Sensing Hydration of Biomimetic Cell Membranes
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. SLB Preparation
2.2.2. SLBs Hydration State Control
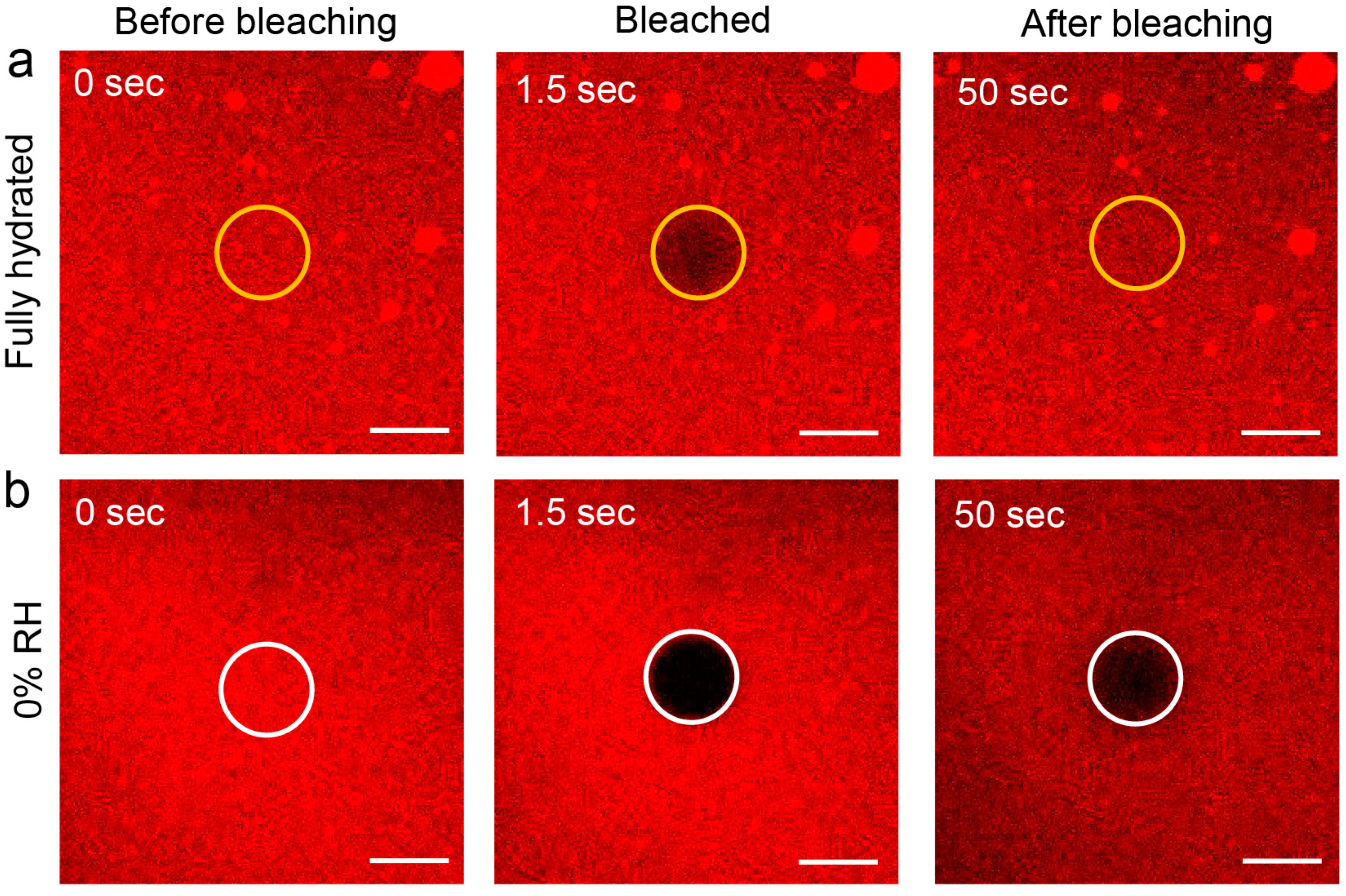
2.2.3. FRAP Experiments
3. Results
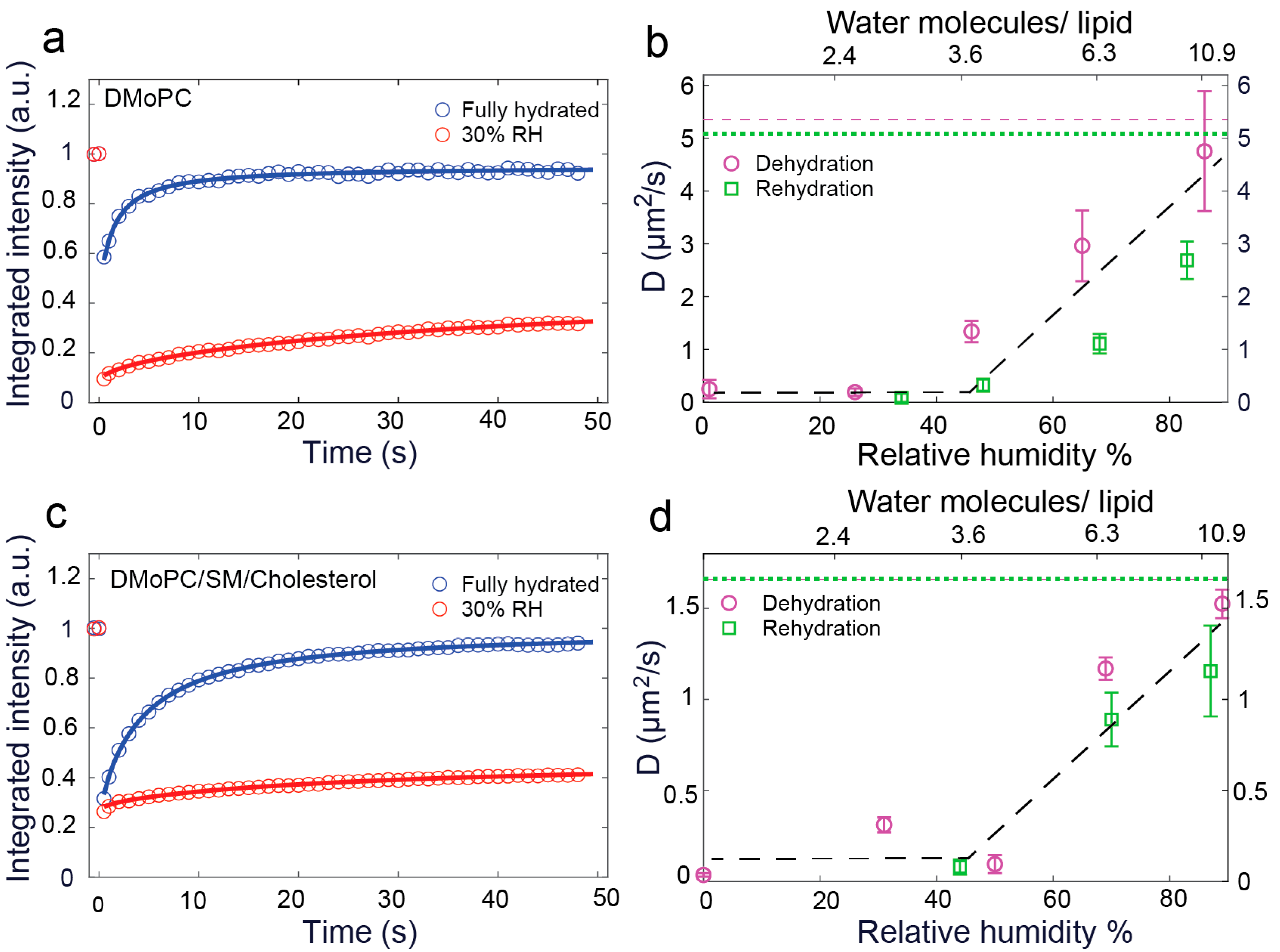
3.1. Measurement and Analysis of Diffusion Coefficient of Lipids at Different Hydration Conditions
3.2. Hydration Sensing
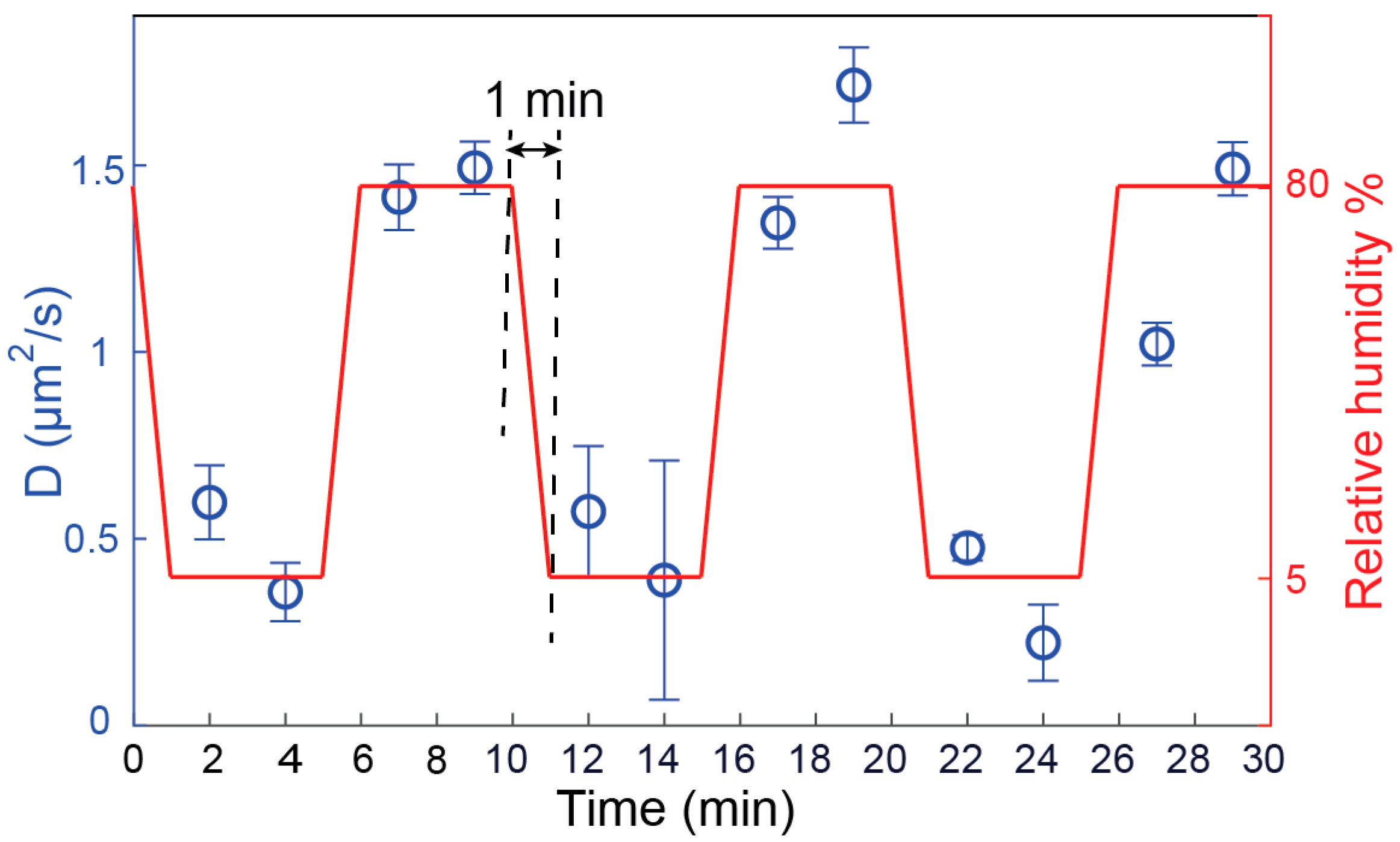
3.3. Validation of Hydration Sensing Approach
4. Discussion
4.1. Applicability and Measurement Criteria
4.2. Perspectives
5. Conclusions
6. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aeffner, S.; Reusch, T.; Weinhausen, B.; Salditt, T. Energetics of Stalk Intermediates in Membrane Fusion Are Controlled by Lipid Composition. Proc. Natl. Acad. Sci. USA 2012, 109, 1609–1618. [Google Scholar] [CrossRef]
- Subczynski, W.K.; Pasenkiewicz-Gierula, M.; Widomska, J.; Mainali, L.; Raguz, M. High Cholesterol/Low Cholesterol: Effects in Biological Membranes: A Review. Cell Biochem. Biophys. 2017, 75, 369–385. [Google Scholar] [CrossRef]
- Jackman, J.A.; Cho, N.J. Supported Lipid Bilayer Formation: Beyond Vesicle Fusion. Langmuir 2020, 36, 1387–1400. [Google Scholar] [CrossRef]
- Richter, R.P.; Bérat, R.; Brisson, A.R. Formation of Solid-Supported Lipid Bilayers: An Integrated View. Langmuir 2006, 22, 3497–3505. [Google Scholar] [CrossRef]
- Chan, Y.H.M.; Boxer, S.G. Model Membrane Systems and Their Applications. Curr. Opin. Chem. Biol. 2007, 11, 581–587. [Google Scholar] [CrossRef]
- Nieciecka, D.; Królikowska, A.; Krysinski, P. Probing the Interactions of Mitoxantrone with Biomimetic Membranes with Electrochemical and Spectroscopic Techniques. Electrochim. Acta 2015, 165, 430–442. [Google Scholar] [CrossRef]
- Khadka, N.K.; Cheng, X.; Ho, C.S.; Katsaras, J.; Pan, J. Interactions of the Anticancer Drug Tamoxifen with Lipid Membranes. Biophys. J. 2015, 108, 2492–2501. [Google Scholar] [CrossRef]
