Biological Responses of Onion-Shaped Carbon Nanoparticles
Abstract
1. Introduction
2. Materials and Methods
2.1. Pyrolysis of Nanodiamond
2.2. Carboxylated Nano-Onion
2.3. Characterization
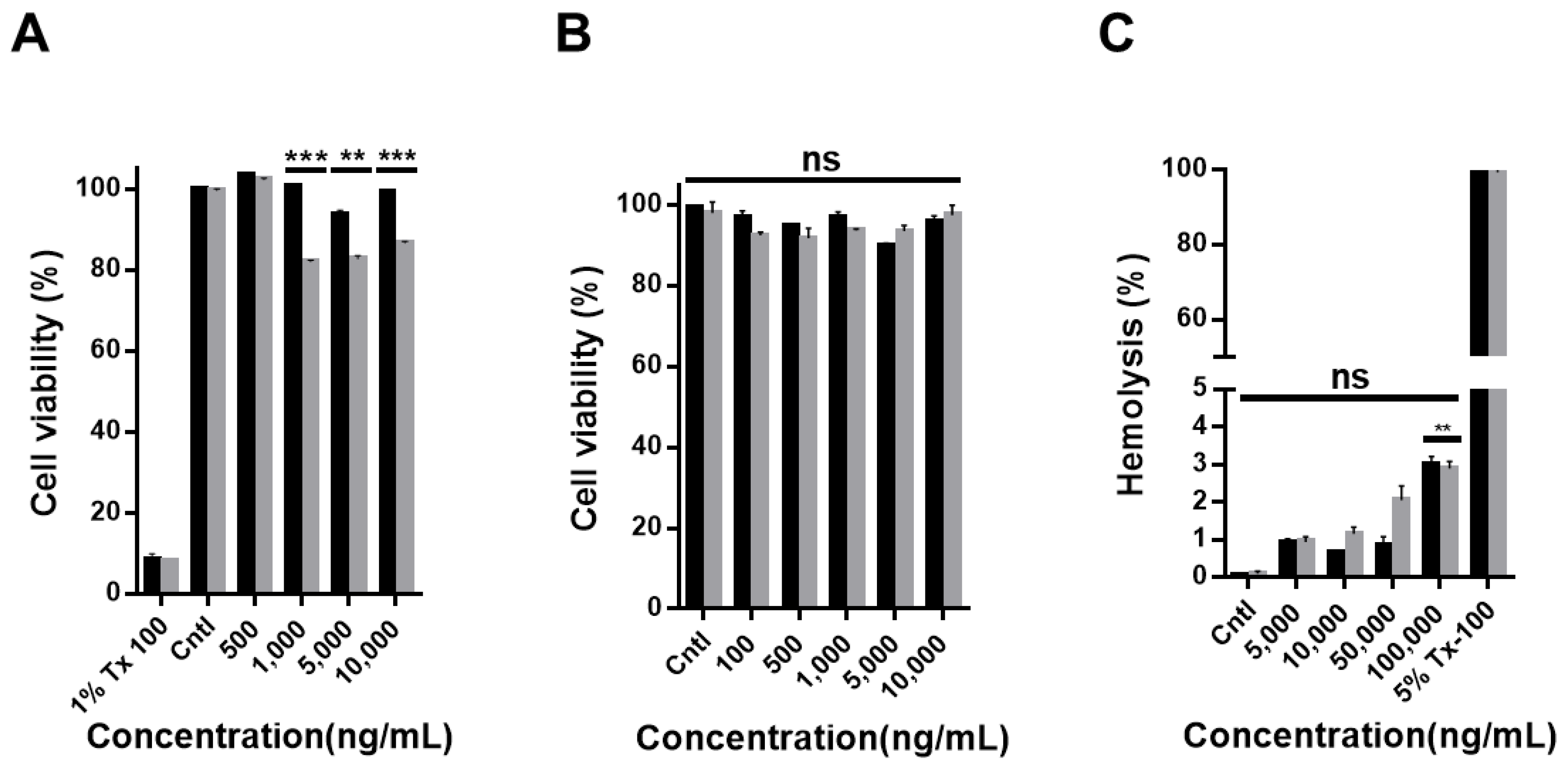
2.4. Cell Viability Assay
2.5. Hemolysis Test
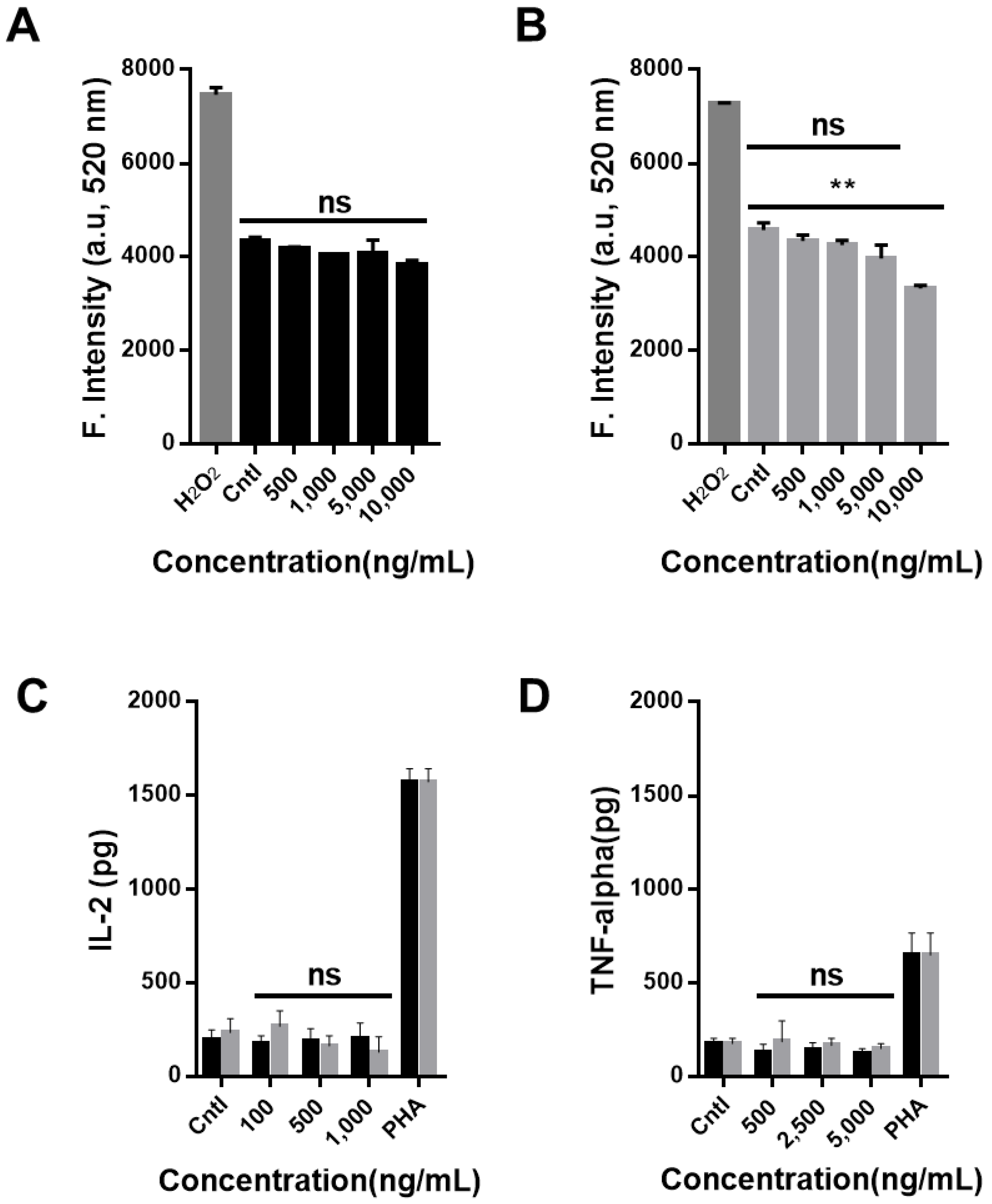
2.6. Intracellular Reactive Oxygen Species Measurement
2.7. Cytokine Profiling Assay
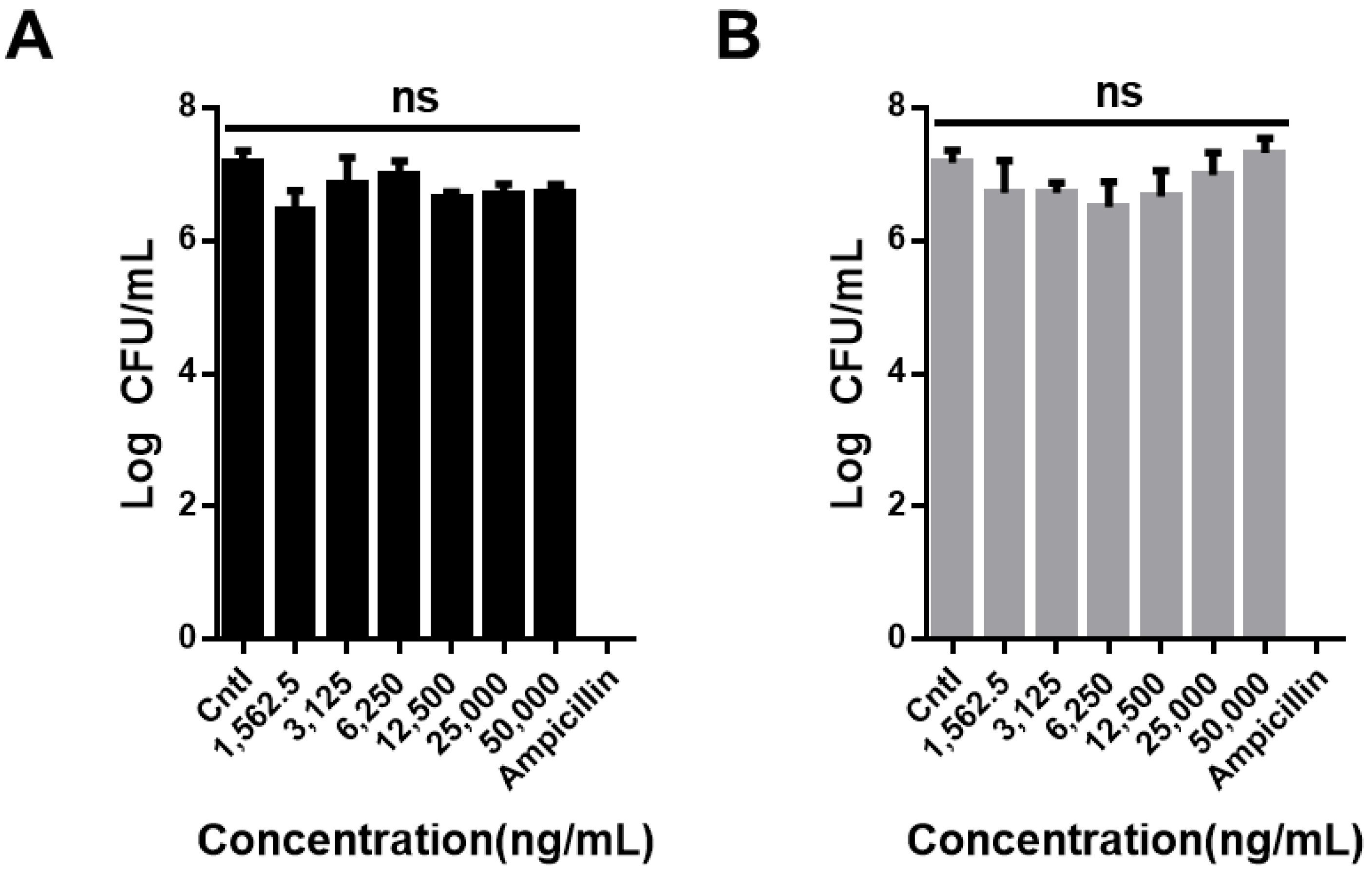
2.8. Bacterial Tests
2.9. Statistical Analysis
3. Results and Discussion
4. Summary
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Farokhzad, O.C.; Langer, R. Nanomedicine: developing smarter therapeutic and diagnostic modalities. Adv. Drug Deliv. Rev. 2006, 58, 1456–1459. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Tung, S.H.; Wang, N.S.; Reipa, V. Small-angle neutron scattering measurement of silicon nanoparticle size. Nanotechnology 2008, 19, 085715. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Lee, E.K.; Choo, J.; Yuh, J.; Hong, J.W. Micro 3D cell culture systems for cellular behavior studies: Culture matrices, devices, substrates, and in-situ sensing methods. Biotechnol. J. 2015, 10, 1682–1688. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Park, S.; Stojanović, Z.; Han, H.-S.; Lee, J.; Seok, H.K.; Uskoković, D.; Lee, K.H. Facile Solvothermal preparation of monodisperse gold nanoparticles and their engineered assembly of ferritin–gold nanoclusters. Langmuir 2013, 29, 15698–15703. [Google Scholar] [CrossRef] [PubMed]
- Reipa, V.; Purdum, G.; Choi, J. Measurement of Nanoparticle Concentration Using Quartz Crystal Microgravimetry. J. Phys. Chem. B 2010, 114, 16112–16117. [Google Scholar] [CrossRef] [PubMed]
