Visual Quantitation of Copper Ions Based on a Microfluidic Particle Dam Reflecting the Cu(II)-Catalyzed Oxidative Damage of DNA
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of Oligonucleotides
2.2. Cu(II)-Catalyzed Fenton Reaction
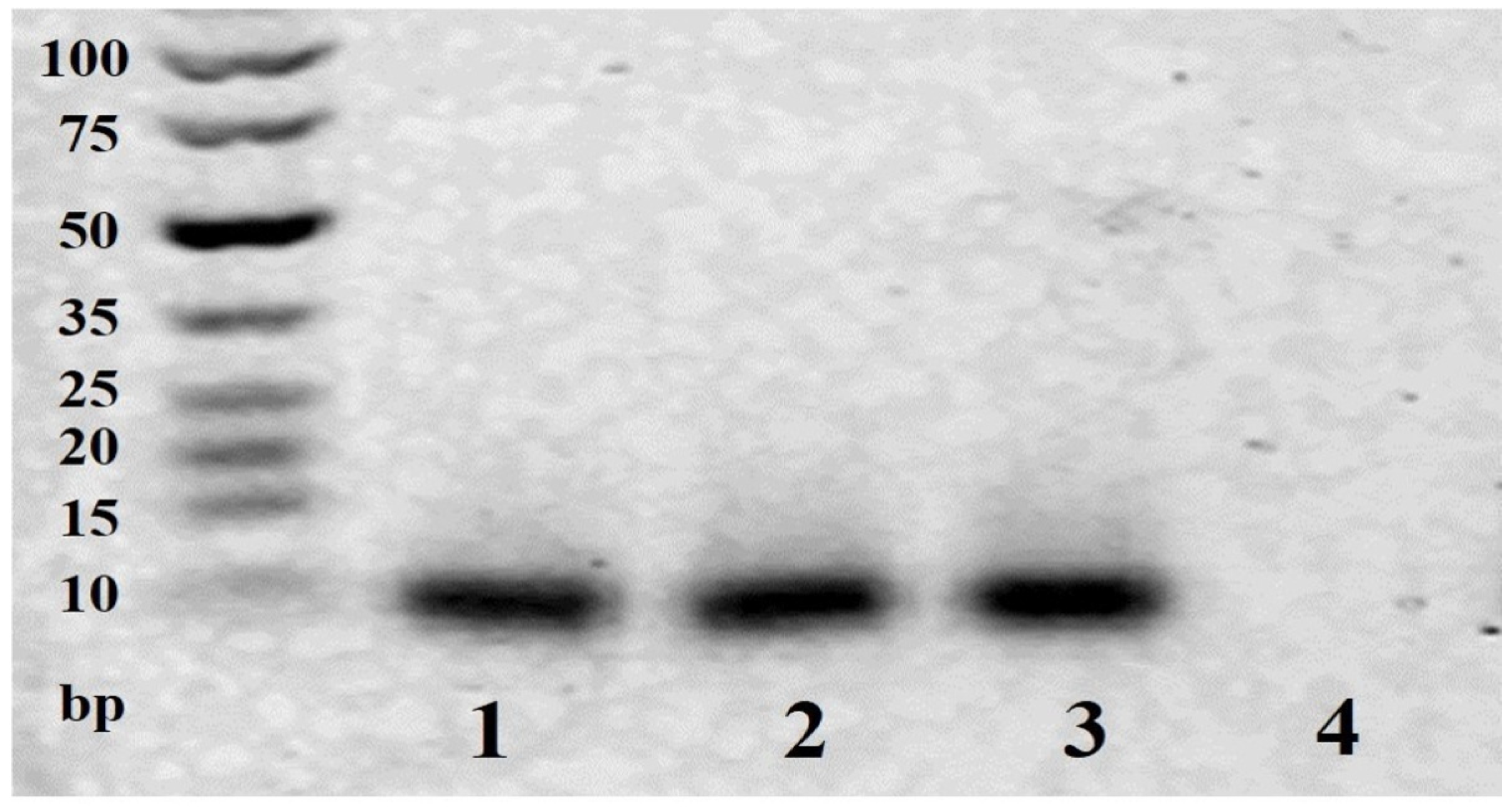
2.3. Agarose Gel Electrophoresis
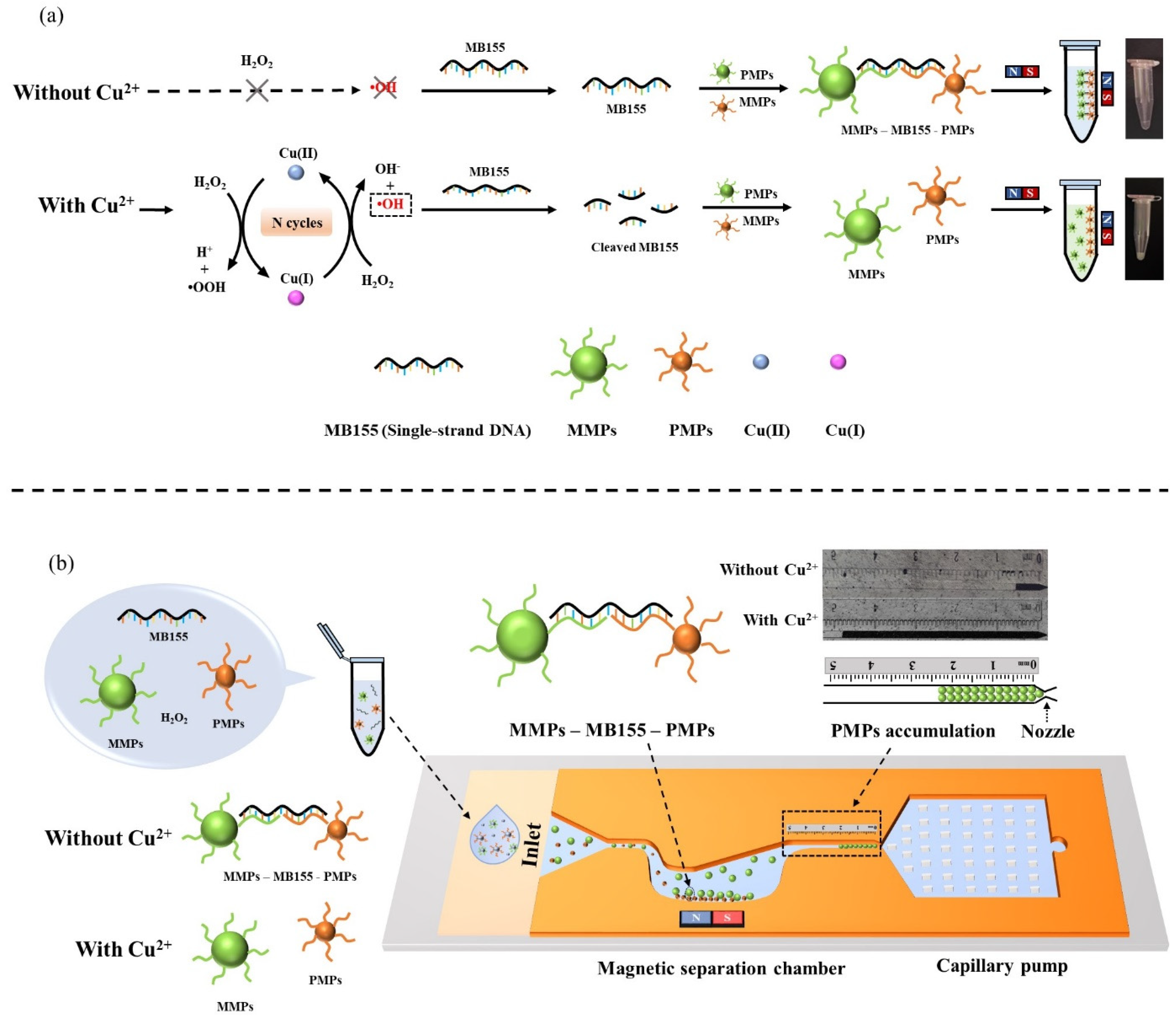
2.4. Magnetophoresis Assay
2.5. Detection on the Microfluidic Chip
3. Results and Discussion
3.1. Agarose Gel Electrophoresis of the Cleaved ssDNA
3.2. Magnetophoresis Assay
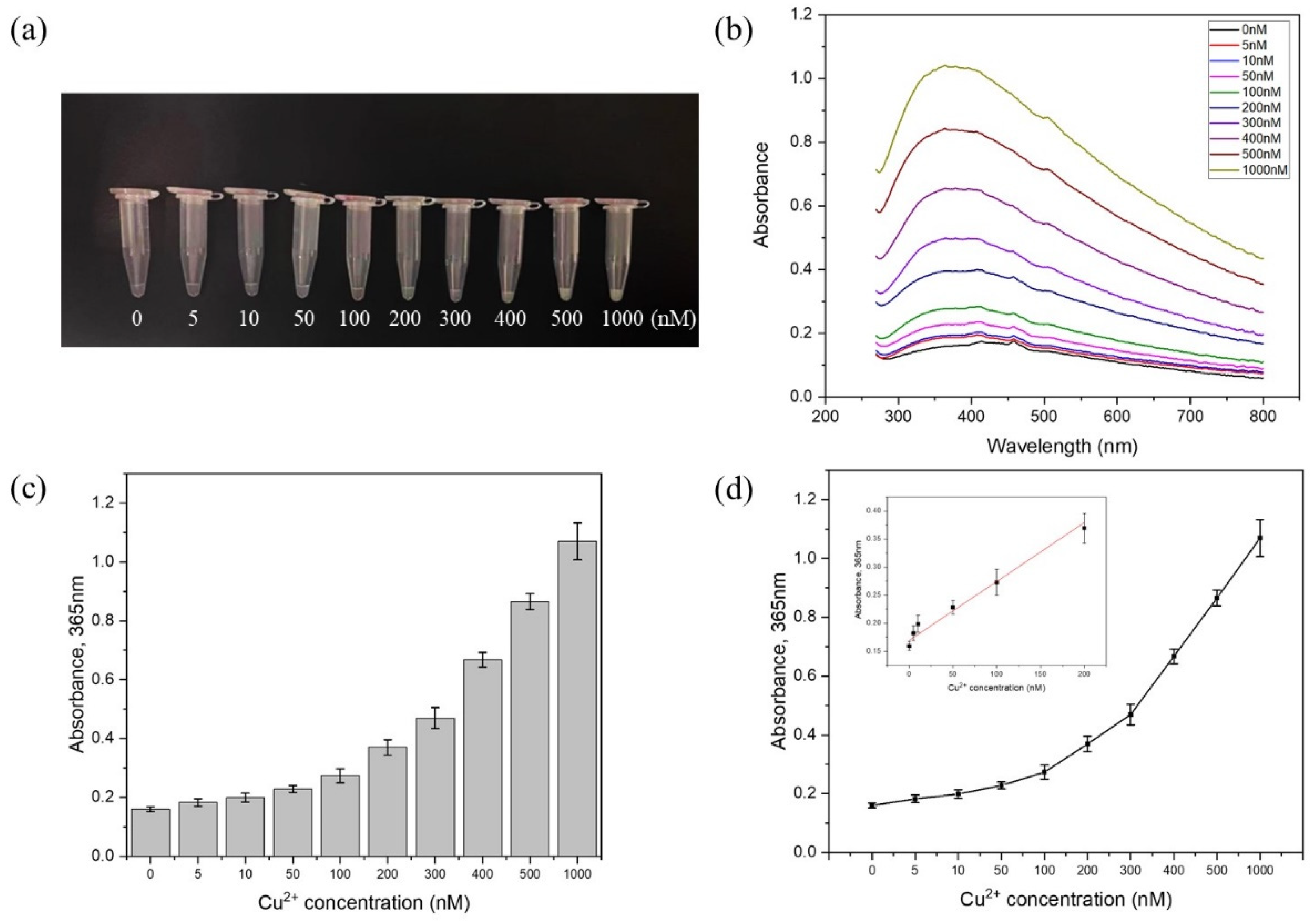
3.3. LOD on the Microfluidic Chip Test
3.4. Tolerance to Environmental Interference
3.5. Detection of Cu2+ in Tap Water
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Desai, V.; Kaler, S.G. Role of copper in human neurological disorders. Am. J. Clin. Nutr. 2008, 88, 855S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Percival, S.S. Copper and immunity. Am. J. Clin. Nutr. 1998, 67, 1064S–1068S. [Google Scholar] [CrossRef] [PubMed]
- Saari, J.T. Copper deficiency and cardiovascular disease: Role of peroxidation, glycation, and nitration. Can. J. Physiol. Pharmacol. 2000, 78, 848–855. [Google Scholar] [CrossRef] [PubMed]
- Lentini, P.; Zanoli, L.; Granata, A.; Signorelli, S.S.; Castellino, P.; Dell’Aquila, R. Kidney and heavy metals—The role of environmental exposure (Review). Mol. Med. Rep. 2017, 15, 3413–3419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patil, M.; Sheth, K.A.; Krishnamurthy, A.C.; Devarbhavi, H. A Review and Current Perspective on Wilson Disease. J. Clin. Exp. Hepatol. 2013, 3, 321–336. [Google Scholar] [CrossRef] [Green Version]
