In-Situ Monitoring of Real-Time Loop-Mediated Isothermal Amplification with QCM: Detecting Listeria monocytogenes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Template DNA Isolation
2.3. QCM Sensor Measurements and Construction of ssDNA Films
2.4. Real-Time LAMP Monitoring with QCM
2.5. Visual Inspection of LAMP Products
2.6. Assessing QCM Electrode Sensitivity toward LAMP Product
2.7. QCM Surface Characterization
3. Results and Discussion
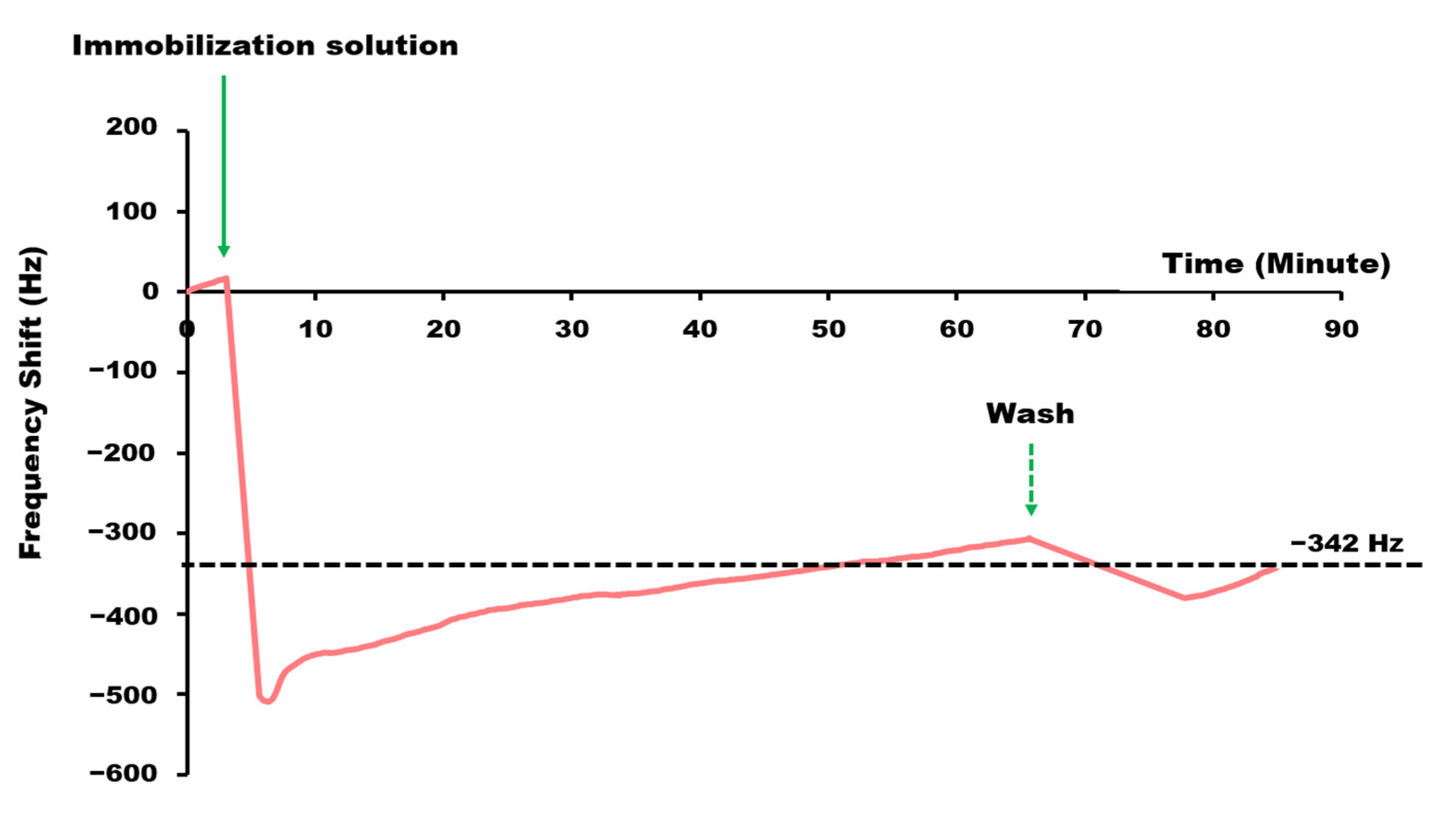
3.1. Sensor Functionalization and ssDNA/L-Cysteine Coverage Measurement
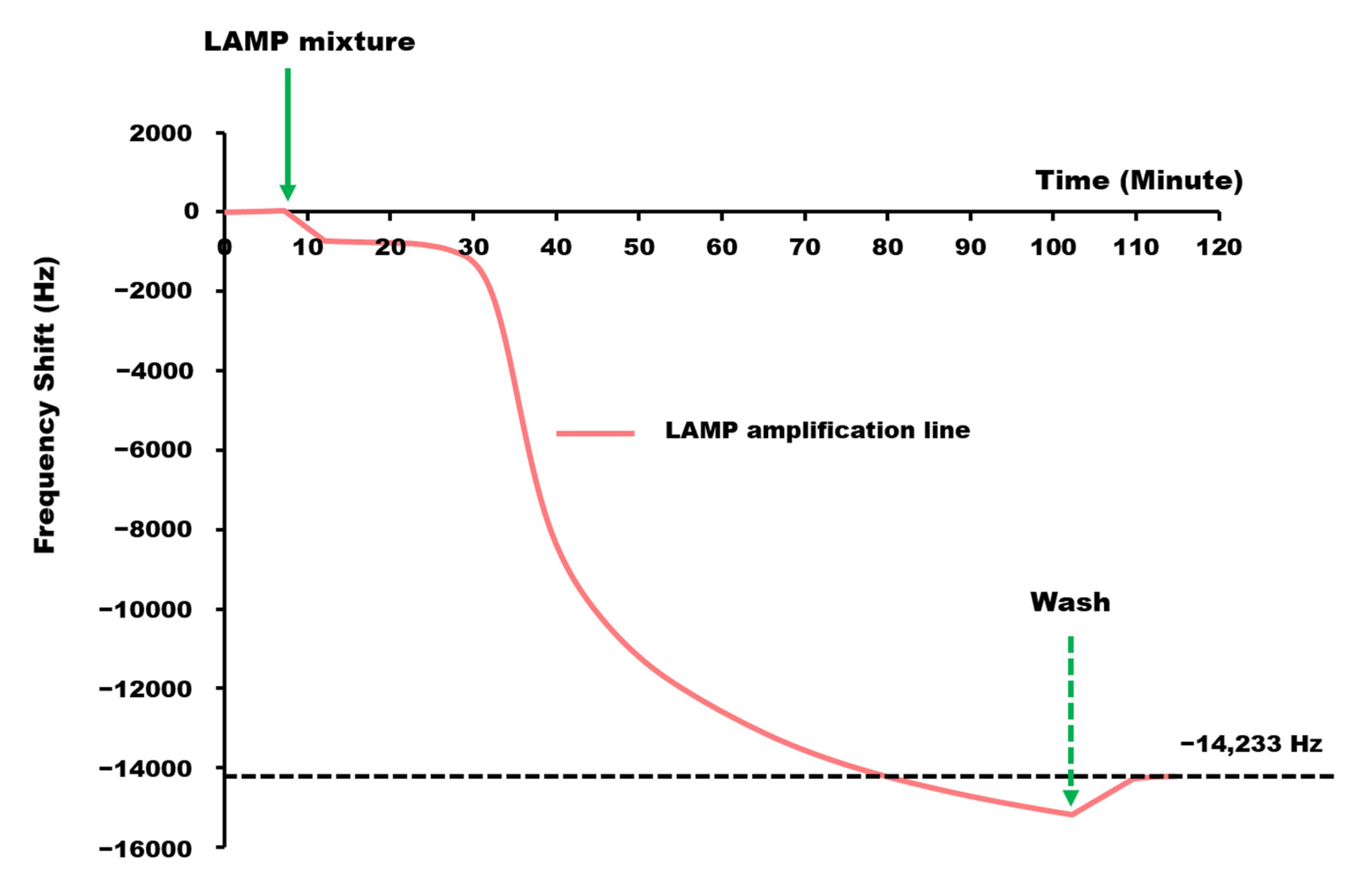
3.2. Real-Time LAMP-QCM Measurement
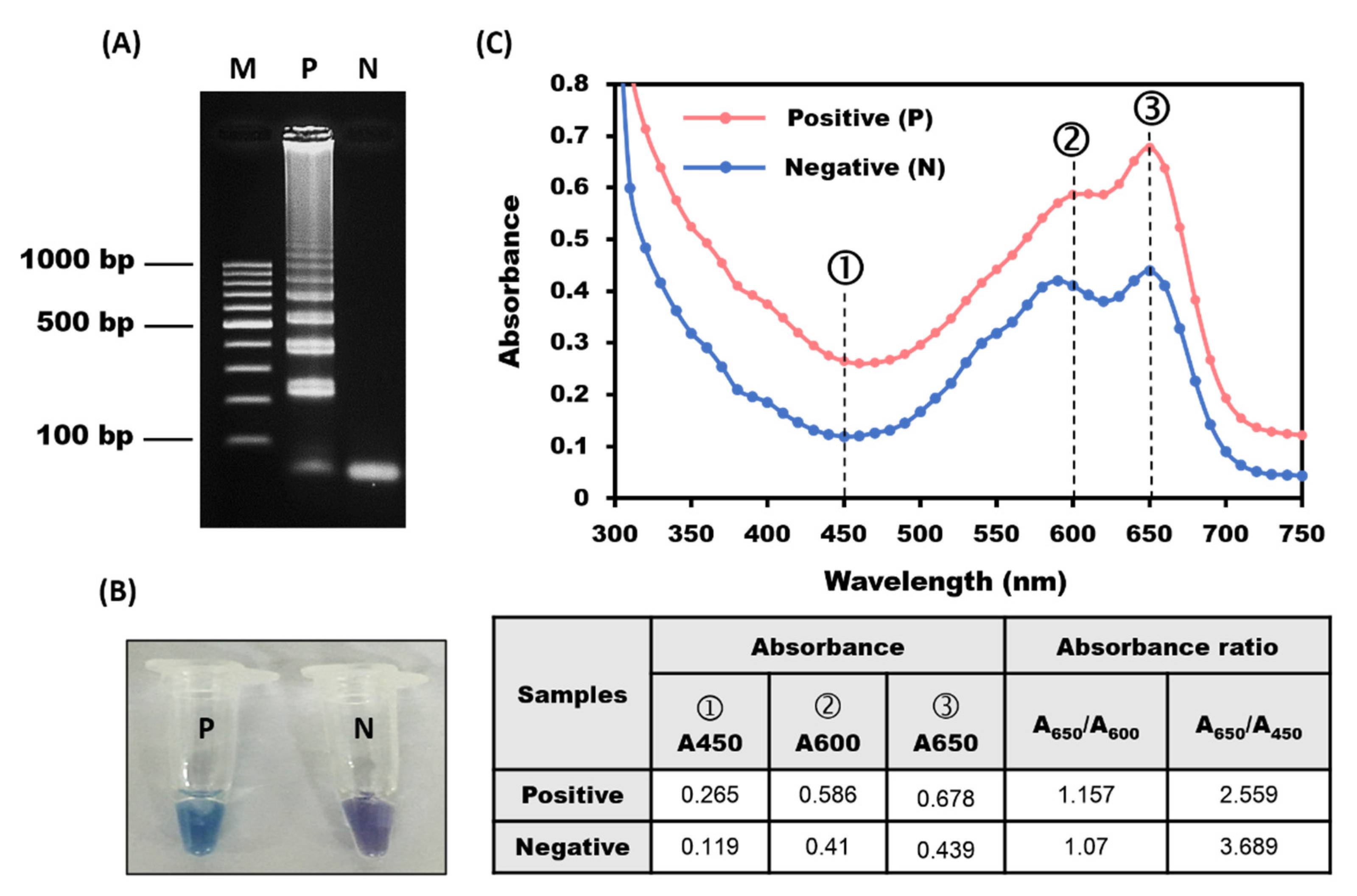
3.3. Rapid Screening of LAMP Products
3.4. Estimating Bacteria Load of the Sample
3.5. X-ray Photoelectron Spectroscopy (XPS) Spectra
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Díaz-Betancor, Z.; Bañuls, M.-J.; Maquieira, Á. Photoclick Chemistry to Create Dextran-Based Nucleic Acid Microarrays. Anal. Bioanal. Chem. 2019, 411, 6745–6754. [Google Scholar] [CrossRef]
- Jin, F.; Li, H.; Xu, D. Enzyme-Free Fluorescence Microarray for Determination of Hepatitis B Virus DNA Based on Silver Nanoparticle Aggregates-Assisted Signal Amplification. Anal. Chim. Acta 2019, 1077, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Peng, X.; Wang, Y.; Bao, T.; Wen, W.; Zhang, X.; Wang, S. An Exonuclease-Assisted Triple-Amplified Electrochemical Aptasensor for Mucin 1 Detection Based on Strand Displacement Reaction and Enzyme Catalytic Strategy. Anal. Chim. Acta 2019, 1086, 75–81. [Google Scholar] [CrossRef]
- Zhou, M.; Feng, C.; Mao, D.; Yang, S.; Ren, L.; Chen, G.; Zhu, X. An Electrochemical Biosensor Integrating Immunoassay and Enzyme Activity Analysis for Accurate Detection of Active Human Apurinic/Apyrimidinic Endonuclease 1. Biosens. Bioelectron. 2019, 142, 111558. [Google Scholar] [CrossRef]
