Oro-Respiratory Dysbiosis and Its Modulatory Effect on Lung Mucosal Toxicity during Exposure or Co-Exposure to Carbon Nanotubes and Cigarette Smoke
Abstract
1. Introduction
2. Materials and Methods
2.1. CNT Preparation and Characterization
2.2. CSE Preparation
2.3. Animals, Microbiota Depletion, and Toxicant Exposures
2.4. Organ Harvesting and Microbial Cell Recovery
2.5. Microbial DNA Isolation and Nextgen Sequencing
2.6. RNA Isolation
2.7. qRT-PCR for Gene Expression Analysis
2.8. IgA Estimation in BAL Fluid
2.9. Histopathological Analysis of Lung Tissue
2.10. Statistical Analyses
3. Results
3.1. Toxicant Exposure and Co-Exposure Led to Oro-Respiratory Microbiota Dysbiosis in WT Mice
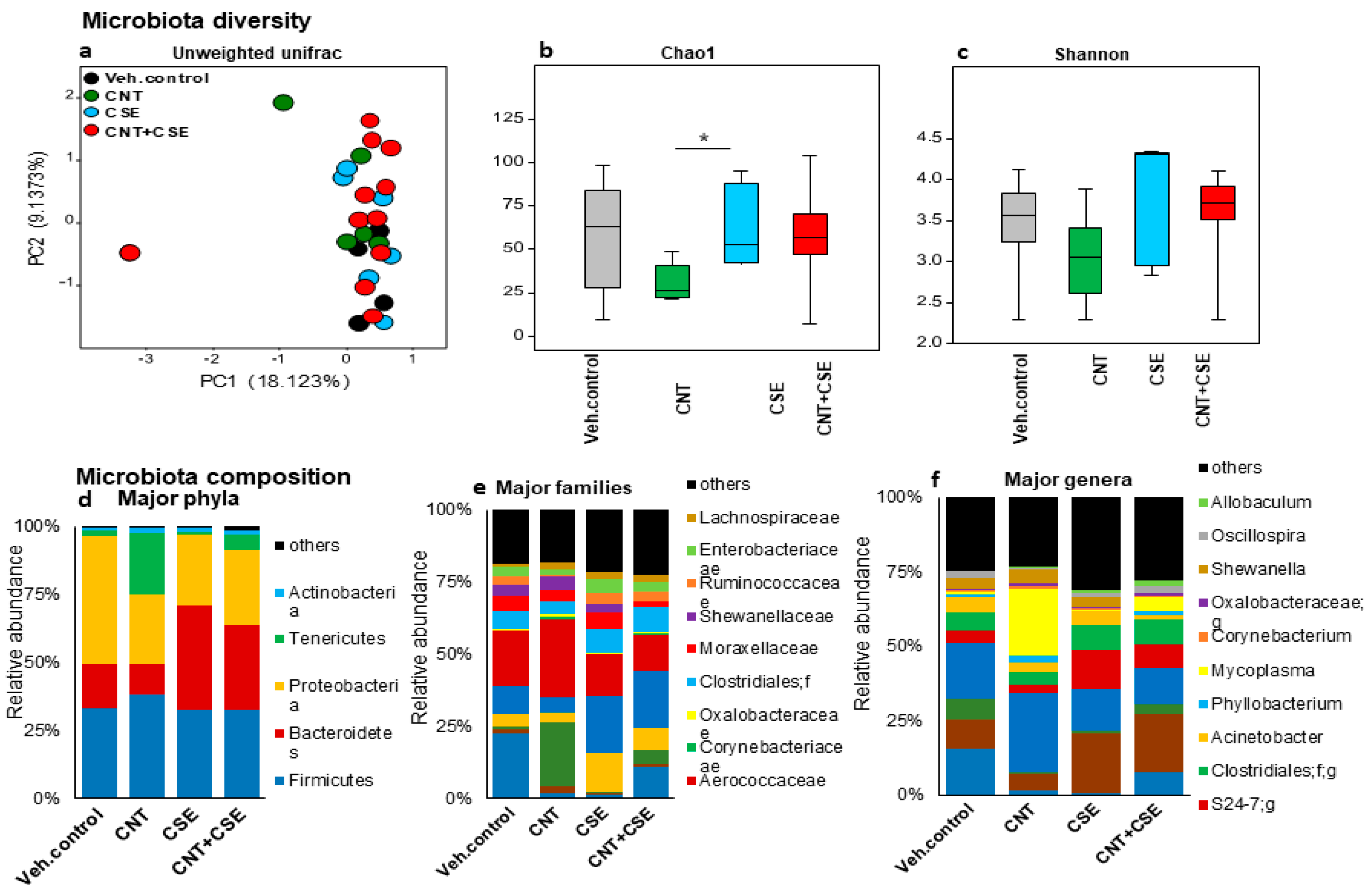
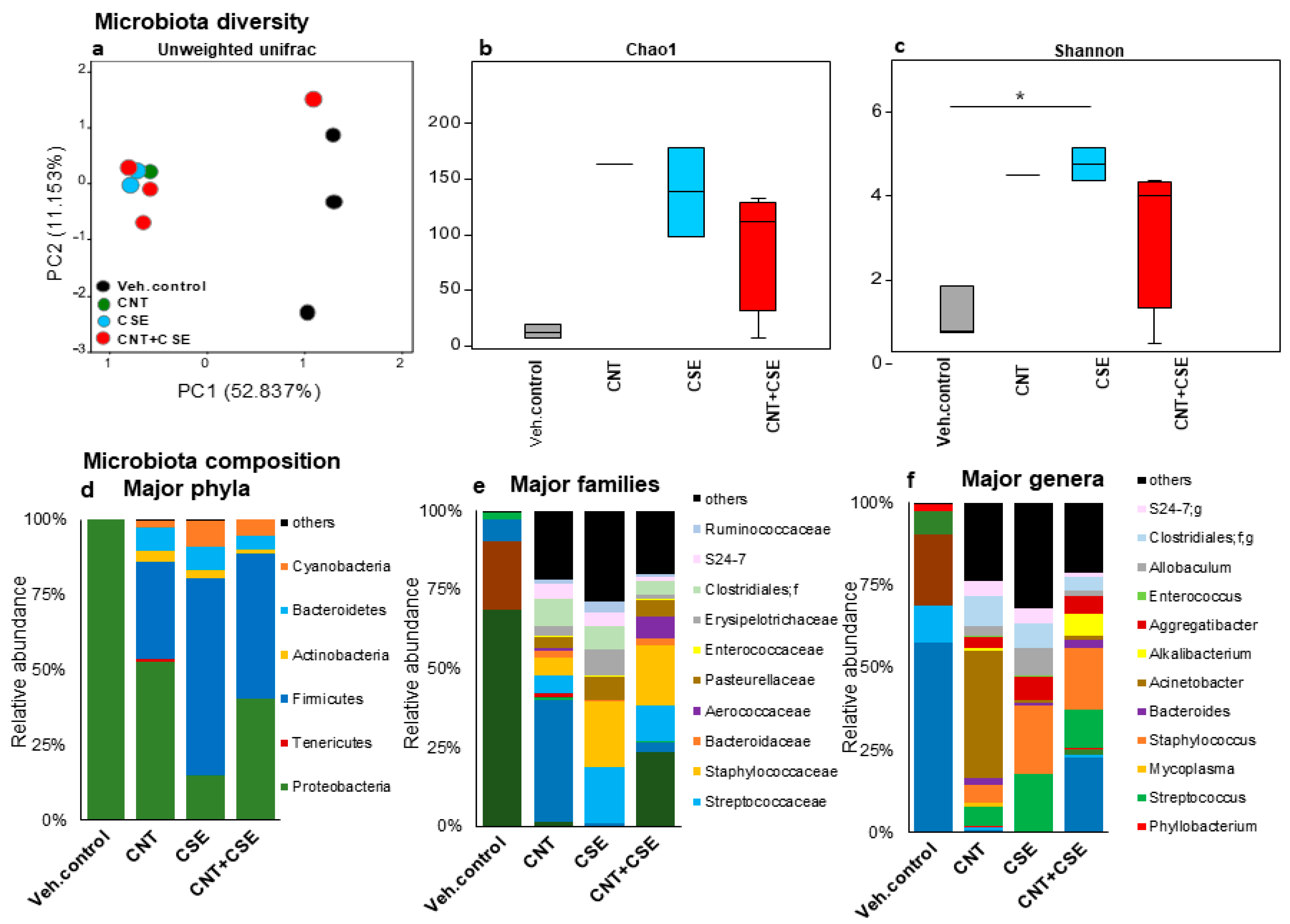
3.1.1. Lung Microbiota Dysbiosis
3.1.2. Oral Microbiota Dysbiosis
3.1.3. Nasal Microbiota Dysbiosis
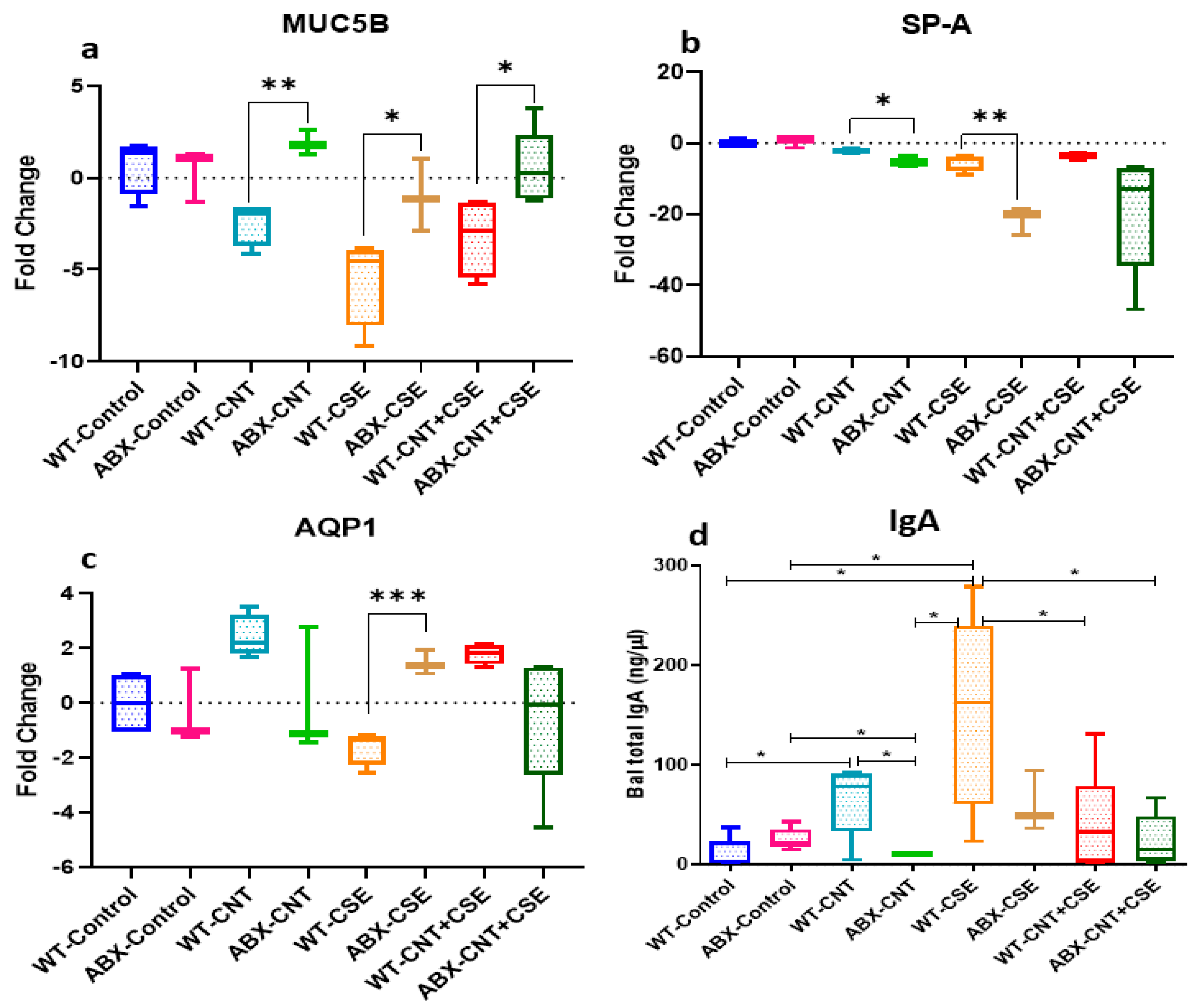
3.2. Microbiota Depletion Differentially Regulated the Lung Mucosal Homeostasis/Defense Proteins
3.2.1. Microbiota Depletion Upregulated Mucin (MUC5B) Gene Expression
3.2.2. Surfactant Protein-A (SP-A) Was Downregulated in Microbiota-Depleted Mice
3.2.3. Microbiota Depletion Exerted a Differential Effect on Aquaporin-1 (AQP-1) Gene Expression
3.2.4. Microbiota Depletion Reduced Total Immunoglobulin A (IgA) Secretion in Lung
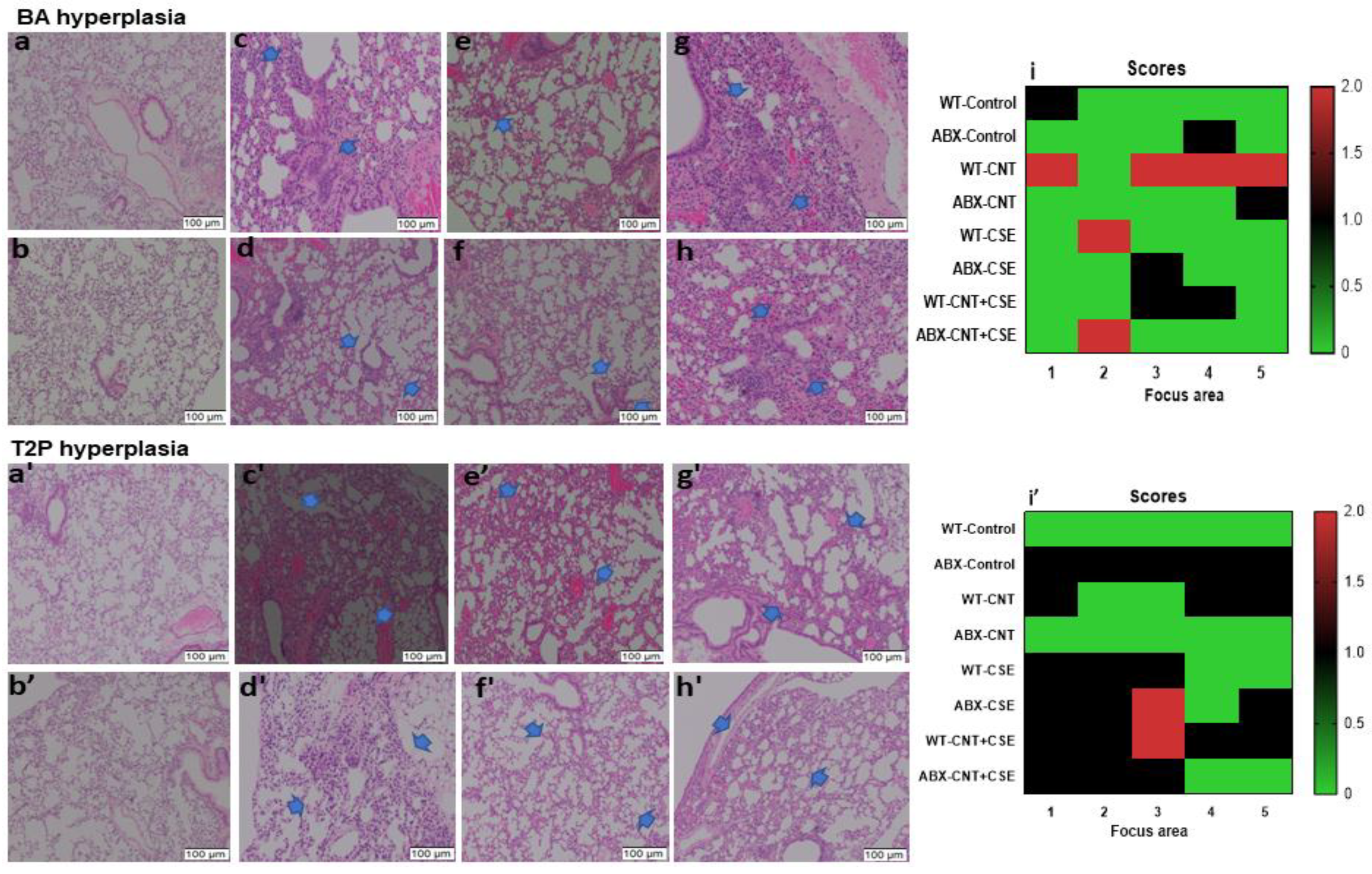
3.3. Microbiota Dysbiosis Modulated the CNT- and/or CSE-Induced Lung Pathology
4. Discussion
4.1. Dysbiosis of Oro-Respiratory Microbiota Due to Respiratory Exposure to CNT and/or Tobacco Smoke Constituents
4.1.1. Lung Microbiota
4.1.2. Oral Microbiota
4.2. CNT- and/or CSE-Induced Lung Damage and Pathology
4.3. Modulatory Effects of Lung Microbiota on Mucosal Homeostasis and Damage in Toxicant-Exposed Lungs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rahman, G.; Najaf, Z.; Mehmood, A.; Bilal, S.; Shah, A.H.; Mian, S.A.; Ali, G. An Overview of the Recent Progress in the Synthesis and Applications of Carbon Nanotubes. C 2019, 5, 3. [Google Scholar] [CrossRef]
- Ravichandran, P.; Baluchamy, S.; Gopikrishnan, R.; Biradar, S.; Ramesh, V.; Goornavar, V.; Thomas, R.; Wilson, B.L.; Jeffers, R.; Hall, J.C.; et al. Pulmonary Biocompatibility Assessment of Inhaled Single-Wall and Multiwall Carbon Nanotubes in BALB/c Mice. J. Biol. Chem. 2011, 286, 29725–29733. [Google Scholar] [CrossRef]