- Bilginer, R.; Arslan Yildiz, A. Biomimetic Model Membranes as Drug Screening Platform. In Biomimetic Lipid Membranes: Fundamentals, Applications, and Commercialization; Springer International Publishing: Cham, Switzerland, 2019; pp. 225–247. [Google Scholar]
- Wang, H.; Liu, Y.; He, R.; Xu, D.; Zang, J.; Weeranoppanant, N.; Dong, H.; Li, Y. Cell Membrane Biomimetic Nanoparticles for Inflammation and Cancer Targeting in Drug Delivery. Biomater. Sci. 2020, 8, 552–568. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Cheng, S.; Jin, Y.; Zhang, N.; Wang, Y. Membrane Engineering of Cell Membrane Biomimetic Nanoparticles for Nanoscale Therapeutics. Clin. Transl. Med. 2021, 11, e292. [Google Scholar] [CrossRef] [PubMed]
- Hindley, J.W.; Law, R.V.; Ces, O. Membrane Functionalization in Artificial Cell Engineering. SN Appl. Sci. 2020, 2, 1–10. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, Z.; Zhang, Y.; Cao, F.; Dong, K.; Ren, J.; Qu, X. Erythrocyte Membrane Cloaked Metal-Organic Framework Nanoparticle as Biomimetic Nanoreactor for Starvation-Activated Colon Cancer Therapy. ACS Nano 2018, 12, 10201–10211. [Google Scholar] [CrossRef]
- Liu, W.; Wu, J.; Ji, X.; Ma, Y.; Liu, L.; Zong, X.; Yang, H.; Dai, J.; Chen, X.; Xue, W. Advanced Biomimetic Nanoreactor for Specifically Killing Tumor Cells through Multi-Enzyme Cascade. Theranostics 2020, 10, 6245–6260. [Google Scholar] [CrossRef] [PubMed]
- Andersson, J.; Knoll, W. Tethered Lipid Membranes as Platforms for Biophysical Studies and Advanced Biosensors. In Biomimetic Lipid Membranes: Fundamentals, Applications, and Commercialization; Springer International Publishing: Cham, Switzerland, 2019; pp. 183–191. [Google Scholar]