- Mitta, S.B.; Reddeppa, M.; Vellampatti, S.; Dugasani, S.R.; Yoo, S.; Lee, S.; Kim, M.-D.; Ha Park, S. Gold Nanoparticle-Embedded DNA Thin Films for Ultraviolet Photodetectors. Sens. Actuators B Chem. 2018, 275, 137–144. [Google Scholar] [CrossRef]
- Yoo, S.; Dugasani, S.R.; Mitta, S.B.; Park, S.H. Layer-Dependent Characterization of Individual and Mixed Ion-Doped Multi-Layered DNA Thin Films. Appl. Surf. Sci. 2019, 479, 47–54. [Google Scholar] [CrossRef]
- Roether, J.; Chu, K.-Y.; Willenbacher, N.; Shen, A.Q.; Bhalla, N. Real-Time Monitoring of DNA Immobilization and Detection of DNA Polymerase Activity by a Microfluidic Nanoplasmonic Platform. Biosens. Bioelectron. 2019, 142, 111528. [Google Scholar] [CrossRef]
- Zhao, Y.; Gosai, A.; Shrotriya, P. Effect of Receptor Attachment on Sensitivity of Label Free Microcantilever Based Biosensor Using Malachite Green Aptamer. Sens. Actuators B Chem. 2019, 300, 126963. [Google Scholar] [CrossRef]
- Wachiralurpan, S.; Chansiri, K.; Lieberzeit, P.A. Direct Detection of Listeria Monocytogenes DNA Amplification Products with Quartz Crystal Microbalances at Elevated Temperatures. Sens. Actuators B Chem. 2020, 308, 127678. [Google Scholar] [CrossRef]
- Lin, Z.; Zhang, Y.; Zhang, H.; Zhou, Y.; Cao, J.; Zhou, J. Comparison of Loop-Mediated Isothermal Amplification (LAMP) and Real-Time PCR Method Targeting a 529-Bp Repeat Element for Diagnosis of Toxoplasmosis. Vet. Parasitol. 2012, 185, 296–300. [Google Scholar] [CrossRef] [PubMed]
- Pilchová, V.; Seinige, D.; Hennig-Pauka, I.; Büttner, K.; Abdulmawjood, A.; Kehrenberg, C. Development and Validation of a Loop-Mediated Isothermal Amplification (LAMP) Assay for Rapid Detection of Glaesserella (Haemophilus) Parasuis. Microorganisms 2021, 9, 41. [Google Scholar] [CrossRef] [PubMed]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-Mediated Isothermal Amplification of DNA. Nucleic Acids Res. 2000, 28, E63. [Google Scholar] [CrossRef] [Green Version]