- Nie, S.; Xing, Y.; Kim, G.J.; Simons, J.W. Nanotechnology applications in cancer. Annu. Rev. Biomed. Eng. 2007, 9, 257–288. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Piao, Y.; Hyeon, T. Multifunctional nanostructured materials for multimodal imaging, and simultaneous imaging and therapy. Chem. Soc. Rev. 2009, 38, 372–390. [Google Scholar] [CrossRef]
- Sun, C.; Lee, J.S.; Zhang, M. Magnetic nanoparticles in MR imaging and drug delivery. Adv. Drug Deliv. Rev. 2008, 60, 1252–1265. [Google Scholar] [CrossRef]
- Yang, K.; Feng, L.Z.; Liu, Z. Stimuli responsive drug delivery systems based on nano-graphene for cancer therapy. Adv. Drug Deliv. Rev. 2016, 105, 228–241. [Google Scholar] [CrossRef]
- Hwang, J.; Lee, E.; Kim, J.; Seo, Y.; Lee, K.H.; Hong, J.W.; Gilad, A.A.; Park, H.; Choi, J. Effective delivery of immunosuppressive drug molecules by silica coated iron oxide nanoparticles. Colloid Surface B 2016, 142, 290–296. [Google Scholar] [CrossRef]
- Morones, J.R.; Elechiguerra, J.L.; Camacho, A.; Holt, K.; Kouri, J.B.; Ramirez, J.T.; Yacaman, M.J. The bactericidal effect of silver nanoparticles. Nanotechnology 2005, 16, 2346–2353. [Google Scholar] [CrossRef] [PubMed]
- Shadjou, N.; Hasanzadeh, M. Graphene and its nanostructure derivatives for use in bone tissue engineering: Recent advances. J. Biomed Mater. Res. A 2016, 104, 1250–1275. [Google Scholar] [CrossRef] [PubMed]
- Gajendiran, M.; Choi, J.; Kim, S.J.; Kim, K.; Shin, H.; Koo, H.J.; Kim, K. Conductive biomaterials for tissue engineering applications. J. Ind. Eng. Chem. 2017, 51, 12–26. [Google Scholar] [CrossRef]
- Tiwari, J.N.; Vij, V.; Kemp, K.C.; Kim, K.S. Engineered Carbon-Nanomaterial-Based Electrochemical Sensors for Biomolecules. Acs Nano 2016, 10, 46–80. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.K.; Na, H.K.; Kim, S.; Jang, H.; Chang, S.J.; Min, D.H. One-pot synthesis of multifunctional Au@graphene oxide nanocolloid core@shell nanoparticles for Raman bioimaging, photothermal, and photodynamic therapy. Small 2015, 11, 2527–2535. [Google Scholar] [CrossRef] [PubMed]