- Alvarez-Malmagro, J.; García-Molina, G.; De Lacey, A.L. Electrochemical Biosensors Based on Membrane-Bound Enzymes in Biomimetic Configurations. Sensors 2020, 20, 3393. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Osaki, T.; Kamiya, K.; Yamada, T.; Miki, N.; Takeuchi, S. Rapid and Resilient Detection of Toxin Pore Formation Using a Lipid Bilayer Array. Small 2020, 16, 2005550. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Qi, S.; Tian, M.; Widjajanti, W.; Wang, R. Fabrication of Aquaporin-Based Biomimetic Membrane for Seawater Desalination. Desalination 2019, 467, 103–112. [Google Scholar] [CrossRef]
- Di Vincenzo, M.; Tiraferri, A.; Musteata, V.E.; Chisca, S.; Sougrat, R.; Huang, L.B.; Nunes, S.P.; Barboiu, M. Biomimetic Artificial Water Channel Membranes for Enhanced Desalination. Nat. Nanotechnol. 2021, 16, 190–196. [Google Scholar] [CrossRef] [PubMed]
- Abaie, E.; Xu, L.; Shen, Y.X. Bioinspired and Biomimetic Membranes for Water Purification and Chemical Separation: A Review. Front. Environ. Sci. Eng. 2021, 15, 124. [Google Scholar] [CrossRef]
- Fukuma, T.; Higgins, M.J.; Jarvis, S.P. Direct Imaging of Individual Intrinsic Hydration Layers on Lipid Bilayers at Ångstrom Resolution. Biophys. J. 2007, 92, 3603–3609. [Google Scholar] [CrossRef]
- Pasenkiewicz-Gierula, M.; Baczynski, K.; Markiewicz, M.; Murzyn, K. Computer Modelling Studies of the Bilayer/Water Interface. Biochim. Biophys. Acta Biomembr. 2016, 1858, 2305–2321. [Google Scholar] [CrossRef]
- Disalvo, E.A.; Lairion, F.; Martini, F.; Tymczyszyn, E.; Frías, M.; Almaleck, H.; Gordillo, G.J. Structural and Functional Properties of Hydration and Confined Water in Membrane Interfaces. Biochim. Biophys. Acta Biomembr. 2008, 1778, 2655–2670. [Google Scholar] [CrossRef]
- Tian, Z.; Gong, J.; Crowe, M.; Lei, M.; Li, D.; Ji, B.; Diao, J. Biochemical Studies of Membrane Fusion at the Single-Particle Level. Prog. Lipid Res. 2019, 73, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Sparrman, T.; Westlund, P.O. An NMR Line Shape and Relaxation Analysis of Heavy Water Powder Spectra of the Lα, Lβ′ and Pβ′ Phases in the DPPC/Water System. Phys. Chem. Chem. Phys. 2003, 5, 2114–2121. [Google Scholar] [CrossRef]