- Chetyrkina, M.R.; Fedorov, F.S.; Nasibulin, A.G. In Vitro Toxicity of Carbon Nanotubes: A Systematic Review. RSC Adv. 2022, 12, 16235–16256. [Google Scholar] [CrossRef] [PubMed]
- Frank, E.A.; Carreira, V.S.; Shanmukhappa, K.; Medvedovic, M.; Prows, D.R.; Yadav, J.S. Genetic Susceptibility to Toxicologic Lung Responses among Inbred Mouse Strains Following Exposure to Carbon Nanotubes and Profiling of Underlying Gene Networks. Toxicol. Appl. Pharmacol. 2017, 327, 59–70. [Google Scholar] [CrossRef]
- Frank, E.A.; Carreira, V.S.; Birch, M.E.; Yadav, J.S. Carbon Nanotube and Asbestos Exposures Induce Overlapping but Distinct Profiles of Lung Pathology in Non-Swiss Albino CF-1 Mice. Toxicol. Pathol. 2016, 44, 211–225. [Google Scholar] [CrossRef]
- Frank, E.A.; Birch, M.E.; Yadav, J.S. MyD88 Mediates in Vivo Effector Functions of Alveolar Macrophages in Acute Lung Inflammatory Responses to Carbon Nanotube Exposure. Toxicol. Appl. Pharmacol. 2015, 288, 322–329. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.S.; Yadav, B.; Rosen, L.; Nagpal, R.; Yadav, H.; Yadav, J.S. Crosstalk between Gut Microbiota and Lung Inflammation in Murine Toxicity Models of Respiratory Exposure or Co-Exposure to Carbon Nanotube Particles and Cigarette Smoke Extract. Toxicol. Appl. Pharmacol. 2022, 447, 116066. [Google Scholar] [CrossRef] [PubMed]
- White, B.; Sterrett, J.D.; Grigoryan, Z.; Lally, L.; Heinze, J.D.; Alikhan, H.; Lowry, C.A.; Perez, L.J.; DeSipio, J.; Phadtare, S. Characterization of Gut Microbiome and Metabolome in Helicobacter Pylori Patients in an Underprivileged Community in the United States. World J. Gastroenterol. 2021, 27, 5575–5594. [Google Scholar] [CrossRef]
- Adibi, S.; Seferovic, D.; Tribble, G.D.; Alcorn, J.L.; Fakhouri, W.D. Surfactant Protein A and Microbiome Composition in Patients with Atraumatic Intraoral Lesions. Front. Oral. Health 2021, 2, 663483. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Q.; Zhang, J.; Ouyang, Y.; Sun, Z.; Liu, X.; Qaio, F.; Xu, L.-Q.; Niu, Y.; Li, J. Exploring the Change of Host and Microorganism in Chronic Obstructive Pulmonary Disease Patients Based on Metagenomic and Metatranscriptomic Sequencing. Front. Microbiol. 2022, 13, 818281. [Google Scholar] [CrossRef]
