.; Wang, W.; Du, T.-E. Rapid quantitative immunochromatographic strip for multiple proteins test. Sens. Actuators B Chem. 2013, 186, 315–320. [Google Scholar] [CrossRef]
- Kang, J.; Kim, M.-G. Advancements in DNA-assisted Immunosensors. BioChip J. 2020, 14, 18–31. [Google Scholar] [CrossRef]
- Ma, K.-S.; Ma, Y.; Chiou, F. Nanotechnology Applications in Polymerase Chain Reaction (PCR) BT-Encyclopedia of Nanotechnology; Bhushan, B., Ed.; Springer: Dordrecht, The Netherlands, 2016; pp. 2869–2876. ISBN 978-94-017-9780-1. [Google Scholar]
- Hønsvall, B.K.; Robertson, L.J. From research lab to standard environmental analysis tool: Will NASBA make the leap? Water Res. 2017, 109, 389–397. [Google Scholar] [CrossRef]
- Salamin, O.; Kuuranne, T.; Saugy, M.; Leuenberger, N. Loop-mediated isothermal amplification (LAMP) as an alternative to PCR: A rapid on-site detection of gene doping. Drug Test. Anal. 2017, 9, 1731–1737. [Google Scholar] [CrossRef]
- Watson, J.D. The Polymerase Chain Reaction; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2012; ISBN 1461202574. [Google Scholar]
- Chin, W.H.; Sun, Y.; Høgberg, J.; Quyen, T.L.; Engelsmann, P.; Wolff, A.; Bang, D.D. Direct PCR–A rapid method for multiplexed detection of different serotypes of Salmonella in enriched pork meat samples. Mol. Cell. Probes 2017, 32, 24–32. [Google Scholar] [CrossRef]
- Vinayaka, A.C.; Ngo, T.A.; Kant, K.; Engelsmann, P.; Dave, V.P.; Shahbazi, M.-A.; Wolff, A.; Bang, D.D. Rapid detection of Salmonella enterica in food samples by a novel approach with combination of sample concentration and direct PCR. Biosens. Bioelectron. 2019, 129, 224–230. [Google Scholar] [CrossRef]
- Park, S.H.; Aydin, M.; Khatiwara, A.; Dolan, M.C.; Gilmore, D.F.; Bouldin, J.L.; Ahn, S.; Ricke, S.C. Current and emerging technologies for rapid detection and characterization of Salmonella in poultry and poultry products. Food Microbiol. 2014, 38, 250–262. [Google Scholar] [CrossRef]
- De Medici, D.; Croci, L.; Delibato, E.; Di Pasquale, S.; Filetici, E.; Toti, L. Evaluation of DNA extraction methods for use in combination with SYBR green I real-time PCR to detect Salmonella enterica serotype enteritidis in poultry. Appl. Environ. Microbiol. 2003, 69, 3456–3461. [Google Scholar] [CrossRef]
- Malorny, B.; Huehn, S.; Dieckmann, R.; Krämer, N.; Helmuth, R. Polymerase chain reaction for the rapid detection and serovar identification of Salmonella in food and feeding stuff. Food Anal. Methods 2009, 2, 81–95. [Google Scholar] [CrossRef]
- Azinheiro, S.; Carvalho, J.; Prado, M.; Garrido-Maestu, A. Multiplex Detection of Salmonella spp., E. coli O157 and L. monocytogenes by qPCR Melt Curve Analysis in Spiked Infant Formula. Microorganisms 2020, 8, 1359. [Google Scholar] [CrossRef] [PubMed]
- Ruan, J.; Wang, W.; Zhang, T.; Zheng, T.; Zheng, J.; Yu, S.; Yu, D.; Huang, Y. Establishment of a duplex real-time qPCR method for detection of Salmonella spp. and Serratia fonticola in fishmeal. AMB Express 2020, 10, 1–8. [Google Scholar] [CrossRef]
- Parker, A.M.; Mohler, V.L.; Gunn, A.A.; House, J.K. Development of a qPCR for the detection and quantification of Salmonella spp. in sheep feces and tissues. J. Vet. Diagn. Investig. 2020, 32, 835–843. [Google Scholar] [CrossRef]
- Dmitric, M.; Vidanovic, D.; Matovic, K.; Sekler, M.; Saric, L.; Arsic, M.; Karabasil, N. In-house validation of real-time PCR methods for detecting the INV A and TTR genes of Salmonella spp. in food. J. Food Process. Preserv. 2018, 42, e13455. [Google Scholar] [CrossRef]
- Kulshreshtha, G.; Borza, T.; Rathgeber, B.; Stratton, G.S.; Thomas, N.A.; Critchley, A.; Hafting, J.; Prithiviraj, B. Red Seaweeds Sarcodiotheca gaudichaudii and Chondrus crispus down Regulate Virulence Factors of Salmonella Enteritidis and Induce Immune Responses in Caenorhabditis elegans. Front. Microbiol. 2016, 7, 421. [Google Scholar] [CrossRef] [PubMed]
- Tatavarthy, A.; Cannons, A. Real-time PCR detection of Salmonella species using a novel target: The outer membrane porin F gene (ompF). Lett. Appl. Microbiol. 2010, 50, 645–652. [Google Scholar] [CrossRef] [PubMed]
- McCabe, E.M.; Burgess, C.M.; O’Regan, E.; McGuinness, S.; Barry, T.; Fanning, S.; Duffy, G. Development and evaluation of DNA and RNA real-time assays for food analysis using the hilA gene of Salmonella enterica subspecies enterica. Food Microbiol. 2011, 28, 447–456. [Google Scholar] [CrossRef]
- Hassena, A.B.; Barkallah, M.; Fendri, I.; Grosset, N.; Neila, I.B.; Gautier, M.; Gdoura, R. Real time PCR gene profiling and detection of Salmonella using a novel target: The siiA gene. J. Microbiol. Methods 2015, 109, 9–15. [Google Scholar] [CrossRef]
- Ganz, K.; Gill, A. Inhibition of polymerase chain reaction for the detection of Escherichia coli O157: H7 and Salmonella enterica on walnut kernels. Food Microbiol. 2013, 35, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Cossu, A.; Levin, R.E. Rapid conventional PCR and real-time-qPCR detection of low numbers of Salmonella enterica from ground beef without enrichment. Food Biotechnol. 2014, 28, 96–105. [Google Scholar] [CrossRef]
- Margot, H.; Stephan, R.; Guarino, S.; Jagadeesan, B.; Chilton, D.; O’Mahony, E.; Iversen, C. Inclusivity, exclusivity and limit of detection of commercially available real-time PCR assays for the detection of Salmonella. Int. J. Food Microbiol. 2013, 165, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Zhang, Z.; Sun, X.; Pan, Y.; Zhao, Y. Development of a quantitative real-time PCR assay for viable Salmonella spp. without enrichment. Food Control 2015, 57, 185–189. [Google Scholar] [CrossRef]
- Li, Y.; Fan, P.; Zhou, S.; Zhang, L. Loop-mediated isothermal amplification (LAMP): A novel rapid detection platform for pathogens. Microb. Pathog. 2017, 107, 54–61. [Google Scholar] [CrossRef]
- Souii, A.; M’hadheb-Gharbi, M.B.; Gharbi, J. Nucleic acid-based biotechnologies for food-borne pathogen detection using routine time-intensive culture-based methods and fast molecular diagnostics. Food Sci. Biotechnol. 2016, 25, 11–20. [Google Scholar] [CrossRef]
- Yang, Q.; Wang, F.; Jones, K.L.; Meng, J.; Prinyawiwatkul, W.; Ge, B. Evaluation of loop-mediated isothermal amplification for the rapid, reliable, and robust detection of Salmonella in produce. Food Microbiol. 2015, 46, 485–493. [Google Scholar] [CrossRef]