- Tristram-Nagle, S.; Nagle, J.F. Lipid Bilayers: Thermodynamics, Structure, Fluctuations, and Interactions. Chem. Phys. Lipids 2004, 127, 3–14. [Google Scholar] [CrossRef]
- Rheinstädter, M.C.; Ollinger, C.; Fragneto, G.; Demmel, F.; Salditt, T. Collective Dynamics of Lipid Membranes Studied by Inelastic Neutron Scattering. Phys. Rev. Lett. 2004, 93, 108107. [Google Scholar] [CrossRef]
- Berntsen, P.; Svanberg, C.; Swenson, J. Interplay between Hydration Water and Headgroup Dynamics in Lipid Bilayers. J. Phys. Chem. B 2011, 115, 1825–1832. [Google Scholar] [CrossRef]
- Tielrooij, K.J.; Paparo, D.; Piatkowski, L.; Bakker, H.J.; Bonn, M. Dielectric Relaxation Dynamics of Water in Model Membranes Probed by Terahertz Spectroscopy. Biophys. J. 2009, 97, 2484–2492. [Google Scholar] [CrossRef]
- Gennaro, A.; Deschaume, O.; Pfeiffer, H.; Bartic, C.; Wagner, P.; Wübbenhorst, M. Understanding the Dehydration Stress in Lipid Vesicles by a Combined Quartz Crystal Microbalance and Dielectric Spectroscopy Study. Phys. Status Solidi 2020, 217, 1900986. [Google Scholar] [CrossRef]
- Piatkowski, L.; De Heij, J.; Bakker, H.J. Probing the Distribution of Water Molecules Hydrating Lipid Membranes with Ultrafast Förster Vibrational Energy Transfer. J. Phys. Chem. B 2013, 117, 1367–1377. [Google Scholar] [CrossRef]
- Volkov, V.V.; Palmer, D.J.; Righini, R. Distinct Water Species Confined at the Interface of a Phospholipid Membrane. Phys. Rev. Lett. 2007, 99, 078302. [Google Scholar] [CrossRef]
- Demchenko, A.P.; Mély, Y.; Duportail, G.; Klymchenko, A.S. Monitoring Biophysical Properties of Lipid Membranes by Environment-Sensitive Fluorescent Probes. Biophys. J. 2009, 96, 3461–3470. [Google Scholar] [CrossRef]