- Yang, J.; Zhou, X.; Liu, X.; Ling, Z.; Ji, F. Role of the Gastric Microbiome in Gastric Cancer: From Carcinogenesis to Treatment. Front. Microbiol. 2021, 12, 641322. [Google Scholar] [CrossRef] [PubMed]
- Allais, L.; Kerckhof, F.-M.; Verschuere, S.; Bracke, K.R.; De Smet, R.; Laukens, D.; Van den Abbeele, P.; De Vos, M.; Boon, N.; Brusselle, G.G.; et al. Chronic Cigarette Smoke Exposure Induces Microbial and Inflammatory Shifts and Mucin Changes in the Murine Gut. Environ. Microbiol. 2016, 18, 1352–1363. [Google Scholar] [CrossRef] [PubMed]
- Gui, X.; Yang, Z.; Li, M.D. Effect of Cigarette Smoke on Gut Microbiota: State of Knowledge. Front. Physiol. 2021, 12, 673341. [Google Scholar] [CrossRef] [PubMed]
- Lahiani, M.H.; Khare, S.; Cerniglia, C.E.; Boy, R.; Ivanov, I.N.; Khodakovskaya, M. The Impact of Tomato Fruits Containing Multi-Walled Carbon Nanotube Residues on Human Intestinal Epithelial Cell Barrier Function and Intestinal Microbiome Composition. Nanoscale 2019, 11, 3639–3655. [Google Scholar] [CrossRef] [PubMed]
- Qu, Z.; Zhang, L.; Hou, R.; Ma, X.; Yu, J.; Zhang, W.; Zhuang, C. Exposure to a Mixture of Cigarette Smoke Carcinogens Disturbs Gut Microbiota and Influences Metabolic Homeostasis in A/J Mice. Chem. Biol. Interact. 2021, 344, 109496. [Google Scholar] [CrossRef]
- Huffnagle, G.B.; Dickson, R.P.; Lukacs, N.W. The Respiratory Tract Microbiome and Lung Inflammation: A Two-Way Street. Mucosal Immunol. 2017, 10, 299–306. [Google Scholar] [CrossRef]
- Ramsheh, M.Y.; Haldar, K.; Esteve-Codina, A.; Purser, L.F.; Richardson, M.; Müller-Quernheim, J.; Greulich, T.; Nowinski, A.; Barta, I.; Stendardo, M.; et al. Lung Microbiome Composition and Bronchial Epithelial Gene Expression in Patients with COPD versus Healthy Individuals: A Bacterial 16S rRNA Gene Sequencing and Host Transcriptomic Analysis. Lancet Microbe 2021, 2, e300–e310. [Google Scholar] [CrossRef]
- O’Dwyer, D.N.; Dickson, R.P.; Moore, B.B. The Lung Microbiome, Immunity, and the Pathogenesis of Chronic Lung Disease. J. Immunol. 2016, 196, 4839–4847. [Google Scholar] [CrossRef]