- Kokkinos, P.A.; Ziros, P.G.; Bellou, M.; Vantarakis, A. Loop-mediated isothermal amplification (LAMP) for the detection of Salmonella in food. Food Anal. Methods 2014, 7, 512–526. [Google Scholar] [CrossRef]
- Techathuvanan, C.; Draughon, F.A.; D’Souza, D.H. Loop-mediated isothermal amplification (LAMP) for the rapid and sensitive detection of Salmonella Typhimurium from pork. J. Food Sci. 2010, 75, M165–M172. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Jiang, X.; Qu, Y.; Pan, R.; Pang, X.; Jiang, Y.; Man, C. Salmonella detection in powdered dairy products using a novel molecular tool. J. Dairy Sci. 2017, 100, 3480–3496. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Zou, D.; Dong, D.; Yang, Z.; Ao, D.; Liu, W.; Huang, L. Development of a multiplex loop-mediated isothermal amplification method for the simultaneous detection of Salmonella spp. and Vibrio parahaemolyticus. Sci. Rep. 2017, 7, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.P.; Chen, S.H.; Levin, R.E. Application of ethidium bromide monoazide for quantification of viable and dead cells of Salmonella enterica by real-time loop-mediated isothermal amplification. J. Microbiol. Methods 2015, 117, 41–48. [Google Scholar] [CrossRef]
- Deiman, B.; van Aarle, P.; Sillekens, P. Characteristics and applications of nucleic acid sequence-based amplification (NASBA). Mol. Biotechnol. 2002, 20, 163–179. [Google Scholar] [CrossRef]
- Zhai, L.; Liu, H.; Chen, Q.; Lu, Z.; Zhang, C.; Lv, F.; Bie, X. Development of a real-time nucleic acid sequence–based amplification assay for the rapid detection of Salmonella spp. from food. Braz. J. Microbiol. 2019, 50, 255–261. [Google Scholar] [CrossRef]
- D’souza, D.H.; Jaykus, L. Nucleic acid sequence based amplification for the rapid and sensitive detection of Salmonella enterica from foods. J. Appl. Microbiol. 2003, 95, 1343–1350. [Google Scholar] [CrossRef]
- Mollasalehi, H.; Yazdanparast, R. An improved non-crosslinking gold nanoprobe-NASBA based on 16S rRNA for rapid discriminative bio-sensing of major salmonellosis pathogens. Biosens. Bioelectron. 2013, 47, 231–236. [Google Scholar] [CrossRef]
- Chen, J.; Liu, X.; Chen, J.; Guo, Z.; Wang, Y.; Chen, G.; Chen, X.; Yan, Q.; Yang, P.; Li, R. Development of a rapid test method for Salmonella enterica detection based on fluorescence probe-based recombinase polymerase amplification. Food Anal. Methods 2019, 12, 1791–1798. [Google Scholar] [CrossRef]
- Rosser, A.; Rollinson, D.; Forrest, M.; Webster, B.L. Isothermal Recombinase Polymerase amplification (RPA) of Schistosoma haematobium DNA and oligochromatographic lateral flow detection. Parasit. Vectors 2015, 8, 1–5. [Google Scholar] [CrossRef]
- Dao, T.N.T.; Lee, E.Y.; Koo, B.; Jin, C.E.; Lee, T.Y.; Shin, Y. A microfluidic enrichment platform with a recombinase polymerase amplification sensor for pathogen diagnosis. Anal. Biochem. 2018, 544, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Huang, H.; Zhu, P.; Yan, X.; Fan, J.; Jiang, J.; Xu, J. Recombinase polymerase amplification combined with lateral flow dipstick for equipment-free detection of Salmonella in shellfish. Bioprocess Biosyst. Eng. 2018, 41, 603–611. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Man, Y.; Li, A.; Liang, G.; Jin, X.; Pan, L. Detection of Salmonella enteritidis and Salmonella typhimurium in foods using a rapid, multiplex real-time recombinase polymerase amplification assay. J. Food Saf. 2020, 40, e12784. [Google Scholar] [CrossRef]
- Chen, Z.; Zhong, H.; Luo, H.; Zhang, R.; Huang, J. Recombinase polymerase amplification combined with unmodified gold nanoparticles for Salmonella detection in milk. Food Anal. Methods 2019, 12, 190–197. [Google Scholar] [CrossRef]
- Gao, W.; Huang, H.; Zhang, Y.; Zhu, P.; Yan, X.; Fan, J.; Chen, X. Recombinase polymerase amplification-based assay for rapid detection of Listeria monocytogenes in food samples. Food Anal. Methods 2017, 10, 1972–1981. [Google Scholar] [CrossRef]
- Schena, M.; Shalon, D.; Davis, R.W.; Brown, P.O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 1995, 270, 467–470. [Google Scholar] [CrossRef]
- Raspoet, R.; Appia-Ayme, C.; Shearer, N.; Martel, A.; Pasmans, F.; Haesebrouck, F.; Ducatelle, R.; Thompson, A.; Van Immerseel, F. Microarray-based detection of Salmonella enterica serovar Enteritidis genes involved in chicken reproductive tract colonization. Appl. Environ. Microbiol. 2014, 80, 7710–7716. [Google Scholar] [CrossRef]
- Litrup, E.; Torpdahl, M.; Malorny, B.; Huehn, S.; Helms, M.; Christensen, H.; Nielsen, E.M. DNA microarray analysis of Salmonella serotype Typhimurium strains causing different symptoms of disease. BMC Microbiol. 2010, 10, 96. [Google Scholar] [CrossRef]
- Koyuncu, S.; Andersson, G.; Vos, P.; Häggblom, P. DNA microarray for tracing Salmonella in the feed chain. Int. J. Food Microbiol. 2011, 145, S18–S22. [Google Scholar] [CrossRef]
- Guo, D.; Liu, B.; Liu, F.; Cao, B.; Chen, M.; Hao, X.; Feng, L.; Wang, L. Development of a DNA microarray for molecular identification of all 46 Salmonella O serogroups. Appl. Environ. Microbiol. 2013, 79, 3392–3399. [Google Scholar] [CrossRef]
- Ronholm, J. Editorial: Game Changer—Next Generation Sequencing and Its Impact on Food Microbiology. Front. Microbiol. 2018, 9, 363. [Google Scholar] [CrossRef] [PubMed]
- Cheng, K.; Chui, H.; Domish, L.; Hernandez, D.; Wang, G. Recent development of mass spectrometry and proteomics applications in identification and typing of bacteria. Proteom.–Clin. Appl. 2016, 10, 346–357. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.-M.; Kim, E.; Kim, D.; Baek, J.; Yoon, H.; Kim, H.-Y. Rapid Detection of Salmonella Enteritidis, Typhimurium, and Thompson by Specific Peak Analysis Using Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry. Foods 2021, 10, 933. [Google Scholar] [CrossRef] [PubMed]
- Callahan, J.H.; McFarland, M.A.; Musser, S.M.; Bell, R.; Andrzejewski, D. Anyl 282-intact protein liquid chromatography mass spectrometry for bacterial identification and differentiation. In Proceedings of the Abstracts of Papers of the American Chemical Society, Washington, DC, USA, 16 August 2009; Volume 238. [Google Scholar]
- McFarland, M.A.; Andrzejewski, D.; Musser, S.M.; Callahan, J.H. Platform for identification of Salmonella serovar differentiating bacterial proteins by top-down mass spectrometry: S. Typhimurium vs S. Heidelberg. Anal. Chem. 2014, 86, 6879–6886. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.; Dent, G. Modern Raman Spectroscopy: A Practical Approach; John Wiley & Sons: Hoboken, NJ, USA, 2019; ISBN 1119440556. [Google Scholar]