- Watanabe, N.; Suga, K.; Slotte, J.P.; Nyholm, T.K.M.; Umakoshi, H. Lipid-Surrounding Water Molecules Probed by Time-Resolved Emission Spectra of Laurdan. Langmuir 2019, 35, 46. [Google Scholar] [CrossRef]
- Jurkiewicz, P.; Olżyńska, A.; Langner, M.; Hof, M. Headgroup hydration and mobility of DOTAP/DOPC bilayers: A fluorescence solvent relaxation study. Langmuir 2006, 22, 8741–8749. [Google Scholar] [CrossRef] [PubMed]
- Calero, C.; Franzese, G. Membranes with Different Hydration Levels: The Interface between Bound and Unbound Hydration Water. J. Mol. Liq. 2019, 273, 488–496. [Google Scholar] [CrossRef]
- Hutterer, R.; Hof, M. Dynamics in Diether Lipid Bilayers and Interdigitated Bilayer Structures Studied by Time-Resolved Emission Spectra, Decay Time and Anisotropy Profiles. J. Fluoresc. 2001, 11, 227–236. [Google Scholar] [CrossRef]
- Sýkora, J.; Kapusta, P.; Fidler, V.; Hof, M. On What Time Scale Does Solvent Relaxation in Phospholipid Bilayers Happen? Langmuir 2002, 18, 571–574. [Google Scholar] [CrossRef]
- Jurkiewicz, P.; Cwiklik, L.; Jungwirth, P.; Hof, M. Lipid Hydration and Mobility: An Interplay between Fluorescence Solvent Relaxation Experiments and Molecular Dynamics Simulations. Biochimie 2012, 94, 26–32. [Google Scholar] [CrossRef]
- Parasassi, T.; De Stasio, G.; d’Ubaldo, A.; Gratton, E. Phase Fluctuation in Phospholipid Membranes Revealed by Laurdan Fluorescence. Biophys. J. 1990, 57, 1179–1186. [Google Scholar] [CrossRef]
- Mukherjee, S.; Chattopadhyay, A. Wavelength-Selective Fluorescence as a Novel Tool to Study Organization and Dynamics in Complex Biological Systems. J. Fluoresc. 1995, 5, 237–246. [Google Scholar] [CrossRef]
- Chattopadhyay, A. Exploring Membrane Organization and Dynamics by the Wavelength-Selective Fluorescence Approach. Chem. Phys. Lipids 2003, 122, 3–17. [Google Scholar] [CrossRef]