- Barcik, W.; Boutin, R.C.T.; Sokolowska, M.; Finlay, B.B. The Role of Lung and Gut Microbiota in the Pathology of Asthma. Immunity 2020, 52, 241–255. [Google Scholar] [CrossRef]
- Thomas, A.M.; Gleber-Netto, F.O.; Fernandes, G.R.; Amorim, M.; Barbosa, L.F.; Francisco, A.L.N.; de Andrade, A.G.; Setubal, J.C.; Kowalski, L.P.; Nunes, D.N.; et al. Alcohol and Tobacco Consumption Affects Bacterial Richness in Oral Cavity Mucosa Biofilms. BMC Microbiol. 2014, 14, 250. [Google Scholar] [CrossRef]
- Moon, J.-H.; Lee, J.-H.; Lee, J.-Y. Subgingival Microbiome in Smokers and Non-Smokers in Korean Chronic Periodontitis Patients. Mol. Oral. Microbiol. 2015, 30, 227–241. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Shi, G. Smoking and Microbiome in Oral, Airway, Gut and Some Systemic Diseases. J. Transl. Med. 2019, 17, 225. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Chen, L.; Cao, L.; Li, K.-J.; Huang, Y.; Luan, X.-Q.; Li, G. Effects of Smoking on the Lower Respiratory Tract Microbiome in Mice. Respir. Res. 2018, 19, 253. [Google Scholar] [CrossRef]
- Erb-Downward, J.R.; Thompson, D.L.; Han, M.K.; Freeman, C.M.; McCloskey, L.; Schmidt, L.A.; Young, V.B.; Toews, G.B.; Curtis, J.L.; Sundaram, B.; et al. Analysis of the Lung Microbiome in the “Healthy” Smoker and in COPD. PLoS ONE 2011, 6, e16384. [Google Scholar] [CrossRef]
- Mason, M.R.; Preshaw, P.M.; Nagaraja, H.N.; Dabdoub, S.M.; Rahman, A.; Kumar, P.S. The Subgingival Microbiome of Clinically Healthy Current and Never Smokers. ISME J. 2015, 9, 268–272. [Google Scholar] [CrossRef]
- Bhattacharya, S.S.; Yadav, B.; Yadav, E.; Hus, A.; Yadav, N.; Kaur, P.; Rosen, L.; Jandarov, R.; Yadav, J.S. Differential Modulation of Lung Aquaporins among Other Pathophysiological Markers in Acute (Cl2 Gas) and Chronic (Carbon Nanoparticles, Cigarette Smoke) Respiratory Toxicity Mouse Models. Front. Physiol. 2022, 13, 880815. [Google Scholar] [CrossRef] [PubMed]
- Berbers, R.-M.; Mohamed Hoesein, F.A.A.; Ellerbroek, P.M.; van Montfrans, J.M.; Dalm, V.A.S.H.; van Hagen, P.M.; Paganelli, F.L.; Viveen, M.C.; Rogers, M.R.C.; de Jong, P.A.; et al. Low IgA Associated with Oropharyngeal Microbiota Changes and Lung Disease in Primary Antibody Deficiency. Front. Immunol. 2020, 11, 1245. [Google Scholar] [CrossRef]