- Duan, N.; Chang, B.; Zhang, H.; Wang, Z.; Wu, S. Salmonella typhimurium detection using a surface-enhanced Raman scattering-based aptasensor. Int. J. Food Microbiol. 2016, 218, 38–43. [Google Scholar] [CrossRef]
- Alexandrakis, D.; Downey, G.; Scannell, A.G.M. Detection and identification of bacteria in an isolated system with near-infrared spectroscopy and multivariate analysis. J. Agric. Food Chem. 2008, 56, 3431–3437. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Gao, X.; Qi, W.-L.; Wang, Y.; Wang, X.; Zhou, J.; Lu, D.; Chen, B. Advances in differentiation and identification of foodborne bacteria using near infrared spectroscopy. Anal. Methods 2021, 13, 2558–2566. [Google Scholar] [CrossRef]
- Gowen, A.A.; Feng, Y.; Gaston, E.; Valdramidis, V. Recent applications of hyperspectral imaging in microbiology. Talanta 2015, 137, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Michael, M.; Phebus, R.K.; Amamcharla, J. Hyperspectral imaging of common foodborne pathogens for rapid identification and differentiation. Food Sci. Nutr. 2019, 7, 2716–2725. [Google Scholar] [CrossRef] [PubMed]
- Seo, Y.W.; Yoon, S.-C.; Park, B.; Windham, W.R.; Lawrence, K.C. Development of Hyperspectral Imaging Technique for Salmonella Enteritidis and Typhimurium on Agar Plates. Appl. Eng. Agric. 2014, 30, 495–506. [Google Scholar]
- Buzalewicz, I.; Suchwałko, A.; Trzciński, P.; Sas-Paszt, L.; Sumorok, B.; Kowal, K.; Kozera, R.; Wieliczko, A.; Podbielska, H. Integrated multi-channel optical system for bacteria characterization and its potential use for monitoring of environmental bacteria. Biomed. Opt. Express 2019, 10, 1165–1183. [Google Scholar] [CrossRef]
- Kim, H.; Singh, A.K.; Bhunia, A.K.; Bae, E. Laser-induced speckle scatter patterns in Bacillus colonies. Front. Microbiol. 2014, 5, 537. [Google Scholar] [CrossRef]
- Singh, A.K.; Bettasso, A.M.; Bae, E.; Rajwa, B.; Dundar, M.M.; Forster, M.D.; Liu, L.; Barrett, B.; Lovchik, J.; Robinson, J.P. Laser optical sensor, a label-free on-plate Salmonella enterica colony detection tool. MBio 2014, 5, e01019-13. [Google Scholar] [CrossRef]
- McNaught, A.D.; Wilkinson, A. Compendium of Chemical Terminology; Blackwell Science Oxford: Oxford, UK, 1997; Volume 1669. [Google Scholar]
- Bagheryan, Z.; Raoof, J.-B.; Golabi, M.; Turner, A.P.F.; Beni, V. Diazonium-based impedimetric aptasensor for the rapid label-free detection of Salmonella typhimurium in food sample. Biosens. Bioelectron. 2016, 80, 566–573. [Google Scholar] [CrossRef]
- Malvano, F.; Pilloton, R.; Albanese, D. A novel impedimetric biosensor based on the antimicrobial activity of the peptide nisin for the detection of Salmonella spp. Food Chem. 2020, 325, 126868. [Google Scholar] [CrossRef] [PubMed]
- Melo, A.M.A.; Furtado, R.F.; de Fatima Borges, M.; Biswas, A.; Cheng, H.N.; Alves, C.R. Performance of an amperometric immunosensor assembled on carboxymethylated cashew gum for Salmonella detection. Microchem. J. 2021, 167, 106268. [Google Scholar] [CrossRef]
- Jia, F.; Duan, N.; Wu, S.; Dai, R.; Wang, Z.; Li, X. Impedimetric Salmonella aptasensor using a glassy carbon electrode modified with an electrodeposited composite consisting of reduced graphene oxide and carbon nanotubes. Microchim. Acta 2016, 183, 337–344. [Google Scholar] [CrossRef]
- Velusamy, V.; Arshak, K.; Korostynska, O.; Oliwa, K.; Adley, C. An overview of foodborne pathogen detection: In the perspective of biosensors. Biotechnol. Adv. 2010, 28, 232–254. [Google Scholar] [CrossRef]
- Cordova-Huaman, A.V.; Jauja-Ccana, V.R.; La Rosa-Toro, A. Low-cost smartphone-controlled potentiostat based on Arduino for teaching electrochemistry fundamentals and applications. Heliyon 2021, 7, e06259. [Google Scholar] [CrossRef]
- Dos Santos Schneid, A.; Rodrigues, K.L.; Chemello, D.; Tondo, E.C.; Ayub, M.A.Z.; Aleixo, J.A.G. Evaluation of an indirect ELISA for the detection of Salmonella in chicken meat. Braz. J. Microbiol. 2006, 37, 350–355. [Google Scholar] [CrossRef]
- Jesudason, M.V.; Sridharan, G.; Mukundan, S.; John, T.J. Vi-specific latex agglutination for early and rapid detection of Salmonella serotype typhi in blood cultures. Diagn. Microbiol. Infect. Dis. 1994, 18, 75–78. [Google Scholar] [CrossRef]
- Singh, J.; Sharma, S.; Nara, S. Nanogold based lateral flow assay for the detection of Salmonella typhi in environmental water samples. Anal. Methods 2015, 7, 9281–9288. [Google Scholar] [CrossRef]
- Kumar, R.; Surendran, P.K.; Thampuran, N. Evaluation of culture, ELISA and PCR assays for the detection of Salmonella in seafood. Lett. Appl. Microbiol. 2008, 46, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Ogunremi, D.; Dupras, A.A.; Naushad, S.; Gao, R.; Duceppe, M.-O.; Omidi, K.; Márquez, I.G.; Huang, H.; Goodridge, L.; Lévesque, R.C.; et al. A New Whole Genome Culture-Independent Diagnostic Test (WG-CIDT) for Rapid Detection of Salmonella in Lettuce. Front. Microbiol. 2020, 11, 602. [Google Scholar] [CrossRef] [PubMed]
- Kuhns, M.; Zautner, A.E.; Rabsch, W.; Zimmermann, O.; Weig, M.; Bader, O.; Groß, U. Rapid discrimination of Salmonella enterica serovar Typhi from other serovars by MALDI-TOF mass spectrometry. PLoS ONE 2012, 7, e40004. [Google Scholar]
- Appaturi, J.N.; Pulingam, T.; Thong, K.L.; Muniandy, S.; Ahmad, N.; Leo, B.F. Rapid and sensitive detection of Salmonella with reduced graphene oxide-carbon nanotube based electrochemical aptasensor. Anal. Biochem. 2020, 589, 113489. [Google Scholar] [CrossRef]
- Bhalla, N.; Jolly, P.; Formisano, N.; Estrela, P. Introduction to biosensors. Essays Biochem. 2016, 60, 1–8. [Google Scholar] [CrossRef]
- Azizi, S.; Gholivand, M.-B.; Amiri, M.; Manouchehri, I. DNA biosensor based on surface modification of ITO by physical vapor deposition of gold and carbon quantum dots modified with neutral red as an electrochemical redox probe. Microchem. J. 2020, 159, 105523. [Google Scholar] [CrossRef]
- Ge, C.; Yuan, R.; Yi, L.; Yang, J.; Zhang, H.; Li, L.; Nian, W.; Yi, G. Target-induced aptamer displacement on gold nanoparticles and rolling circle amplification for ultrasensitive live Salmonella typhimurium electrochemical biosensing. J. Electroanal. Chem. 2018, 826, 174–180. [Google Scholar] [CrossRef]