- Mori, Y.; Notomi, T. Loop-Mediated Isothermal Amplification (LAMP): A Rapid, Accurate, and Cost-Effective Diagnostic Method for Infectious Diseases. J. Infect. Chemother. 2009, 15, 62–69. [Google Scholar] [CrossRef]
- Mori, Y.; Nagamine, K.; Tomita, N.; Notomi, T. Detection of Loop-Mediated Isothermal Amplification Reaction by Turbidity Derived from Magnesium Pyrophosphate Formation. Biochem. Biophys. Res. Commun. 2001, 289, 150–154. [Google Scholar] [CrossRef] [PubMed]
- Levicky, R.; Herne, T.M.; Tarlov, M.J.; Satija, S.K. Using Self-Assembly To Control the Structure of DNA Monolayers on Gold: A Neutron Reflectivity Study. J. Am. Chem. Soc. 1998, 120, 9787–9792. [Google Scholar] [CrossRef]
- Pingle, H.; Wang, P.-Y.; Cavaliere, R.; Whitchurch, C.B.; Thissen, H.; Kingshott, P. Minimal Attachment of Pseudomonas Aeruginosa to DNA Modified Surfaces. Biointerphases 2018, 13, 06E405. [Google Scholar] [CrossRef] [Green Version]
- Moses, S.; Brewer, S.H.; Lowe, L.B.; Lappi, S.E.; Gilvey, L.B.G.; Sauthier, M.; Tenent, R.C.; Feldheim, D.L.; Franzen, S. Characterization of Single- and Double-Stranded DNA on Gold Surfaces. Langmuir 2004, 20, 11134–11140. [Google Scholar] [CrossRef] [PubMed]
- Petrovykh, D.Y.; Kimura-Suda, H.; Whitman, L.J.; Tarlov, M.J. Quantitative Analysis and Characterization of DNA Immobilized on Gold. J. Am. Chem. Soc. 2003, 125, 5219–5226. [Google Scholar] [CrossRef]
- Petrovykh, D.Y.; Kimura-Suda, H.; Tarlov, M.J.; Whitman, L.J. Quantitative Characterization of DNA Films by X-ray Photoelectron Spectroscopy. Langmuir 2004, 20, 429–440. [Google Scholar] [CrossRef]
- Jia, Y.; Li, F. Studies of Functional Nucleic Acids Modified Light Addressable Potentiometric Sensors: X-ray Photoelectron Spectroscopy, Biochemical Assay, and Simulation. Anal. Chem. 2018, 90, 5153–5161. [Google Scholar] [CrossRef] [PubMed]
- Wachiralurpan, S.; Sriyapai, T.; Areekit, S.; Sriyapai, P.; Thongphueak, D.; Santiwatanakul, S.; Chansiri, K. A One-Step Rapid Screening Test of Listeria Monocytogenes in Food Samples Using a Real-Time Loop-Mediated Isothermal Amplification Turbidity Assay. Anal. Methods 2017, 9, 6403–6410. [Google Scholar] [CrossRef] [Green Version]