- Fadrosh, D.W.; Ma, B.; Gajer, P.; Sengamalay, N.; Ott, S.; Brotman, R.M.; Ravel, J. An Improved Dual-Indexing Approach for Multiplexed 16S rRNA Gene Sequencing on the Illumina MiSeq Platform. Microbiome 2014, 2, 6. [Google Scholar] [CrossRef]
- Kuczynski, J.; Stombaugh, J.; Walters, W.A.; González, A.; Caporaso, J.G.; Knight, R. Using QIIME to Analyze 16S rRNA Gene Sequences from Microbial Communities. Curr. Protoc. Bioinform. 2011, 36, 10.7.1–10.7.20. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing Real-Time PCR Data by the Comparative C(T) Method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Chen, Y.-W.; Li, S.-W.; Lin, C.-D.; Huang, M.-Z.; Lin, H.-J.; Chin, C.-Y.; Lai, Y.-R.; Chiu, C.-H.; Yang, C.-Y.; Lai, C.-H. Fine Particulate Matter Exposure Alters Pulmonary Microbiota Composition and Aggravates Pneumococcus-Induced Lung Pathogenesis. Front. Cell Dev. Biol. 2020, 8, 570484. [Google Scholar] [CrossRef]
- Peters, B.A.; Wu, J.; Pei, Z.; Yang, L.; Purdue, M.P.; Freedman, N.D.; Jacobs, E.J.; Gapstur, S.M.; Hayes, R.B.; Ahn, J. Oral Microbiome Composition Reflects Prospective Risk for Esophageal Cancers. Cancer Res. 2017, 77, 6777–6787. [Google Scholar] [CrossRef]
- Hu, J.; Iragavarapu, S.; Nadkarni, G.N.; Huang, R.; Erazo, M.; Bao, X.; Verghese, D.; Coca, S.; Ahmed, M.K.; Peter, I. Location-Specific Oral Microbiome Possesses Features Associated with CKD. Kidney Int. Rep. 2018, 3, 193–204. [Google Scholar] [CrossRef]
- Yang, D.; Chen, X.; Wang, J.; Lou, Q.; Lou, Y.; Li, L.; Wang, H.; Chen, J.; Wu, M.; Song, X.; et al. Dysregulated Lung Commensal Bacteria Drive Interleukin-17B Production to Promote Pulmonary Fibrosis through Their Outer Membrane Vesicles. Immunity 2019, 50, 692–706.e7. [Google Scholar] [CrossRef] [PubMed]
- Dickson, R.P.; Erb-Downward, J.R.; Falkowski, N.R.; Hunter, E.M.; Ashley, S.L.; Huffnagle, G.B. The Lung Microbiota of Healthy Mice Are Highly Variable, Cluster by Environment, and Reflect Variation in Baseline Lung Innate Immunity. Am. J. Respir. Crit. Care Med. 2018, 198, 497–508. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Vats, A.; Sharma, A.; Arora, A.; Kumar, A. The Development of Lower Respiratory Tract Microbiome in Mice. Microbiome 2017, 5, 61. [Google Scholar] [CrossRef]