- Purcell, R.V.; Pearson, J.; Frizelle, F.A.; Keenan, J.I. Comparison of standard, quantitative and digital PCR in the detection of enterotoxigenic Bacteroides fragilis. Sci. Rep. 2016, 6, 34554. [Google Scholar] [CrossRef] [PubMed]
- Hyeon, J.-Y.; Chon, J.-W.; Choi, I.-S.; Park, C.; Kim, D.-E.; Seo, K.-H. Development of RNA aptamers for detection of Salmonella Enteritidis. J. Microbiol. Methods 2012, 89, 79–82. [Google Scholar] [CrossRef] [PubMed]
- Sheikhzadeh, E.; Chamsaz, M.; Turner, A.P.F.; Jager, E.W.H.; Beni, V. Label-free impedimetric biosensor for Salmonella Typhimurium detection based on poly [pyrrole-co-3-carboxyl-pyrrole] copolymer supported aptamer. Biosens. Bioelectron. 2016, 80, 194–200. [Google Scholar] [CrossRef]
- Duan, Y.F.; Ning, Y.; Song, Y.; Deng, L. Fluorescent aptasensor for the determination of Salmonella typhimurium based on a graphene oxide platform. Microchim. Acta 2014, 181, 647–653. [Google Scholar] [CrossRef]
- Clark, L.C., Jr.; Lyons, C. Electrode systems for continuous monitoring in cardiovascular surgery. Ann. N. Y. Acad. Sci. 1962, 102, 29–45. [Google Scholar] [CrossRef]
- Yoo, E.-H.; Lee, S.-Y. Glucose Biosensors: An Overview of Use in Clinical Practice. Sensors 2010, 10, 4558–4576. [Google Scholar] [CrossRef]
- Jayanthi, V.S.A.; Das, A.B.; Saxena, U. Recent advances in biosensor development for the detection of cancer biomarkers. Biosens. Bioelectron. 2017, 91, 15–23. [Google Scholar] [CrossRef]
- Ameri, M.; Shabaninejad, Z.; Movahedpour, A.; Sahebkar, A.; Mohammadi, S.; Hosseindoost, S.; Ebrahimi, M.S.; Savardashtaki, A.; Karimipour, M.; Mirzaei, H. Biosensors for detection of Tau protein as an Alzheimer’s disease marker. Int. J. Biol. Macromol. 2020, 162, 1100–1108. [Google Scholar] [CrossRef]
- Lowe, C.R. Biosensors. Trends Biotechnol. 1984, 2, 59–65. [Google Scholar] [CrossRef]
- Grieshaber, D.; MacKenzie, R.; Vörös, J.; Reimhult, E. Electrochemical Biosensors—Sensor Principles and Architectures. Sensors 2008, 8, 1400–1458. [Google Scholar] [CrossRef]
- Kumar, A.; Malinee, M.; Dhiman, A.; Kumar, A.; Sharma, T.K. Chapter 2—Aptamer Technology for the Detection of Foodborne Pathogens and Toxins. In Advance Biosensors for Healthcare Application; Elsevier: Amsterdam, The Netherlands, 2019; pp. 45–69. ISBN 978-0-12-815743-5. [Google Scholar]
- Shen, Y.; Xu, L.; Li, Y. Biosensors for rapid detection of Salmonella in food: A review. Compr. Rev. Food Sci. Food Saf. 2021, 20, 149–197. [Google Scholar] [CrossRef]
- Nayak, M.; Kotian, A.; Marathe, S.; Chakravortty, D. Detection of microorganisms using biosensors—A smarter way towards detection techniques. Biosens. Bioelectron. 2009, 25, 661–667. [Google Scholar] [CrossRef] [PubMed]
- Evtugyn, G. Biosensors: Essentials; Springer: Berlin/Heidelberg, Germany, 2014; Volume 84, pp. 21–97. [Google Scholar]
- Singh, R.; Mukherjee, M.D.; Sumana, G.; Gupta, R.K.; Sood, S.; Malhotra, B.D. Biosensors for pathogen detection: A smart approach towards clinical diagnosis. Sens. Actuators B Chem. 2014, 197, 385–404. [Google Scholar] [CrossRef]
- Bergveld, P. A critical evaluation of direct electrical protein detection methods. Biosens. Bioelectron. 1991, 6, 55–72. [Google Scholar] [CrossRef]
- De Moraes, A.C.; Kubota, L.T. Recent Trends in Field-Effect Transistors-Based Immunosensors. Chemosens. 2016, 4, 20. [Google Scholar] [CrossRef]
- Atta, N.F.; Galal, A.; El-Ads, E.H. Graphene—a platform for sensor and biosensor applications. Biosens.-Micro Nanoscale Appl. 2015, 9, 38–84. [Google Scholar]
- Shanker, A.; Lee, K.; Kim, J. Chapter 12: Synthetic Hybrid Biosensors. In Nature Reviews Molecular Cell Biology; Wiley-VCH Verlag GmbH & Co. KgaA: Weinheim, Germany, 2014; pp. 359–386. [Google Scholar]
- Malhotra, B.D.; Ali, M.A. Nanomaterials in biosensors: Fundamentals and applications. Nanomater. Biosens. 2018, 1, 1–74. [Google Scholar]
- Zelada-Guillén, G.A.; Blondeau, P.; Rius, F.X.; Riu, J. Carbon nanotube-based aptasensors for the rapid and ultrasensitive detection of bacteria. Methods 2013, 63, 233–238. [Google Scholar] [CrossRef]
- Silva, N.F.D.; Magalhães, J.M.C.S.; Oliva-Teles, M.T.; Delerue-Matos, C. A potentiometric magnetic immunoassay for rapid detection of Salmonella typhimurium. Anal. Methods 2015, 7, 4008–4011. [Google Scholar] [CrossRef]
- Silva, N.F.D.; Almeida, C.M.R.; Magalhães, J.M.C.S.; Gonçalves, M.P.; Freire, C.; Delerue-Matos, C. Development of a disposable paper-based potentiometric immunosensor for real-time detection of a foodborne pathogen. Biosens. Bioelectron. 2019, 141, 111317. [Google Scholar] [CrossRef]
- Punbusayakul, N.; Talapatra, S.; Ajayan, P.M.; Surareungchai, W. Label-free as-grown double wall carbon nanotubes bundles for Salmonella typhimurium immunoassay. Chem. Cent. J. 2013, 7, 102. [Google Scholar] [CrossRef]
- Melo, A.M.A.; Alexandre, D.L.; Oliveira, M.R.F.; Furtado, R.F.; Borges, M.F.; Ribeiro, P.R.V.; Biswas, A.; Cheng, H.N.; Alves, C.R.; Figueiredo, E.A.T. Optimization and characterization of a biosensor assembly for detection of Salmonella Typhimurium. J. Solid State Electrochem. 2018, 22, 1321–1330. [Google Scholar] [CrossRef]
- Singh, C.; Ali, M.A.; Kumar, V.; Ahmad, R.; Sumana, G. Functionalized MoS2 nanosheets assembled microfluidic immunosensor for highly sensitive detection of food pathogen. Sens. Actuators B Chem. 2018, 259, 1090–1098. [Google Scholar] [CrossRef]
- Pal, N.; Sharma, S.; Gupta, S. Sensitive and rapid detection of pathogenic bacteria in small volumes using impedance spectroscopy technique. Biosens. Bioelectron. 2016, 77, 270–276. [Google Scholar] [CrossRef]
- Wang, L.; Huo, X.; Qi, W.; Xia, Z.; Li, Y.; Lin, J. Rapid and sensitive detection of Salmonella Typhimurium using nickel nanowire bridge for electrochemical impedance amplification. Talanta 2020, 211, 120715. [Google Scholar] [CrossRef]
- Afonso, A.S.; Pérez-López, B.; Faria, R.C.; Mattoso, L.H.C.; Hernández-Herrero, M.; Roig-Sagués, A.X.; Maltez-da Costa, M.; Merkoçi, A. Electrochemical detection of Salmonella using gold nanoparticles. Biosens. Bioelectron. 2013, 40, 121–126. [Google Scholar] [CrossRef]
- Fathi, S.; Saber, R.; Adabi, M.; Rasouli, R.; Douraghi, M.; Morshedi, M.; Farid-Majidi, R. Novel Competitive Voltammetric Aptasensor Based on Electrospun Carbon Nanofibers-Gold Nanoparticles Modified Graphite Electrode for Salmonella enterica serovar Detection. Biointerface Res. Appl. Chem. 2021, 11, 8702–8715. [Google Scholar]