- Barucha-Kraszewska, J.; Kraszewski, S.; Jurkiewicz, P.; Ramseyer, C.; Hof, M. Numerical Studies of the Membrane Fluorescent Dyes Dynamics in Ground and Excited States. Biochim. Biophys. Acta Biomembr. 2010, 1798, 1724–1734. [Google Scholar] [CrossRef] [PubMed]
- Jurkiewicz, P.; Sýkora, J.; Olzyńska, A.; Humpolíčková, J.; Hof, M. Solvent Relaxation in Phospholipid Bilayers: Principles and Recent Applications. J. Fluoresc. 2005, 15, 883–894. [Google Scholar] [CrossRef]
- Matysik, A.; Kraut, R.S. Preparation of Mica Supported Lipid Bilayers for High Resolution Optical Microscopy Imaging. J. Vis. Exp. 2014, 88, 52054. [Google Scholar] [CrossRef]
- Soumpasis, D.M. Theoretical Analysis of Fluorescence Photobleaching Recovery Experiments. Biophys. J. 1983, 41, 95–97. [Google Scholar] [CrossRef]
- Chattopadhyay, M.; Krok, E.; Orlikowska, H.; Schwille, P.; Franquelim, H.G.; Piatkowski, L. Hydration Layer of Only Few Molecules Controls Lipid Mobility in Biomimetic Membranes. bioRxiv 2021. [Google Scholar] [CrossRef]
- Honigmann, A.; Mueller, V.; Hell, S.W.; Eggeling, C. STED Microscopy Detects and Quantifies Liquid Phase Separation in Lipid Membranes Using a New Far-Red Emitting Fluorescent Phosphoglycerolipid Analogue. Faraday Discuss. 2012, 161, 77–89. [Google Scholar] [CrossRef]
- Böckmann, R.A.; Hac, A.; Heimburg, T.; Grubmüller, H. Effect of Sodium Chloride on a Lipid Bilayer. Biophys. J. 2003, 85, 1647–1655. [Google Scholar] [CrossRef]
- Hristova, K.; White, S.H. Determination of the Hydrocarbon Core Structure of Fluid Dioleoylphosphocholine (DOPC) Bilayers by X-ray Diffraction Using Specific Bromination of the Double-Bonds: Effect of Hydration. Biophys. J. 1998, 74, 2419–2433. [Google Scholar] [CrossRef]
- Leikin, S.L.; Kozlov, M.M.; Chernomordik, L.V.; Markin, V.S.; Chizmadzhev, Y.A. Membrane Fusion: Overcoming of the Hydration Barrier and Local Restructuring. J. Theor. Biol. 1987, 129, 411–425. [Google Scholar] [CrossRef]