- Montes, S.; Rivera-Mancia, S.; Diaz-Ruiz, A.; Tristan-Lopez, L.; Rios, C. Copper and Copper Proteins in Parkinson’s Disease. Oxidative Med. Cell. Longev. 2014, 2014, 147251. [Google Scholar] [CrossRef] [Green Version]
- Waggoner, D.J.; Bartnikas, T.B.; Gitlin, J.D. The Role of Copper in Neurodegenerative Disease. Neurobiol. Dis. 1999, 6, 221–230. [Google Scholar] [CrossRef] [Green Version]
- Edition, F. Guidelines for drinking-water quality. WHO Chron. 2011, 38, 104–108. [Google Scholar]
- Dawson, J.B.; Ellis, D.J.; Newton-John, H. Direct estimation of copper in serum and urine by atomic absorption spectroscopy. Clin. Chim. Acta 1968, 21, 33–42. [Google Scholar] [CrossRef]
- List, G.R.; Evans, C.D.; Kwolek, W.F. Copper in edible oils: Trace amounts determined by atomic absorption spectroscopy. J. Am. Oil Chem. Soc. 1971, 48, 438–441. [Google Scholar] [CrossRef]
- Ferreira, S.L.C.; Lemos, V.A.; Moreira, B.C.; Costa, A.C.S.; Santelli, R.E. An on-line continuous flow system for copper enrichment and determination by flame atomic absorption spectroscopy. Anal. Chim. Acta 2000, 403, 259–264. [Google Scholar] [CrossRef]
- Townsend, A.T.; Miller, K.A.; McLean, S.; Aldous, S. The determination of copper, zinc, cadmium and lead in urine by high resolution ICP-MS. J. Anal. At. Spectrom. 1998, 13, 1213–1219. [Google Scholar] [CrossRef]
- Hu, Q.; Yang, G.; Zhao, Y.; Yin, J. Determination of copper, nickel, cobalt, silver, lead, cadmium, and mercury ions in water by solid-phase extraction and the RP-HPLC with UV-Vis detection. Anal. Bioanal. Chem. 2003, 375, 831–835. [Google Scholar] [CrossRef] [PubMed]