- De Oliveira, T.R.; Martucci, D.H.; Faria, R.C. Simple disposable microfluidic device for Salmonella typhimurium detection by magneto-immunoassay. Sens. Actuators B Chem. 2018, 255, 684–691. [Google Scholar] [CrossRef]
- Dai, G.; Li, Z.; Luo, F.; Ai, S.; Chen, B.; Wang, Q. Electrochemical determination of Salmonella typhimurium by using aptamer-loaded gold nanoparticles and a composite prepared from a metal-organic framework (type UiO-67) and graphene. Microchim. Acta 2019, 186, 620. [Google Scholar] [CrossRef] [PubMed]
- Anany, H.; Chou, Y.; Cucic, S.; Derda, R.; Evoy, S.; Griffiths, M.W. From bits and pieces to whole phage to nanomachines: Pathogen detection using bacteriophages. Annu. Rev. Food Sci. Technol. 2017, 8, 305–329. [Google Scholar] [CrossRef]
- Morales, M.A.; Halpern, J.M. Guide to Selecting a Biorecognition Element for Biosensors. Bioconjug. Chem. 2018, 29, 3231–3239. [Google Scholar] [CrossRef] [PubMed]
- Melo, A.M.A.; Alexandre, D.L.; Furtado, R.F.; Borges, M.F.; Figueiredo, E.A.T.; Biswas, A.; Cheng, H.N.; Alves, C.R. Electrochemical immunosensors for Salmonella detection in food. Appl. Microbiol. Biotechnol. 2016, 100, 5301–5312. [Google Scholar] [CrossRef]
- Gizeli, E.; Lowe, C.R. Immunosensors. Curr. Opin. Biotechnol. 1996, 7, 66–71. [Google Scholar] [CrossRef]
- Alexandre, D.L.; Melo, A.M.A.; Furtado, R.F.; Borges, M.F.; Figueiredo, E.A.T.; Biswas, A.; Cheng, H.N.; Alves, C.R. A Rapid and Specific Biosensor for Salmonella Typhimurium Detection in Milk. Food Bioprocess Technol. 2018, 11, 748–756. [Google Scholar] [CrossRef]
- Sannigrahi, S.; Arumugasamy, S.K.; Mathiyarasu, J.; Suthindhiran, K. Magnetosome-anti-Salmonella antibody complex based biosensor for the detection of Salmonella typhimurium. Mater. Sci. Eng. C 2020, 114, 111071. [Google Scholar] [CrossRef] [PubMed]
- Bu, S.-J.; Wang, K.-Y.; Liu, X.; Ma, L.; Wei, H.-G.; Zhang, W.-G.; Liu, W.-S.; Wan, J.-Y. Ferrocene-functionalized nanocomposites as signal amplification probes for electrochemical immunoassay of Salmonella typhimurium. Microchim. Acta 2020, 187, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kasman, L.M.; Porter, L.D. Bacteriophages; [Updated 1 October 2020]. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2020. Available online: https://www.ncbi.nlm.nih.gov/books/NBK493185/ (accessed on 8 September 2021).
- Vinay, M.; Franche, N.; Grégori, G.; Fantino, J.-R.; Pouillot, F.; Ansaldi, M. Phage-based fluorescent biosensor prototypes to specifically detect enteric bacteria such as E. coli and Salmonella enterica Typhimurium. PLoS ONE 2015, 10, e0131466. [Google Scholar] [CrossRef]
- Chai, Y.; Li, S.; Horikawa, S.; Park, M.-K.; Vodyanoy, V.; Chin, B.A. Rapid and Sensitive Detection of Salmonella Typhimurium on Eggshells by Using Wireless Biosensors. J. Food Prot. 2012, 75, 631–636. [Google Scholar] [CrossRef]
- Qiao, Z.; Lei, C.; Fu, Y.; Li, Y. Rapid and sensitive detection of E. coli O157: H7 based on antimicrobial peptide functionalized magnetic nanoparticles and urease-catalyzed signal amplification. Anal. Methods 2017, 9, 5204–5210. [Google Scholar] [CrossRef]
- Patel, S.; Akhtar, N. Antimicrobial peptides (AMPs): The quintessential ‘offense and defense’molecules are more than antimicrobials. Biomed. Pharmacother. 2017, 95, 1276–1283. [Google Scholar] [CrossRef]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef]
- Hoyos-Nogués, M.; Gil, F.J.; Mas-Moruno, C. Antimicrobial Peptides: Powerful Biorecognition Elements to Detect Bacteria in Biosensing Technologies. Molecules 2018, 23, 1683. [Google Scholar] [CrossRef]
- Mannoor, M.S.; Zhang, S.; Link, A.J.; McAlpine, M.C. Electrical detection of pathogenic bacteria via immobilized antimicrobial peptides. Proc. Natl. Acad. Sci. USA 2010, 107, 19207–19212. [Google Scholar] [CrossRef]
- Datta, M.; Desai, D.; Kumar, A. Gene Specific DNA Sensors for Diagnosis of Pathogenic Infections. Indian J. Microbiol. 2017, 57, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Vanegas, D.C.; Gomes, C.L.; Cavallaro, N.D.; Giraldo-Escobar, D.; McLamore, E.S. Emerging biorecognition and transduction schemes for rapid detection of pathogenic bacteria in food. Compr. Rev. Food Sci. Food Saf. 2017, 16, 1188–1205. [Google Scholar] [CrossRef]
- Das, R.; Sharma, M.K.; Rao, V.K.; Bhattacharya, B.K.; Garg, I.; Venkatesh, V.; Upadhyay, S. An electrochemical genosensor for Salmonella typhi on gold nanoparticles-mercaptosilane modified screen printed electrode. J. Biotechnol. 2014, 188, 9–16. [Google Scholar] [CrossRef]
- Park, K.S. Nucleic acid aptamer-based methods for diagnosis of infections. Biosens. Bioelectron. 2018, 102, 179–188. [Google Scholar] [CrossRef]
- Robati, R.Y.; Arab, A.; Ramezani, M.; Langroodi, F.A.; Abnous, K.; Taghdisi, S.M. Aptasensors for quantitative detection of kanamycin. Biosens. Bioelectron. 2016, 82, 162–172. [Google Scholar] [CrossRef]
- Davis, K.A.; Abrams, B.; Lin, Y.; Jayasena, S.D. Use of a high affinity DNA ligand in flow cytometry. Nucleic Acids Res. 1996, 24, 702–706. [Google Scholar] [CrossRef]
- Bayat, P.; Nosrati, R.; Alibolandi, M.; Rafatpanah, H.; Abnous, K.; Khedri, M.; Ramezani, M. SELEX methods on the road to protein targeting with nucleic acid aptamers. Biochimie 2018, 154, 132–155. [Google Scholar] [CrossRef]
- Wolter, A.C.; Weickhmann, A.K.; Nasiri, A.H.; Hantke, K.; Ohlenschläger, O.; Wunderlich, C.H.; Kreutz, C.; Duchardt-Ferner, E.; Wöhnert, J. A Stably Protonated Adenine Nucleotide with a Highly Shifted pKa Value Stabilizes the Tertiary Structure of a GTP-Binding RNA Aptamer. Angew. Chem. Int. Ed. 2017, 56, 401–404. [Google Scholar] [CrossRef]
- Zhou, J.; Satheesan, S.; Li, H.; Weinberg, M.S.; Morris, K.V.; Burnett, J.C.; Rossi, J.J. Cell-specific RNA aptamer against human CCR5 specifically targets HIV-1 susceptible cells and inhibits HIV-1 infectivity. Chem. Biol. 2015, 22, 379–390. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Lin, B.; Chen, C. An aptamer targeting shared tumor-specific peptide antigen of MAGE-A3 in multiple cancers. Int. J. Cancer 2016, 138, 918–926. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Chen, X.; Duan, N.; Wu, S.; Wang, Z.; Wei, X.; Wang, Y. Selection and characterization of DNA aptamers against Staphylococcus aureus enterotoxin C1. Food Chem. 2015, 166, 623–629. [Google Scholar] [CrossRef] [PubMed]