- Wilschut, J.; Hoekstra, D. Membrane Fusion: Lipid Vesicles as a Model System. Chem. Phys. Lipids 1986, 40, 145–166. [Google Scholar] [CrossRef]
- Schneider, F.; Waithe, D.; Galiani, S.; Bernardino De La Serna, J.; Sezgin, E.; Eggeling, C. Nanoscale Spatiotemporal Diffusion Modes Measured by Simultaneous Confocal and Stimulated Emission Depletion Nanoscopy Imaging. Nano Lett. 2018, 18, 4233–4240. [Google Scholar] [CrossRef] [PubMed]
- Manzo, C.; Van Zanten, T.S.; Garcia-Parajo, M.F. Nanoscale Fluorescence Correlation Spectroscopy on Intact Living Cell Membranes with NSOM Probes. Biophys. J. 2011, 100, L8. [Google Scholar] [CrossRef]
- Sezgin, E.; Schneider, F.; Galiani, S.; Urbančič, I.; Waithe, D.; Lagerholm, B.C.; Eggeling, C. Measuring Nanoscale Diffusion Dynamics in Cellular Membranes with Super-Resolution STED–FCS. Nat. Protoc. 2019, 14, 1054–1083. [Google Scholar] [CrossRef]
- Eggeling, C. Super-Resolution Optical Microscopy of Lipid Plasma Membrane Dynamics. Essays Biochem. 2015, 57, 69–80. [Google Scholar]
- Sarangi, N.K.; Roobala, C.; Basu, J.K. Unraveling Complex Nanoscale Lipid Dynamics in Simple Model Biomembranes: Insights from Fluorescence Correlation Spectroscopy in Super-Resolution Stimulated Emission Depletion Mode. Methods 2018, 140–141, 198–211. [Google Scholar] [CrossRef]
- Regmi, R.; Winkler, P.M.; Flauraud, V.; Borgman, K.J.E.; Manzo, C.; Brugger, J.; Rigneault, H.; Wenger, J.; García-Parajo, M.F. Planar Optical Nanoantennas Resolve Cholesterol-Dependent Nanoscale Heterogeneities in the Plasma Membrane of Living Cells. Nano Lett. 2017, 17, 6295–6302. [Google Scholar] [CrossRef]
- Mudumbi, K.C.; Schirmer, E.C.; Yang, W. Single-Point Single-Molecule FRAP Distinguishes Inner and Outer Nuclear Membrane Protein Distribution. Nat. Commun. 2016, 7, 1–6. [Google Scholar] [CrossRef]