- Yun, Y.; Srinivas, G.; Kuenzel, S.; Linnenbrink, M.; Alnahas, S.; Bruce, K.D.; Steinhoff, U.; Baines, J.F.; Schaible, U.E. Environmentally Determined Differences in the Murine Lung Microbiota and Their Relation to Alveolar Architecture. PLoS ONE 2014, 9, e113466. [Google Scholar] [CrossRef]
- Morris, A.; Beck, J.M.; Schloss, P.D.; Campbell, T.B.; Crothers, K.; Curtis, J.L.; Flores, S.C.; Fontenot, A.P.; Ghedin, E.; Huang, L.; et al. Comparison of the Respiratory Microbiome in Healthy Nonsmokers and Smokers. Am. J. Respir. Crit. Care Med. 2013, 187, 1067–1075. [Google Scholar] [CrossRef]
- Hilty, M.; Burke, C.; Pedro, H.; Cardenas, P.; Bush, A.; Bossley, C.; Davies, J.; Ervine, A.; Poulter, L.; Pachter, L.; et al. Disordered Microbial Communities in Asthmatic Airways. PLoS ONE 2010, 5, e8578. [Google Scholar] [CrossRef]
- Huang, Y.J.; Nariya, S.; Harris, J.M.; Lynch, S.V.; Choy, D.F.; Arron, J.R.; Boushey, H. The Airway Microbiome in Patients with Severe Asthma: Associations with Disease Features and Severity. J. Allergy Clin. Immunol. 2015, 136, 874–884. [Google Scholar] [CrossRef]
- Sze, M.A.; Dimitriu, P.A.; Hayashi, S.; Elliott, W.M.; McDonough, J.E.; Gosselink, J.V.; Cooper, J.; Sin, D.D.; Mohn, W.W.; Hogg, J.C. The Lung Tissue Microbiome in Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 2012, 185, 1073–1080. [Google Scholar] [CrossRef]
- Gollwitzer, E.S.; Saglani, S.; Trompette, A.; Yadava, K.; Sherburn, R.; McCoy, K.D.; Nicod, L.P.; Lloyd, C.M.; Marsland, B.J. Lung Microbiota Promotes Tolerance to Allergens in Neonates via PD-L1. Nat. Med. 2014, 20, 642–647. [Google Scholar] [CrossRef]
- Hirao, S.; Wada, H.; Nakagaki, K.; Saraya, T.; Kurai, D.; Mikura, S.; Yasutake, T.; Higaki, M.; Yokoyama, T.; Ishii, H.; et al. Inflammation Provoked by Mycoplasma Pneumoniae Extract: Implications for Combination Treatment with Clarithromycin and Dexamethasone. FEMS Immunol. Med. Microbiol. 2011, 62, 182–189. [Google Scholar] [CrossRef]
- Yu, G.; Phillips, S.; Gail, M.H.; Goedert, J.J.; Humphrys, M.S.; Ravel, J.; Ren, Y.; Caporaso, N.E. The Effect of Cigarette Smoking on the Oral and Nasal Microbiota. Microbiome 2017, 5, 3. [Google Scholar] [CrossRef]