- Mehlhorn, A.; Rahimi, P.; Joseph, Y. Aptamer-based biosensors for antibiotic detection: A review. Biosensors 2018, 8, 54. [Google Scholar] [CrossRef]
- Shrivastava, S.; Sohn, I.-Y.; Son, Y.-M.; Lee, W.-I.; Lee, N.-E. Real-time label-free quantitative fluorescence microscopy-based detection of ATP using a tunable fluorescent nano-aptasensor platform. Nanoscale 2015, 7, 19663–19672. [Google Scholar] [CrossRef] [PubMed]
- Lavu, P.S.R.; Mondal, B.; Ramlal, S.; Murali, H.S.; Batra, H.V. Selection and characterization of aptamers using a modified whole cell bacterium SELEX for the detection of Salmonella enterica serovar typhimurium. ACS Comb. Sci. 2016, 18, 292–301. [Google Scholar] [CrossRef]
- Park, H.-C.; Baig, I.A.; Lee, S.-C.; Moon, J.-Y.; Yoon, M.-Y. Development of ssDNA Aptamers for the Sensitive Detection of Salmonella typhimurium and Salmonella enteritidis. Appl. Biochem. Biotechnol. 2014, 174, 793–802. [Google Scholar] [CrossRef]
- Renuka, R.; Maroli, N.; Achuth, J.; Ponmalai, K.; Kadirvelu, K. Highly adaptable and sensitive FRET-based aptamer assay for the detection of Salmonella paratyphi A. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 243, 118662. [Google Scholar] [CrossRef]
- Dinshaw, I.J.; Muniandy, S.; Teh, S.J.; Ibrahim, F.; Leo, B.F.; Thong, K.L. Development of an aptasensor using reduced graphene oxide chitosan complex to detect Salmonella. J. Electroanal. Chem. 2017, 806, 88–96. [Google Scholar] [CrossRef]
- Ma, X.; Jiang, Y.; Jia, F.; Yu, Y.; Chen, J.; Wang, Z. An aptamer-based electrochemical biosensor for the detection of Salmonella. J. Microbiol. Methods 2014, 98, 94–98. [Google Scholar] [CrossRef]
- Fang, S.; Song, D.; Zhuo, Y.; Chen, Y.; Zhu, A.; Long, F. Simultaneous and sensitive determination of Escherichia coli O157:H7 and Salmonella Typhimurium using evanescent wave dual-color fluorescence aptasensor based on micro/nano size effect. Biosens. Bioelectron. 2021, 185, 113288. [Google Scholar] [CrossRef]
- Muniandy, S.; Teh, S.J.; Appaturi, J.N.; Thong, K.L.; Lai, C.W.; Ibrahim, F.; Leo, B.F. A reduced graphene oxide-titanium dioxide nanocomposite based electrochemical aptasensor for rapid and sensitive detection of Salmonella enterica. Bioelectrochemistry 2019, 127, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Pathania, P.K.; Saini, J.K.; Vij, S.; Tewari, R.; Sabherwal, P.; Rishi, P.; Suri, C.R. Aptamer functionalized MoS2-rGO nanocomposite based biosensor for the detection of Vi antigen. Biosens. Bioelectron. 2018, 122, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.R.; Pulingam, T.; Appaturi, J.N.; Zifruddin, A.N.; Teh, S.J.; Lim, T.W.; Ibrahim, F.; Leo, B.F.; Thong, K.L. Carbon nanotube-based aptasensor for sensitive electrochemical detection of whole-cell Salmonella. Anal. Biochem. 2018, 554, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, N.; Tiwari, I. Recent build outs in electroanalytical biosensors based on carbon-nanomaterial modified screen printed electrode platforms. Anal. Methods 2017, 9, 3895–3907. [Google Scholar] [CrossRef]
- Barton, J.; García, M.B.G.; Santos, D.H.; Fanjul-Bolado, P.; Ribotti, A.; McCaul, M.; Diamond, D.; Magni, P. Screen-printed electrodes for environmental monitoring of heavy metal ions: A review. Microchim. Acta 2016, 183, 503–517. [Google Scholar] [CrossRef]
- Shi, Z.; Lu, Y.; Chen, Z.; Cheng, C.; Xu, J.; Zhang, Q.; Yan, Z.; Luo, Z.; Liu, Q. Electrochemical non-enzymatic sensing of glycoside toxins by boronic acid functionalized nano-composites on screen-printed electrode. Sens. Actuators B Chem. 2021, 329, 129197. [Google Scholar] [CrossRef]
- Munteanu, F.-D.; Titoiu, A.M.; Marty, J.-L.; Vasilescu, A. Detection of Antibiotics and Evaluation of Antibacterial Activity with Screen-Printed Electrodes. Sensors 2018, 18, 901. [Google Scholar] [CrossRef] [PubMed]
- Torre, R.; Costa-Rama, E.; Nouws, H.P.A.; Delerue-Matos, C. Screen-Printed Electrode-Based Sensors for Food Spoilage Control: Bacteria and Biogenic Amines Detection. Biosens. 2020, 10, 139. [Google Scholar] [CrossRef]
- Alonso-Lomillo, M.A.; Domínguez-Renedo, O.; Arcos-Martínez, M.J. Screen-printed biosensors in microbiology; a review. Talanta 2010, 82, 1629–1636. [Google Scholar] [CrossRef]
- Ferrari, A.G.-M.; Carrington, P.; Rowley-Neale, S.J.; Banks, C.E. Recent advances in portable heavy metal electrochemical sensing platforms. Environ. Sci. Water Res. Technol. 2020, 6, 2676–2690. [Google Scholar] [CrossRef]
- Choudhry, N.A.; Kampouris, D.K.; Kadara, R.O.; Jenkinson, N.; Banks, C.E. Next generation screen printed electrochemical platforms: Non-enzymatic sensing of carbohydrates using copper (II) oxide screen printed electrodes. Anal. Methods 2009, 1, 183–187. [Google Scholar] [CrossRef] [PubMed]
- Choudhry, N.A.; Kadara, R.O.; Jenkinson, N.; Banks, C.E. Screen printed electrodes provide micro-domain sites for fabricating disposable electro-catalytic ensembles. Electrochem. Commun. 2010, 12, 406–409. [Google Scholar] [CrossRef]
- Fei, J.; Dou, W.; Zhao, G. A sandwich electrochemical immunosensor for Salmonella pullorum and Salmonella gallinarum based on a screen-printed carbon electrode modified with an ionic liquid and electrodeposited gold nanoparticles. Microchim. Acta 2015, 182, 2267–2275. [Google Scholar] [CrossRef]
- Salam, F.; Tothill, I.E. Detection of Salmonella typhimurium using an electrochemical immunosensor. Biosens. Bioelectron. 2009, 24, 2630–2636. [Google Scholar] [CrossRef] [PubMed]
- Farka, Z.; Juřík, T.; Pastucha, M.; Kovář, D.; Lacina, K.; Skládal, P. Rapid Immunosensing of Salmonella Typhimurium Using Electrochemical Impedance Spectroscopy: The Effect of Sample Treatment. Electroanalysis 2016, 28, 1803–1809. [Google Scholar] [CrossRef]
- Fei, J.; Dou, W.; Zhao, G. A sandwich electrochemical immunoassay for Salmonella pullorum and Salmonella gallinarum based on a AuNPs/SiO2/Fe3O4 adsorbing antibody and 4 channel screen printed carbon electrode electrodeposited gold nanoparticles. RSC Adv. 2015, 5, 74548–74556. [Google Scholar] [CrossRef]
- Dahlin, A.B. Size Matters: Problems and Advantages Associated with Highly Miniaturized Sensors. Sensors 2012, 12, 3018–3036. [Google Scholar] [CrossRef] [PubMed]