- De Faria, A.F.; Martinez, D.S.T.; Meira, S.M.M.; de Moraes, A.C.M.; Brandelli, A.; Souza, A.G.; Alves, O.L. Anti-adhesion and antibacterial activity of silver nanoparticles supported on graphene oxide sheets. Colloid Surface B 2014, 113, 115–124. [Google Scholar] [CrossRef]
- Jia, G.; Wang, H.; Yan, L.; Wang, X.; Pei, R.; Yan, T.; Zhao, Y.; Guo, X. Cytotoxicity of carbon nanomaterials: single-wall nanotube, multi-wall nanotube, and fullerene. Environ. Sci. Technol. 2005, 39, 1378–1383. [Google Scholar] [CrossRef]
- Warheit, D.B.; Laurence, B.R.; Reed, K.L.; Roach, D.H.; Reynolds, G.A.; Webb, T.R. Comparative pulmonary toxicity assessment of single-wall carbon nanotubes in rats. Toxicol. Sci. 2004, 77, 117–125. [Google Scholar] [CrossRef]
- Lam, C.W.; James, J.T.; McCluskey, R.; Hunter, R.L. Pulmonary toxicity of single-wall carbon nanotubes in mice 7 and 90 days after intratracheal instillation. Toxicol. Sci. 2004, 77, 126–134. [Google Scholar] [CrossRef]
- Schrand, A.M.; Dai, L.; Schlager, J.J.; Hussain, S.M.; Osawa, E. Differential biocompatibility of carbon nanotubes and nanodiamonds. Diam. Relat. Mater. 2007, 16, 2118–2123. [Google Scholar] [CrossRef]
- Li, Y.; Liu, Y.; Fu, Y.J.; Wei, T.T.; Le Guyader, L.; Gao, G.; Liu, R.S.; Chang, Y.Z.; Chen, C.Y. The triggering of apoptosis in macrophages by pristine graphene through the MAPK and TGF-beta signaling pathways. Biomaterials 2012, 33, 402–411. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.H.; Ren, Y.P.; Zhang, Y.Y.; Chen, Y.; Zhang, C.; Bai, Z.X.; Ma, R.; Chen, Q. Synthesis of Pb nanowires-Au nanoparticles nanostructure decorated with reduced graphene oxide for electrochemical sensing. Talanta 2017, 165, 604–611. [Google Scholar] [CrossRef] [PubMed]