- Yin, B.-C.; Ye, B.-C.; Tan, W.; Wang, H.; Xie, C.-C. An Allosteric Dual-DNAzyme Unimolecular Probe for Colorimetric Detection of Copper(II). J. Am. Chem. Soc. 2009, 131, 14624–14625. [Google Scholar] [CrossRef] [Green Version]
- Lou, T.; Chen, L.; Chen, Z.; Wang, Y.; Chen, L.; Li, J. Colorimetric Detection of Trace Copper Ions Based on Catalytic Leaching of Silver-Coated Gold Nanoparticles. ACS Appl. Mater. Interfaces 2011, 3, 4215–4220. [Google Scholar] [CrossRef]
- Ma, Y.-R.; Niu, H.-Y.; Zhang, X.-L.; Cai, Y.-Q. Colorimetric detection of copper ions in tap water during the synthesis of silver/dopamine nanoparticles. Chem. Commun. 2011, 47, 12643–12645. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Cho, M.S.; Choe, W.S.; Lee, Y. Electrochemical analysis of copper ion using a Gly–Gly–His tripeptide modified poly(3-thiopheneacetic acid) biosensor. Biosens. Bioelectron. 2009, 25, 28–33. [Google Scholar] [CrossRef]
- Qing, M.; Xie, S.; Cai, W.; Tang, D.; Tang, Y.; Zhang, J.; Yuan, R. Click Chemistry Reaction-Triggered 3D DNA Walking Machine for Sensitive Electrochemical Detection of Copper Ion. Anal. Chem. 2018, 90, 11439–11445. [Google Scholar] [CrossRef]
- Wang, J.; Zong, Q. A new turn-on fluorescent probe for the detection of copper ion in neat aqueous solution. Sens. Actuators B Chem. 2015, 216, 572–577. [Google Scholar] [CrossRef]
- Luo, M.; Di, J.; Li, L.; Tu, Y.; Yan, J. Copper ion detection with improved sensitivity through catalytic quenching of gold nanocluster fluorescence. Talanta 2018, 187, 231–236. [Google Scholar] [CrossRef]