- Wachiralurpan, S.; Sriyapai, T.; Areekit, S.; Kaewphinit, T.; Sriyapai, P.; Santiwatanakul, S.; Chansiri, K. Development of a Rapid Screening Test for Listeria Monocytogenes in Raw Chicken Meat Using Loop-Mediated Isothermal Amplification (LAMP) and Lateral Flow Dipstick (LFD). Food Anal. Methods 2017, 1–10. [Google Scholar] [CrossRef]
- Goto, M.; Honda, E.; Ogura, A.; Nomoto, A.; Hanaki, K.-I. Colorimetric Detection of Loop-Mediated Isothermal Amplification Reaction by Using Hydroxy Naphthol Blue. BioTechniques 2009, 46, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.H.; Yuan, S.K.; Hu, X.R.; Zhang, C.Q. Shift of Sensitivity in Botrytis Cinerea to Benzimidazole Fungicides in Strawberry Greenhouse Ascribing to the Rising-Lowering of E198A Subpopulation and Its Visual, On-Site Monitoring by Loop-Mediated Isothermal Amplification. Sci. Rep. 2019, 9, 11644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varona, M.; Anderson, J.L. Visual Detection of Single-Nucleotide Polymorphisms Using Molecular Beacon Loop-Mediated Isothermal Amplification with Centrifuge-Free DNA Extraction. Anal. Chem. 2019, 91, 6991–6995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chahar, M.; Anvikar, A.; Valecha, N. Development and Evaluation of a Novel HNB Based Isothermal Amplification Assay for Fast Detection of Pyrimethamine Resistance (S108N) in Plasmodium Falciparum. Int. J. Environ. Res. Public Health 2019, 16, 1635. [Google Scholar] [CrossRef] [Green Version]
- Wong, I.Y.; Melosh, N.A. An Electrostatic Model for DNA Surface Hybridization. Biophys. J. 2010, 98, 2954–2963. [Google Scholar] [CrossRef] [Green Version]
- Tymoczko, J.; Schuhmann, W.; Gebala, M. Electrical Potential-Assisted DNA Hybridization. How to Mitigate Electrostatics for Surface DNA Hybridization. ACS Appl. Mater. Interfaces 2014, 6, 21851–21858. [Google Scholar] [CrossRef] [PubMed]