- Sannomiya, T.; Vörös, J. Single plasmonic nanoparticles for biosensing. Trends Biotechnol. 2011, 29, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Muniandy, S.; Teh, S.J.; Thong, K.L.; Thiha, A.; Dinshaw, I.J.; Lai, C.W.; Ibrahim, F.; Leo, B.F. Carbon nanomaterial-based electrochemical biosensors for foodborne bacterial detection. Crit. Rev. Anal. Chem. 2019, 49, 510–533. [Google Scholar] [CrossRef]
- Cho, I.-H.; Kim, D.H.; Park, S. Electrochemical biosensors: Perspective on functional nanomaterials for on-site analysis. Biomater. Res. 2020, 24, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Tirado, E.; González-Cortés, A.; Yáñez-Sedeño, P.; Pingarrón, J.M. Carbon nanotubes functionalized by click chemistry as scaffolds for the preparation of electrochemical immunosensors. Application to the determination of TGF-beta 1 cytokine. Analyst 2016, 141, 5730–5737. [Google Scholar] [CrossRef] [PubMed]
- Eatemadi, A.; Daraee, H.; Karimkhanloo, H.; Kouhi, M.; Zarghami, N.; Akbarzadeh, A.; Abasi, M.; Hanifehpour, Y.; Joo, S.W. Carbon nanotubes: Properties, synthesis, purification, and medical applications. Nanoscale Res. Lett. 2014, 9, 393. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Xu, T.; Zhang, X. Graphene-Based Biosensors for Detection of Biomarkers. Micromachines 2020, 11, 60. [Google Scholar] [CrossRef] [PubMed]
- Novoselov, K.S.; Geim, A.K.; Morozov, S.V.; Jiang, D.; Zhang, Y.; Dubonos, S.V.; Grigorieva, I.V.; Firsov, A.A. Electric field effect in atomically thin carbon films. Science 2004, 306, 666–669. [Google Scholar] [CrossRef] [PubMed]
- Randviir, E.P.; Brownson, D.A.C.; Banks, C.E. A decade of graphene research: Production, applications and outlook. Mater. Today 2014, 17, 426–432. [Google Scholar] [CrossRef]
- Cho, I.-H.; Lee, J.; Kim, J.; Kang, M.; Paik, J.K.; Ku, S.; Cho, H.-M.; Irudayaraj, J.; Kim, D.-H. Current technologies of electrochemical immunosensors: Perspective on signal amplification. Sensors 2018, 18, 207. [Google Scholar] [CrossRef]
- Kuila, T.; Bose, S.; Khanra, P.; Mishra, A.K.; Kim, N.H.; Lee, J.H. Recent advances in graphene-based biosensors. Biosens. Bioelectron. 2011, 26, 4637–4648. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Murali, S.; Cai, W.; Li, X.; Suk, J.W.; Potts, J.R.; Ruoff, R.S. Graphene and Graphene Oxide: Synthesis, Properties, and Applications. Adv. Mater. 2010, 22, 3906–3924. [Google Scholar] [CrossRef]
- Smith, A.T.; LaChance, A.M.; Zeng, S.; Liu, B.; Sun, L. Synthesis, properties, and applications of graphene oxide/reduced graphene oxide and their nanocomposites. Nano Mater. Sci. 2019, 1, 31–47. [Google Scholar] [CrossRef]
- Xue, Y. Chapter 11—Carbon Nanotubes for Biomedical Applications. In Micro and Nano Technologies; Elsevier: Amsterdam, The Netherlands, 2017; pp. 323–346. ISBN 978-0-323-41481-4. [Google Scholar]
- Zhang, X.; Li, C.-R.; Wang, W.-C.; Xue, J.; Huang, Y.-L.; Yang, X.-X.; Tan, B.; Zhou, X.-P.; Shao, C.; Ding, S.-J. A novel electrochemical immunosensor for highly sensitive detection of aflatoxin B1 in corn using single-walled carbon nanotubes/chitosan. Food Chem. 2016, 192, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Soares, R.R.A.; Hjort, R.G.; Pola, C.C.; Parate, K.; Reis, E.L.; Soares, N.F.F.; McLamore, E.S.; Claussen, J.C.; Gomes, C.L. Laser-Induced Graphene Electrochemical Immunosensors for Rapid and Label-Free Monitoring of Salmonella enterica in Chicken Broth. ACS Sens. 2020, 5, 1900–1911. [Google Scholar] [CrossRef]
- Ye, Y.; Yan, W.; Liu, Y.; He, S.; Cao, X.; Xu, X.; Zheng, H.; Gunasekaran, S. Electrochemical detection of Salmonella using an invA genosensor on polypyrrole-reduced graphene oxide modified glassy carbon electrode and AuNPs-horseradish peroxidase-streptavidin as nanotag. Anal. Chim. Acta 2019, 1074, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Duo, H.; Zheng, C.; Zhao, S.; Song, S.; Simon, G. Novel approach to fabrication of DNA Biosensor Based on a Carboxylated Graphene Oxide Decorated with Fe3O4 NPs for the Detection of Typhoidal Salmonella. Int. J. Electrochem. Sci 2019, 14, 1248–1269. [Google Scholar] [CrossRef]
- Quiton, P.A.; Carreon, B.M.; Cruz-Papa, D.M.D.; Bergantin, J., Jr. Bacteriophage-modified graphene oxide screen-printed electrodes for the impedimetric biosensing of Salmonella enterica serovar Typhimurium. Sens. Transducers 2018, 28, 38–42. [Google Scholar]
- Weber, J.E.; Pillai, S.; Ram, M.K.; Kumar, A.; Singh, S.R. Electrochemical impedance-based DNA sensor using a modified single walled carbon nanotube electrode. Mater. Sci. Eng. C 2011, 31, 821–825. [Google Scholar] [CrossRef]
- Yang, M.; Peng, Z.; Ning, Y.; Chen, Y.; Zhou, Q.; Deng, L. Highly specific and cost-efficient detection of Salmonella Paratyphi A combining aptamers with single-walled carbon nanotubes. Sensors 2013, 13, 6865–6881. [Google Scholar] [CrossRef] [PubMed]
- Quintela, I.A.; de los Reyes, B.G.; Lin, C.-S.; Wu, V.C.H. Simultaneous Colorimetric Detection of a Variety of Salmonella spp. in Food and Environmental Samples by Optical Biosensing Using Oligonucleotide-Gold Nanoparticles. Front. Microbiol. 2019, 10, 1138. [Google Scholar] [CrossRef] [PubMed]
- Xiang, C.; Li, R.; Adhikari, B.; She, Z.; Li, Y.; Kraatz, H.-B. Sensitive electrochemical detection of Salmonella with chitosan–gold nanoparticles composite film. Talanta 2015, 140, 122–127. [Google Scholar] [CrossRef]
- Ye, Y.; Liu, Y.; He, S.; Xu, X.; Cao, X.; Ye, Y.; Zheng, H. Ultrasensitive electrochemical DNA sensor for virulence invA gene of Salmonella using silver nanoclusters as signal probe. Sens. Actuators B Chem. 2018, 272, 53–59. [Google Scholar] [CrossRef]
- Huang, F.; Guo, R.; Xue, L.; Cai, G.; Wang, S.; Li, Y.; Liao, M.; Wang, M.; Lin, J. An Acid-Responsive Microfluidic Salmonella Biosensor Using Curcumin as Signal Reporter and ZnO-Capped Mesoporous Silica Nanoparticles for Signal Amplification. Sens. Actuators B Chem. 2020, 312, 127958. [Google Scholar] [CrossRef]
- Appaturi, J.N.; Pulingam, T.; Muniandy, S.; Dinshaw, I.J.; Fen, L.B.; Johan, M.R. Supported cobalt nanoparticles on graphene oxide/mesoporous silica for oxidation of phenol and electrochemical detection of H2O2 and Salmonella spp. Mater. Chem. Phys. 2019, 232, 493–505. [Google Scholar] [CrossRef]
- Lin, D.; Harris, K.D.; Chan, N.W.C.; Jemere, A.B. Nanostructured indium tin oxide electrodes immobilized with toll-like receptor proteins for label-free electrochemical detection of pathogen markers. Sens. Actuators B Chem. 2018, 257, 324–330. [Google Scholar] [CrossRef]