- Wan, B.; Wang, Z.X.; Lv, Q.Y.; Dong, P.X.; Zhao, L.X.; Yang, Y.; Guo, L.H. Single-walled carbon nanotubes and graphene oxides induce autophagosome accumulation and lysosome impairment in primarily cultured murine peritoneal macrophages. Toxicol. Lett. 2013, 221, 118–127. [Google Scholar] [CrossRef]
- Zeng, Y.P.; Yang, Z.Y.; Luo, S.L.; Li, H.; Liu, C.; Hao, Y.H.; Liu, J.; Wang, W.D.; Li, R. Fast and facile preparation of PEGylated graphene from graphene oxide by lysosome targeting delivery of photosensitizer to efficiently enhance photodynamic therapy. Rsc Adv. 2015, 5, 57725–57734. [Google Scholar] [CrossRef]
- Dong, P.X.; Wan, B.; Wang, Z.X.; Guo, L.H.; Yang, Y.; Zhao, L. Exposure of single-walled carbon nanotubes impairs the functions of primarily cultured murine peritoneal macrophages. Nanotoxicology 2013, 7, 1028–1042. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Liu, Y.; Song, W.M.; Hayashi, Y.; Ding, X.C.; Li, W.H. In Vitro Evaluation of Cytotoxicity and Oxidative Stress Induced by Multiwalled Carbon Nanotubes in Murine RAW 264.7 Macrophages and Human A549 Lung Cells. Biomed. Environ. Sci. 2011, 24, 593–601. [Google Scholar]
- Wang, X.; Guo, J.; Chen, T.; Nie, H.; Wang, H.; Zang, J.; Cui, X.; Jia, G. Multi-walled carbon nanotubes induce apoptosis via mitochondrial pathway and scavenger receptor. Toxicol. In Vitro 2012, 26, 799–806. [Google Scholar] [CrossRef]
- Chen, H.Q.; Wang, B.; Gao, D.; Guan, M.; Zheng, L.N.; Ouyang, H.; Chai, Z.F.; Zhao, Y.L.; Feng, W.Y. Broad-Spectrum Antibacterial Activity of Carbon Nanotubes to Human Gut Bacteria. Small 2013, 9, 2735–2746. [Google Scholar] [CrossRef]