- Wong, I.Y.; Melosh, N.A. Directed Hybridization and Melting of DNA Linkers Using Counterion-Screened Electric Fields. Nano Lett. 2009, 9, 3521–3526. [Google Scholar] [CrossRef]
- Capaldo, P.; Zilio, S.D.; Torre, V.; Yang, Y. Semi-Analytical Dynamic Modeling of DNA Surface-Hybridization via AC Electro-Kinetic Steering. bioRxiv 2020, 2020, 964809. [Google Scholar] [CrossRef]
- Sauerbrey, G. The use of quartz oscillators for weighing thin layers and for microweighing. Z. Phys. 1959, 155, 206–222. [Google Scholar] [CrossRef]
- United Nations Office on Drugs and Crime; Laboratory and Scientific Section. Guidance for the Validation of Analytical Methodology and Calibration of Equipment Used for Testing of Illicit Drugs in Seized Materials and Biological Specimens: A Commitment to Quality and Continuous Improvement; United Nations Office on Drugs and Crime (UNODC): Vienna, Austria, 2009; ISBN 978-92-1-148243-0. [Google Scholar]
- Plekan, O.; Feyer, V.; Ptasińska, S.; Tsud, N.; Cháb, V.; Matolín, V.; Prince, K.C. Photoemission Study of Thymidine Adsorbed on Au(111) and Cu(110). J. Phys. Chem. C 2010, 114, 15036–15041. [Google Scholar] [CrossRef]
- Kick, A.; Bönsch, M.; Kummer, K.; Vyalikh, D.V.; Molodtsov, S.L.; Mertig, M. Controlling Structural Properties of Self-Assembled Oligonucleotide–Mercaptohexanol Monolayers. J. Electr. Spectrosc. Relat. Phenom. 2009, 172, 36–41. [Google Scholar] [CrossRef]
- Battocchio, C.; Porcaro, F.; Mukherjee, S.; Magnano, E.; Nappini, S.; Fratoddi, I.; Quintiliani, M.; Russo, M.V.; Polzonetti, G. Gold Nanoparticles Stabilized with Aromatic Thiols: Interaction at the Molecule–Metal Interface and Ligand Arrangement in the Molecular Shell Investigated by SR-XPS and NEXAFS. J. Phys. Chem. C 2014, 118, 8159–8168. [Google Scholar] [CrossRef]