- Nayak, A.; Dodagatta-Marri, E.; Tsolaki, A.G.; Kishore, U. An Insight into the Diverse Roles of Surfactant Proteins, SP-A and SP-D in Innate and Adaptive Immunity. Front. Immunol. 2012, 3, 131. [Google Scholar] [CrossRef] [PubMed]
- Rahman, L.; Jacobsen, N.R.; Aziz, S.A.; Wu, D.; Williams, A.; Yauk, C.L.; White, P.; Wallin, H.; Vogel, U.; Halappanavar, S. Multi-Walled Carbon Nanotube-Induced Genotoxic, Inflammatory, and pro-Fibrotic Responses in Mice: Investigating the Mechanisms of Pulmonary Carcinogenesis. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2017, 823, 28–44. [Google Scholar] [CrossRef] [PubMed]
- Juge, P.-A.; Lee, J.S.; Ebstein, E.; Furukawa, H.; Dobrinskikh, E.; Gazal, S.; Kannengiesser, C.; Ottaviani, S.; Oka, S.; Tohma, S.; et al. MUC5B Promoter Variant and Rheumatoid Arthritis with Interstitial Lung Disease. N. Engl. J. Med. 2018, 379, 2209–2219. [Google Scholar] [CrossRef] [PubMed]
- Ash, S.Y.; Harmouche, R.; Putman, R.K.; Ross, J.C.; Martinez, F.J.; Choi, A.M.; Bowler, R.P.; Regan, E.A.; Curtis, J.L.; Han, M.K.; et al. Association between Acute Respiratory Disease Events and the MUC5B Promoter Polymorphism in Smokers. Thorax 2018, 73, 1071–1074. [Google Scholar] [CrossRef]
- Shakoor, S.; Sun, L.; Wang, D. Multi-Walled Carbon Nanotubes Enhanced Fungal Colonization and Suppressed Innate Immune Response to Fungal Infection in Nematodes. Toxicol. Res. 2016, 5, 492–499. [Google Scholar] [CrossRef]
- Lugade, A.A.; Bogner, P.N.; Thatcher, T.H.; Sime, P.J.; Phipps, R.P.; Thanavala, Y. Cigarette Smoke Exposure Exacerbates Lung Inflammation and Compromises Immunity to Bacterial Infection. J. Immunol. 2014, 192, 5226–5235. [Google Scholar] [CrossRef]
- LeVine, A.M.; Kurak, K.E.; Bruno, M.D.; Stark, J.M.; Whitsett, J.A.; Korfhagen, T.R. Surfactant Protein-A-Deficient Mice Are Susceptible to Pseudomonas Aeruginosa Infection. Am. J. Respir. Cell Mol. Biol. 1998, 19, 700–708. [Google Scholar] [CrossRef] [PubMed]
- LeVine, A.M.; Bruno, M.D.; Huelsman, K.M.; Ross, G.F.; Whitsett, J.A.; Korfhagen, T.R. Surfactant Protein A-Deficient Mice Are Susceptible to Group B Streptococcal Infection. J. Immunol. 1997, 158, 4336–4340. [Google Scholar] [CrossRef] [PubMed]