- Fernandes, A.M.; Abdalhai, M.H.; Ji, J.; Xi, B.W.; Xie, J.; Sun, J.; Noeline, R.; Lee, B.H.; Sun, X. Development of highly sensitive electrochemical genosensor based on multiwalled carbon nanotubes-chitosan-bismuth and lead sulfide nanoparticles for the detection of pathogenic Aeromonas. Biosens Bioelectron 2015, 63, 399–406. [Google Scholar] [CrossRef]
- Zhang, X.K.; Meng, L.J.; Lu, Q.H.; Fei, Z.F.; Dyson, P.J. Targeted delivery and controlled release of doxorubicin to cancer cells using modified single wall carbon nanotubes. Biomaterials 2009, 30, 6041–6047. [Google Scholar] [CrossRef]
- Ali, M.A.; Srivastava, S.; Solanki, P.R.; Reddy, V.; Agrawal, V.V.; Kim, C.; John, R.; Malhotra, B.D. Highly efficient bienzyme functionalized nanocomposite-based microfluidics biosensor platform for biomedical application. Sci. Rep. 2013, 3, 2661. [Google Scholar] [CrossRef] [PubMed]
- Flavin, K.; Chaur, M.N.; Echegoyen, L.; Giordani, S. Functionalization of multilayer fullerenes (carbon nano-onions) using diazonium compounds and "click" chemistry. Org. Lett. 2010, 12, 840–843. [Google Scholar] [CrossRef] [PubMed]
- Bartelmess, J.; Giordani, S. Carbon nano-onions (multi-layer fullerenes): chemistry and applications. Beilstein J. Nanotechnol. 2014, 5, 1980–1998. [Google Scholar] [CrossRef] [PubMed]
- Marchesano, V.; Ambrosone, A.; Bartelmess, J.; Strisciante, F.; Tino, A.; Echegoyen, L.; Tortiglione, C.; Giordani, S. Impact of Carbon Nano-Onions on Hydra vulgaris as a Model Organism for Nanoecotoxicology. Nanomater. Basel 2015, 5, 1331–1350. [Google Scholar] [CrossRef] [PubMed]
- Palkar, A.; Melin, F.; Cardona, C.M.; Elliott, B.; Naskar, A.K.; Edie, D.D.; Kumbhar, A.; Echegoyen, L. Reactivity differences between carbon nano onions (CNOs) prepared by different methods. Chem-Asian J. 2007, 2, 625–633. [Google Scholar] [CrossRef]