- Jia, J.; Kara, A.; Pasquali, L.; Bendounan, A.; Sirotti, F.; Esaulov, V.A. On Sulfur Core Level Binding Energies in Thiol Self-Assembly and Alternative Adsorption Sites: An Experimental and Theoretical Study. J. Chem. Phys. 2015, 143, 104702. [Google Scholar] [CrossRef]
- Porcaro, F.; Carlini, L.; Ugolini, A.; Visaggio, D.; Visca, P.; Fratoddi, I.; Venditti, I.; Meneghini, C.; Simonelli, L.; Marini, C.; et al. Synthesis and Structural Characterization of Silver Nanoparticles Stabilized with 3-Mercapto-1-Propansulfonate and 1-Thioglucose Mixed Thiols for Antibacterial Applications. Materials 2016, 9, 1028. [Google Scholar] [CrossRef]
- Lee, C.-Y.; Gong, P.; Harbers, G.M.; Grainger, D.W.; Castner, D.G.; Gamble, L.J. Surface Coverage and Structure of Mixed DNA/Alkylthiol Monolayers on Gold: Characterization by XPS, NEXAFS, and Fluorescence Intensity Measurements. Anal. Chem. 2006, 78, 3326–3334. [Google Scholar] [CrossRef] [Green Version]
- Lavrich, D.J.; Wetterer, S.M.; Bernasek, S.L.; Scoles, G. Physisorption and Chemisorption of Alkanethiols and Alkyl Sulfides on Au(111). J. Phys. Chem. B 1998, 102, 3456–3465. [Google Scholar] [CrossRef]
- Schwartz, D.K. Mechanisms and Kinetics of Self-Assembled Monolayer Formation. Annu. Rev. Phys. Chem. 2001, 52, 107–137. [Google Scholar] [CrossRef]
- Bain, C.D.; Troughton, E.B.; Tao, Y.T.; Evall, J.; Whitesides, G.M.; Nuzzo, R.G. Formation of Monolayer Films by the Spontaneous Assembly of Organic Thiols from Solution onto Gold. Adv. Ceram. Mater. 1989, 111, 321–335. [Google Scholar] [CrossRef]
- Schreiber, F.; Eberhardt, A.; Leung, T.Y.B.; Schwartz, P.; Wetterer, S.M.; Lavrich, D.J.; Berman, L.; Fenter, P.; Eisenberger, P.; Scoles, G. Adsorption Mechanisms, Structures, and Growth Regimes of an Archetypal Self-Assembling System: Decanethiol on Au(111). Phys. Rev. B 1998, 57, 12476–12481. [Google Scholar] [CrossRef] [Green Version]
- Rei Vilar, M.; Botelho do Rego, A.M. High-Resolution Electron Energy Loss Spectroscopy as a Probe of SAMs. J. Electr. Spectrosc. Relat. Phenom. 2009, 172, 27–35. [Google Scholar] [CrossRef]