- Zhang, L.-P.; Xing, Y.-P.; Liu, C.; Zhou, X.-H.; Shi, H.-C. Label-free colorimetric detection of Cu2+ on the basis of Fenton reaction-assisted signal amplification with unmodified gold nanoparticles as indicator. Sens. Actuators B Chem. 2015, 215, 561–567. [Google Scholar] [CrossRef]
- Shan, Z.; Lu, M.; Wang, L.; MacDonald, B.; MacInnis, J.; Mkandawire, M.; Zhang, X.; Oakes, K.D. Chloride accelerated Fenton chemistry for the ultrasensitive and selective colorimetric detection of copper. Chem. Commun. 2016, 52, 2087–2090. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Deng, J.; Yi, Y.; Li, H.; Zhang, Y.; Yao, S. Label-free silicon quantum dots as fluorescent probe for selective and sensitive detection of copper ions. Talanta 2014, 125, 372–377. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, L.; Xue, J.; Dong, J.; Cai, J.; Hua, X.; Wang, M.; Zhang, C.; Liu, F. Signal-Amplified Lateral Flow Test Strip for Visual Detection of Cu2. PLoS ONE 2017, 12, e0169345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clayton, C.A.; Hines, J.W.; Elkins, P.D. Detection limits with specified assurance probabilities. Anal. Chem. 1987, 59, 2506–2514. [Google Scholar] [CrossRef]
- Lavagnini, I.; Magno, F. A statistical overview on univariate calibration, inverse regression, and detection limits: Application to gas chromatography/mass spectrometry technique. Mass Spectrom. Rev. 2007, 26, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhao, Y.; Zhang, Y. One-step green synthesized fluorescent carbon nanodots from bamboo leaves for copper(II) ion detection. Sens. Actuators B Chem. 2014, 196, 647–652. [Google Scholar] [CrossRef]