- Liong, M.; Lu, J.; Kovochich, M.; Xia, T.; Ruehm, S.G.; Nel, A.E.; Tamanoi, F.; Zink, J.I. Multifunctional inorganic nanoparticles for imaging, targeting, and drug delivery. Acs Nano 2008, 2, 889–896. [Google Scholar] [CrossRef]
- Lee, S.C.; Kim, C.; Kwon, I.C.; Chung, H.; Jeong, S.Y. Polymeric micelles of poly (2-ethyl-2-oxazoline)-block-poly (ε-caprolactone) copolymer as a carrier for paclitaxel. J. Controll. Release 2003, 89, 437–446. [Google Scholar]
- Karlsson, H.L.; Cronholm, P.; Gustafsson, J.; Moller, L. Copper oxide nanoparticles are highly toxic: a comparison between metal oxide nanoparticles and carbon nanotubes. Chem. Res. Toxicol. 2008, 21, 1726–1732. [Google Scholar] [CrossRef]
- Fritz, J.M.; Weaver, T.E. The BiP cochaperone ERdj4 is required for B cell development and function. PLoS ONE 2014, 9, e107473. [Google Scholar] [CrossRef]
- Harwig, S.; Tan, L.; Qu, X.-D.; Cho, Y.; Eisenhauer, P.B.; Lehrer, R.I. Bactericidal properties of murine intestinal phospholipase A2. J. Clin. Investig. 1995, 95, 603–610. [Google Scholar] [CrossRef]
- Das, S.; Singh, S.; Singh, V.; Joung, D.; Dowding, J.M.; Reid, D.; Anderson, J.; Zhai, L.; Khondaker, S.I.; Self, W.T. Oxygenated functional group density on graphene oxide: its effect on cell toxicity. Part. Part. Syst. Charact. 2013, 30, 148–157. [Google Scholar] [CrossRef]
- Arthur, J.C.; Gharaibeh, R.Z.; Muhlbauer, M.; Perez-Chanona, E.; Uronis, J.M.; McCafferty, J.; Fodor, A.A.; Jobin, C. Microbial genomic analysis reveals the essential role of inflammation in bacteria-induced colorectal cancer. Nat. Commun. 2014, 5, 4724. [Google Scholar] [CrossRef] [PubMed]
- Fei, N.; Zhao, L. An opportunistic pathogen isolated from the gut of an obese human causes obesity in germfree mice. ISME J. 2013, 7, 880–884. [Google Scholar] [CrossRef] [PubMed]