- Cho, E.; Brown, A.; Kuech, T.F. Chemical Characterization of DNA-Immobilized InAs Surfaces Using X-Ray Photoelectron Spectroscopy and Near-Edge X-Ray Absorption Fine Structure. Langmuir 2012, 28, 11890–11898. [Google Scholar] [CrossRef]
- Alshaaer, M.; Issa, K.; Alanazi, A.; Mallouh, S.A.; Afify, A.S.; Moustapha, M.E.; Komnitsas, K. Gradual Replacement of Ca2+ with Mg2+ Ions in Brushite for the Production of Ca1−xMgxHPO4·nH2O Materials. Minerals 2021, 11, 284. [Google Scholar] [CrossRef]
- Beamson, G.; Briggs, D. High Resolution XPS of Organic Polymers: The Scienta ESCA300 Database; John Wiley & Sons: New York, NY, USA, 1993; Volume 70. [Google Scholar]
- Saprigin, A.V.; Thomas, C.W.; Dulcey, C.S.; Patterson, C.H.; Spector, M.S. Spectroscopic Quantification of Covalently Immobilized Oligonucleotides. Surf. Interface Anal. 2005, 37, 24–32. [Google Scholar] [CrossRef]
- Wagner, C.D. NIST X-Ray Photoelectron Spectroscopy Database. NIST Stand. Ref. Database 2000. [Google Scholar] [CrossRef]
- Laiho, T.; Leiro, J.A. Influence of Initial Oxygen on the Formation of Thiol Layers. Appl. Surf. Sci. 2006, 252, 6304–6312. [Google Scholar] [CrossRef]
- May, C.J.; Canavan, H.E.; Castner, D.G. Quantitative X-Ray Photoelectron Spectroscopy and Time-of-Flight Secondary Ion Mass Spectrometry Characterization of the Components in DNA. Anal. Chem. 2004, 76, 1114–1122. [Google Scholar] [CrossRef]
- Roelfs, B.; Bunge, E.; Schröter, C.; Solomun, T.; Meyer, H.; Nichols, R.J.; Baumgärtel, H. Adsorption of Thymine on Gold Single-Crystal Electrodes. J. Phys. Chem. B 1997, 101, 754–765. [Google Scholar] [CrossRef]
- Haycock, D.E.; Kasrai, M.; Nicholls, C.J.; Urch, D.S. The Electronic Structure of Magnesium Hydroxide (Brucite) Using X-Ray Emission, X-Ray Photoelectron, and Auger Spectroscopy. J. Chem. Soc. Dalton Trans. 1978, 1791–1796. [Google Scholar] [CrossRef]


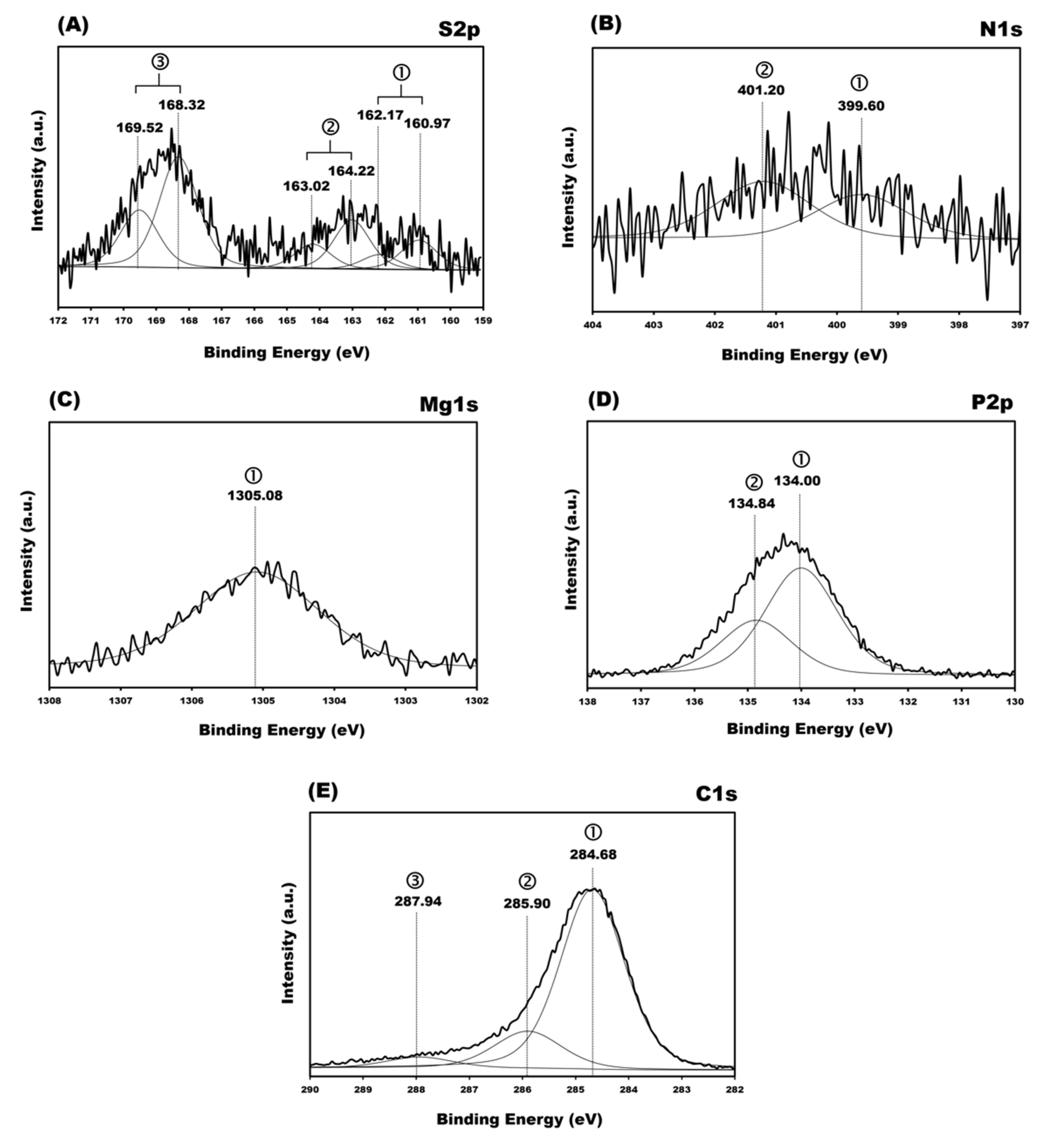