- Lobo-Castañón, M.J.; Barreda-Garcia, S.; Miranda-Castro, R.; Estrela, P. Electrochemical Detection of Salmonella via On-surface Isothermal Amplification of its Genetic Material onto Highly Stable and Reproducible Indium Tin Oxide Platforms. In Proceedings of the 3rd International Electronic Conference on Medical Chemistry, Online. 1–30 November 2017. [Google Scholar]
- Thiha, A.; Ibrahim, F.; Muniandy, S.; Dinshaw, I.J.; Teh, S.J.; Thong, K.L.; Leo, B.F.; Madou, M. All-carbon suspended nanowire sensors as a rapid highly-sensitive label-free chemiresistive biosensing platform. Biosens. Bioelectron. 2018, 107, 145–152. [Google Scholar] [CrossRef]
- Mustafa, M.R.B.; Dhahi, T.S.; Ehfaed, N.A.K.H.; Adam, T.; Hashim, U.; Azizah, N.; Mohammed, M.; Noriman, N.Z. Specific and selective target detection of supra-genome 21 Mers Salmonella via silicon nanowires biosensor. In Proceedings of 2017 International Conference on Biotechnology and Bioengineering (ICBB-2017), Offenburg, Germany, 26–28 September 2017; AIP Publishing LLC: New York, NY, USA, 2017; Volume 1885, p. 20196. [Google Scholar]
- Cao, X.; Ye, Y.; Liu, S. Gold nanoparticle-based signal amplification for biosensing. Anal. Biochem. 2011, 417, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, P.A.; Sandhyarani, N. Electrochemical DNA sensors based on the use of gold nanoparticles: A review on recent developments. Microchim. Acta 2017, 184, 981–1000. [Google Scholar] [CrossRef]
- Pingarrón, J.M.; Yáñez-Sedeño, P.; González-Cortés, A. Gold nanoparticle-based electrochemical biosensors. Electrochim. Acta 2008, 53, 5848–5866. [Google Scholar] [CrossRef]
- Lepoitevin, M.; Lemouel, M.; Bechelany, M.; Janot, J.-M.; Balme, S. Gold nanoparticles for the bare-eye based and spectrophotometric detection of proteins, polynucleotides and DNA. Microchim. Acta 2015, 182, 1223–1229. [Google Scholar] [CrossRef]
- Elghanian, R.; Storhoff, J.J.; Mucic, R.C.; Letsinger, R.L.; Mirkin, C.A. Selective Colorimetric Detection of Polynucleotides Based on the Distance-Dependent Optical Properties of Gold Nanoparticles. Science 1997, 277, 1078–1081. [Google Scholar] [CrossRef]
- Yang, Z.; Chai, Y.; Yuan, R.; Zhuo, Y.; Li, Y.; Han, J.; Liao, N. Hollow platinum decorated Fe3O4 nanoparticles as peroxidase mimetic couple with glucose oxidase for pseudobienzyme electrochemical immunosensor. Sens. Actuators B Chem. 2014, 193, 461–466. [Google Scholar] [CrossRef]
- Wang, S.-F.; Tan, Y.-M. A novel amperometric immunosensor based on Fe 3 O 4 magnetic nanoparticles/chitosan composite film for determination of ferritin. Anal. Bioanal. Chem. 2007, 387, 703–708. [Google Scholar] [CrossRef]
- Aznar, E.; Oroval, M.; Pascual, L.; Murguía, J.R.; Martinez-Manez, R.; Sancenon, F. Gated materials for on-command release of guest molecules. Chem. Rev. 2016, 116, 561–718. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Liu, S.; Feng, Q.; Wang, N. Silver nanowire-based electrochemical immunoassay for sensing immunoglobulin G with signal amplification using strawberry-like ZnO nanostructures as labels. Biosens. Bioelectron. 2013, 49, 256–262. [Google Scholar] [CrossRef]
- Jimenez-Falcao, S.; Parra-Nieto, J.; Pérez-Cuadrado, H.; Martínez-Máñez, R.; Martínez-Ruiz, P.; Villalonga, R. Avidin-gated mesoporous silica nanoparticles for signal amplification in electrochemical biosensor. Electrochem. Commun. 2019, 108, 106556. [Google Scholar] [CrossRef]
- Chopra, K.L.; Major, S.; Pandya, D.K. Transparent conductors—A status review. Thin Solid Films 1983, 102, 1–46. [Google Scholar] [CrossRef]
- Tran, D.-P.; Lu, H.-I.; Lin, C.-K. Conductive characteristics of indium tin oxide thin film on polymeric substrate under long-term static deformation. Coatings 2018, 8, 212. [Google Scholar] [CrossRef]
- Rabbani, M.; Hoque, M.E.; Mahbub, Z. Bin Nanosensors in biomedical and environmental applications: Perspectives and prospects. In Nanofabrication for Smart Nanosensor Applications; Elsevier: Amsterdam, The Netherlands, 2020; pp. 163–186. [Google Scholar]
- He, B.; Morrow, T.J.; Keating, C.D. Nanowire sensors for multiplexed detection of biomolecules. Curr. Opin. Chem. Biol. 2008, 12, 522–528. [Google Scholar] [CrossRef]
- Nambiar, S.; Yeow, J.T.W. Conductive polymer-based sensors for biomedical applications. Biosens. Bioelectron. 2011, 26, 1825–1832. [Google Scholar] [CrossRef]
- Liu, J.; Agarwal, M.; Varahramyan, K. Glucose sensor based on organic thin film transistor using glucose oxidase and conducting polymer. Sens. Actuators B Chem. 2008, 135, 195–199. [Google Scholar] [CrossRef]
- Gniadek, M.; Modzelewska, S.; Donten, M.; Stojek, Z. Modification of electrode surfaces: Deposition of thin layers of Polypyrrole− au nanoparticle materials using a combination of interphase synthesis and dip-in method. Anal. Chem. 2010, 82, 469–472. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Cordero, J.L.; Ricco, A.J. Lab-on-a-Chip (General Philosophy) BT—Encyclopedia of Microfluidics and Nanofluidics; Springer: Boston, MA, USA, 2008; pp. 962–969. ISBN 978-0-387-48998-8. [Google Scholar]
- Mauk, M.; Song, J.; Bau, H.H.; Gross, R.; Bushman, F.D.; Collman, R.G.; Liu, C. Miniaturized devices for point of care molecular detection of HIV. Lab Chip 2017, 17, 382–394. [Google Scholar] [CrossRef]
- Xiao, J.; Liu, Y.; Su, L.; Zhao, D.; Zhao, L.; Zhang, X. Microfluidic chip-based wearable colorimetric sensor for simple and facile detection of sweat glucose. Anal. Chem. 2019, 91, 14803–14807. [Google Scholar] [CrossRef]
- Tsougeni, K.; Kaprou, G.; Loukas, C.M.; Papadakis, G.; Hamiot, A.; Eck, M.; Rabus, D.; Kokkoris, G.; Chatzandroulis, S.; Papadopoulos, V.; et al. Lab-on-Chip platform and protocol for rapid foodborne pathogen detection comprising on-chip cell capture, lysis, DNA amplification and surface-acoustic-wave detection. Sens. Actuators B Chem. 2020, 320, 128345. [Google Scholar] [CrossRef]
- Wang, S.; Liu, N.; Zheng, L.; Cai, G.; Lin, J. A lab-on-chip device for the sample-in-result-out detection of viable Salmonella using loop-mediated isothermal amplification and real-time turbidity monitoring. Lab Chip 2020, 20, 2296–2305. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.Q.; Sato, M.; Tan, W.L.; Khoo, G.P.W.; Fisher, K.; Meibers, H.; Wei Foo, D.G.; Sen Phang, H.C.; Bird, P.; Joseph Benzinger, M. The Validation of the VereBeefTM Detection Kit. J. AOAC Int. 2019, 102, 508–524. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).