- Jie, Z.; Xia, H.; Zhong, S.L.; Feng, Q.; Li, S.; Liang, S.; Zhong, H.; Liu, Z.; Gao, Y.; Zhao, H.; et al. The gut microbiome in atherosclerotic cardiovascular disease. Nat. Commun. 2017, 8, 845. [Google Scholar] [CrossRef]

| Nanomaterial | Target Cell | Effect in Cells | Application | Ref. |
|---|---|---|---|---|
| Neat graphene | RAW 264.7 | ROS generation, apoptosis induction | Electrochemical sensing | [21,22] |
| Graphene oxides (GO) | Peritoneal macrophages | LDH release, Autophagosome accumulation, | Photodynamic therapy (PDT) | [23,24] |
| SWCNTs | Mouse peritoneal macrophages | Mitochondrial damage | Antibacterial material | [25,28] |
| Acid functionalized SWCNTs | Peritoneal macrophages | LDH release, Autophagosome accumulation | Drug delivery carrier | [23,30] |
| MWCNTs | RAW264.7, A549 | LDH release and oxidative stress | Genosensor | [26,29] |
| Acid functionalized MWCNTs | RAW 264.7 | Apoptosis via the mitochondrial pathway | Biosensor | [27,31] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jang, J.; Kim, Y.; Hwang, J.; Choi, Y.; Tanaka, M.; Kang, E.; Choi, J. Biological Responses of Onion-Shaped Carbon Nanoparticles. Nanomaterials 2019, 9, 1016. https://doi.org/10.3390/nano9071016
Jang J, Kim Y, Hwang J, Choi Y, Tanaka M, Kang E, Choi J. Biological Responses of Onion-Shaped Carbon Nanoparticles. Nanomaterials. 2019; 9(7):1016. https://doi.org/10.3390/nano9071016
Chicago/Turabian StyleJang, Jaehee, Youngjun Kim, Jangsun Hwang, Yonghyun Choi, Masayoshi Tanaka, Eunah Kang, and Jonghoon Choi. 2019. "Biological Responses of Onion-Shaped Carbon Nanoparticles" Nanomaterials 9, no. 7: 1016. https://doi.org/10.3390/nano9071016
APA StyleJang, J., Kim, Y., Hwang, J., Choi, Y., Tanaka, M., Kang, E., & Choi, J. (2019). Biological Responses of Onion-Shaped Carbon Nanoparticles. Nanomaterials, 9(7), 1016. https://doi.org/10.3390/nano9071016