| REGIONS | BE Exp a, eV | BE Literature [16,35,36,37,38,46,47,48,49,50,51,52], eV | Relative Area Intensity | Assignment | |
|---|---|---|---|---|---|
| S2p | 2p3/2 | 160.97 | ~161–162 | 91 | Au–S |
| 163.02 | ~163 | 150 | S–H | ||
| 168.32 | ~168–169 | 363 | Sulfonate group | ||
| 2p1/2 | 162.17 | 45 | |||
| 164.22 | 75 | ||||
| 169.52 | 182 | ||||
| N1s | 399.60 | 399.0–399.3 | 186 | N= conjugated | |
| 401.2 | 399.9–401.00 | 245 | >N–, N–(C=O)–N, –NH2 | ||
| Mg1s | 1305.08 | 1305 | 1749 | Mg–O | |
| P2p | 2p3/2 | 134.00 | 133.9–134.2 | 1800 | (backbone), PBS buffer |
| 2p1/2 | 134.84 | 134.5 | 900 | ||
| C1s | 284.68 | 284.7–286.2 | 8846 | –C–C–, –C=, –CH, –CH3, carbons of aliphatic contamination. | |
| 285.90 | 286.0–287.0 | 1921 | C–N, N– C=N, C–O | ||
| 287.94 | 287.8–289.1 | 564 | N–(CO)–N, N–(CO)–C | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wachiralurpan, S.; Phung-On, I.; Chanlek, N.; Areekit, S.; Chansiri, K.; Lieberzeit, P.A. In-Situ Monitoring of Real-Time Loop-Mediated Isothermal Amplification with QCM: Detecting Listeria monocytogenes. Biosensors 2021, 11, 308. https://doi.org/10.3390/bios11090308
Wachiralurpan S, Phung-On I, Chanlek N, Areekit S, Chansiri K, Lieberzeit PA. In-Situ Monitoring of Real-Time Loop-Mediated Isothermal Amplification with QCM: Detecting Listeria monocytogenes. Biosensors. 2021; 11(9):308. https://doi.org/10.3390/bios11090308
Chicago/Turabian StyleWachiralurpan, Sirirat, Isaratat Phung-On, Narong Chanlek, Supatra Areekit, Kosum Chansiri, and Peter A. Lieberzeit. 2021. "In-Situ Monitoring of Real-Time Loop-Mediated Isothermal Amplification with QCM: Detecting Listeria monocytogenes" Biosensors 11, no. 9: 308. https://doi.org/10.3390/bios11090308
APA StyleWachiralurpan, S., Phung-On, I., Chanlek, N., Areekit, S., Chansiri, K., & Lieberzeit, P. A. (2021). In-Situ Monitoring of Real-Time Loop-Mediated Isothermal Amplification with QCM: Detecting Listeria monocytogenes. Biosensors, 11(9), 308. https://doi.org/10.3390/bios11090308