- Tatsuta, M.; Kan-O, K.; Ishii, Y.; Yamamoto, N.; Ogawa, T.; Fukuyama, S.; Ogawa, A.; Fujita, A.; Nakanishi, Y.; Matsumoto, K. Effects of Cigarette Smoke on Barrier Function and Tight Junction Proteins in the Bronchial Epithelium: Protective Role of Cathelicidin LL-37. Respir. Res. 2019, 20, 251. [Google Scholar] [CrossRef] [PubMed]
- Roy, M.G.; Livraghi-Butrico, A.; Fletcher, A.A.; McElwee, M.M.; Evans, S.E.; Boerner, R.M.; Alexander, S.N.; Bellinghausen, L.K.; Song, A.S.; Petrova, Y.M.; et al. Muc5b Is Required for Airway Defence. Nature 2014, 505, 412–416. [Google Scholar] [CrossRef]
- Kuczma, M.P.; Szurek, E.A.; Cebula, A.; Chassaing, B.; Jung, Y.-J.; Kang, S.-M.; Fox, J.G.; Stecher, B.; Ignatowicz, L. Commensal Epitopes Drive Differentiation of Colonic Tregs. Sci. Adv. 2020, 6, eaaz3186. [Google Scholar] [CrossRef]
- López-Campos, J.L.; Sánchez Silva, R.; Gómez Izquierdo, L.; Márquez, E.; Ortega Ruiz, F.; Cejudo, P.; Barrot Cortés, E.; Toledo Aral, J.J.; Echevarría, M. Overexpression of Aquaporin-1 in Lung Adenocarcinomas and Pleural Mesotheliomas. Histol. Histopathol. 2011, 26, 451–459. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yadav, B.; Bhattacharya, S.S.; Rosen, L.; Nagpal, R.; Yadav, H.; Yadav, J.S. Oro-Respiratory Dysbiosis and Its Modulatory Effect on Lung Mucosal Toxicity during Exposure or Co-Exposure to Carbon Nanotubes and Cigarette Smoke. Nanomaterials 2024, 14, 314. https://doi.org/10.3390/nano14030314
Yadav B, Bhattacharya SS, Rosen L, Nagpal R, Yadav H, Yadav JS. Oro-Respiratory Dysbiosis and Its Modulatory Effect on Lung Mucosal Toxicity during Exposure or Co-Exposure to Carbon Nanotubes and Cigarette Smoke. Nanomaterials. 2024; 14(3):314. https://doi.org/10.3390/nano14030314
Chicago/Turabian StyleYadav, Brijesh, Sukanta S. Bhattacharya, Lauren Rosen, Ravinder Nagpal, Hariom Yadav, and Jagjit S. Yadav. 2024. "Oro-Respiratory Dysbiosis and Its Modulatory Effect on Lung Mucosal Toxicity during Exposure or Co-Exposure to Carbon Nanotubes and Cigarette Smoke" Nanomaterials 14, no. 3: 314. https://doi.org/10.3390/nano14030314
APA StyleYadav, B., Bhattacharya, S. S., Rosen, L., Nagpal, R., Yadav, H., & Yadav, J. S. (2024). Oro-Respiratory Dysbiosis and Its Modulatory Effect on Lung Mucosal Toxicity during Exposure or Co-Exposure to Carbon Nanotubes and Cigarette Smoke. Nanomaterials, 14(3), 314. https://doi.org/10.3390/nano14030314