- Manzo, C.; Garcia-Parajo, M.F. A Review of Progress in Single Particle Tracking: From Methods to Biophysical Insights. Rep. Prog. Phys. 2015, 78, 124601. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.M.; Lin, Y.H.; Yen, T.C.; Hsieh, C.L. Nanoscopic Substructures of Raft-Mimetic Liquid-Ordered Membrane Domains Revealed by High-Speed Single-Particle Tracking. Sci. Rep. 2016, 6, 1–10. [Google Scholar]
- Spillane, K.M.; Ortega-Arroyo, J.; De Wit, G.; Eggeling, C.; Ewers, H.; Wallace, M.I.; Kukura, P. High-Speed Single-Particle Tracking of Gm1 in Model Membranes Reveals Anomalous Diffusion Due to Interleaflet Coupling and Molecular Pinning. Nano Lett. 2014, 14, 5390–5397. [Google Scholar] [CrossRef]
- Hsieh, C.L.; Spindler, S.; Ehrig, J.; Sandoghdar, V. Tracking Single Particles on Supported Lipid Membranes: Multimobility Diffusion and Nanoscopic Confinement. J. Phys. Chem. B 2014, 118, 1545–1554. [Google Scholar] [CrossRef]
- Pohl, P.; Saparov, S.M.; Pohl, E.E.; Evtodienko, V.Y.; Agapov, I.I.; Tonevitsky, A.G. Dehydration of Model Membranes Induced by Lectins from Ricinus Communis and Viscum Album. Biophys. J. 1998, 75, 2868–2876. [Google Scholar] [CrossRef][Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chattopadhyay, M.; Orlikowska, H.; Krok, E.; Piatkowski, L. Sensing Hydration of Biomimetic Cell Membranes. Biosensors 2021, 11, 241. https://doi.org/10.3390/bios11070241
Chattopadhyay M, Orlikowska H, Krok E, Piatkowski L. Sensing Hydration of Biomimetic Cell Membranes. Biosensors. 2021; 11(7):241. https://doi.org/10.3390/bios11070241
Chicago/Turabian StyleChattopadhyay, Madhurima, Hanna Orlikowska, Emilia Krok, and Lukasz Piatkowski. 2021. "Sensing Hydration of Biomimetic Cell Membranes" Biosensors 11, no. 7: 241. https://doi.org/10.3390/bios11070241
APA StyleChattopadhyay, M., Orlikowska, H., Krok, E., & Piatkowski, L. (2021). Sensing Hydration of Biomimetic Cell Membranes. Biosensors, 11(7), 241. https://doi.org/10.3390/bios11070241