- Kashima-Tanaka, M.; Tsujimoto, Y.; Yamazaki, M. Quantification of Hydroxyl Radical Generated from the Fe2+-H2O2 and Cu2+-H2O2 Reaction Systems by Electron Spin Resonance Stop and Flow Technique. Int. J. Oral-Med Sci. 2002, 1, 67–73. [Google Scholar] [CrossRef]


| Method | LOD | Linear Range | Quantification | Ref. |
|---|---|---|---|---|
| Fluorescence | 0.15 µM | 0.1–0.6 µM | Fluorescence spectra | [19] |
| Colorimetric | 23 nM | 0.1–10 μM | Absorption spectra | [21] |
| Fluorescence | 115 nM | 0.333–66.6 μM | Fluorescence spectra | [27] |
| Visualization | 70.1 nM | 0–300 nM | The naked eye | This work |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, C.; Chen, T.-H. Visual Quantitation of Copper Ions Based on a Microfluidic Particle Dam Reflecting the Cu(II)-Catalyzed Oxidative Damage of DNA. Biosensors 2021, 11, 487. https://doi.org/10.3390/bios11120487
Cui C, Chen T-H. Visual Quantitation of Copper Ions Based on a Microfluidic Particle Dam Reflecting the Cu(II)-Catalyzed Oxidative Damage of DNA. Biosensors. 2021; 11(12):487. https://doi.org/10.3390/bios11120487
Chicago/Turabian StyleCui, Chenyu, and Ting-Hsuan Chen. 2021. "Visual Quantitation of Copper Ions Based on a Microfluidic Particle Dam Reflecting the Cu(II)-Catalyzed Oxidative Damage of DNA" Biosensors 11, no. 12: 487. https://doi.org/10.3390/bios11120487
APA StyleCui, C., & Chen, T.-H. (2021). Visual Quantitation of Copper Ions Based on a Microfluidic Particle Dam Reflecting the Cu(II)-Catalyzed Oxidative Damage of DNA. Biosensors, 11(12), 487. https://doi.org/10.3390/bios11120487





