Data-Driven and Machine Learning to Screen Metal–Organic Frameworks for the Efficient Separation of Methane
Abstract
1. Introduction
2. Model and Methods
2.1. Molecular Model
2.2. Molecular Simulations
2.3. Machine Learning
3. Results and Discussion
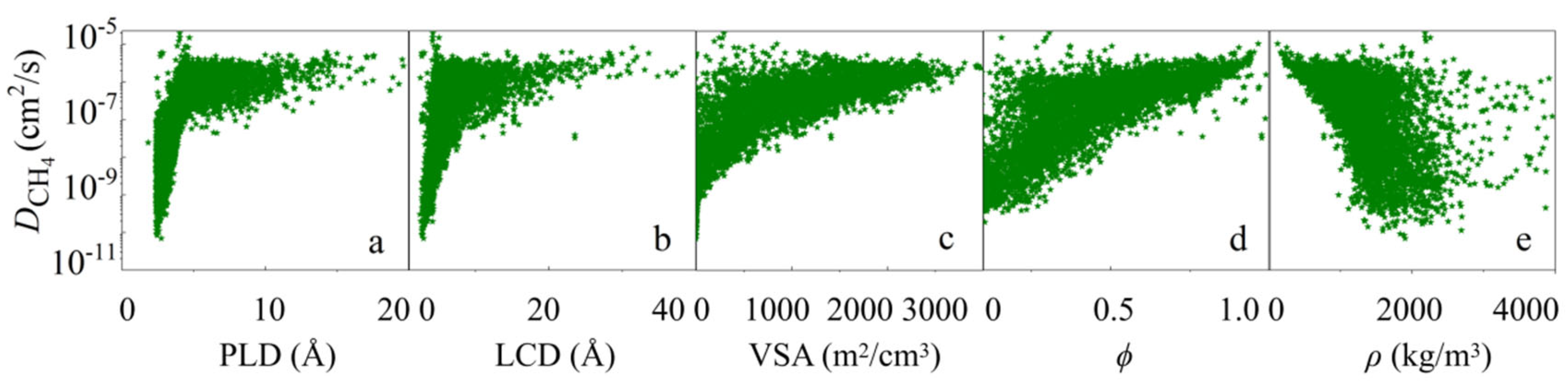
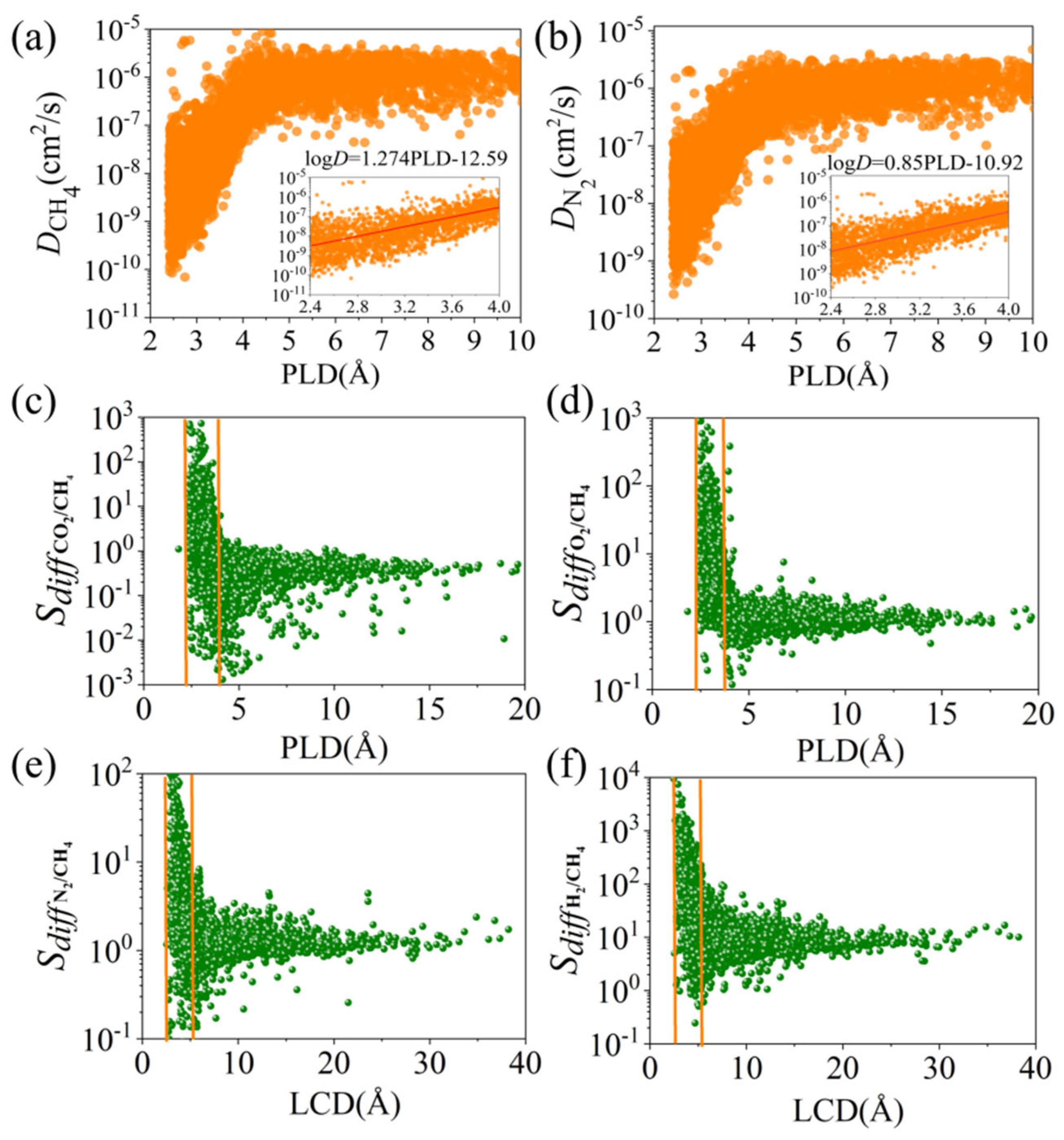
3.1. Statistical Analysis
3.2. Machine Learning
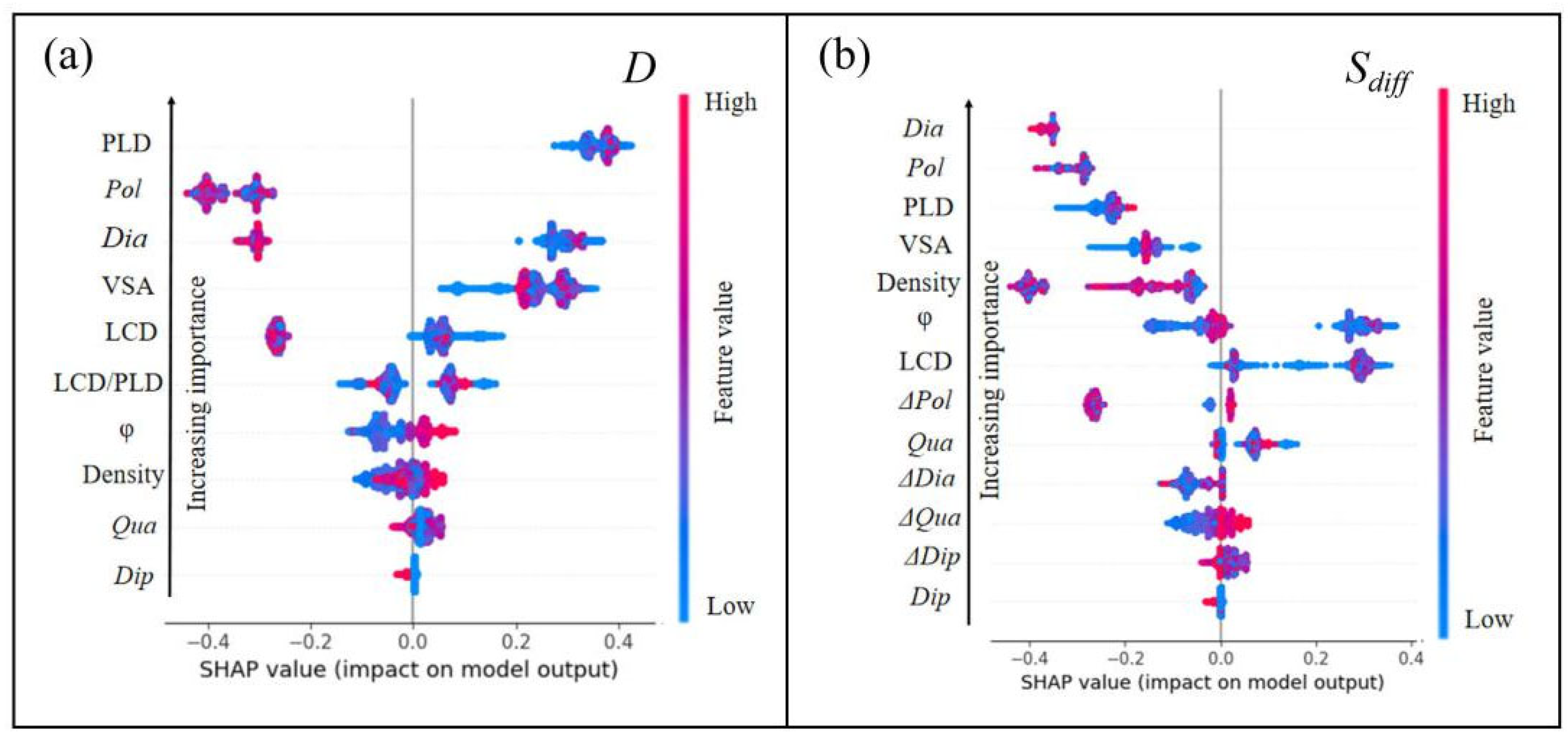
3.3. SHAP Analysis
3.4. Top-Performing Metal–Organic Frameworks (MOFs)
3.5. Design Strategies of MOFs with High Performances
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Warmuzinski, K. Harnessing methane emissions from coal mining. Process Saf. Environ. Prot. 2008, 86, 315–320. [Google Scholar] [CrossRef]
- Moore, T.A. Coalbed methane: A review. Int. J. Coal Geol. 2012, 101, 36–81. [Google Scholar] [CrossRef]
- Thielemann, T.; Cramer, B.; Schippers, A. Coalbed methane in the Ruhr Basin, Germany: A renewable energy resource. Org. Geochem. 2004, 35, 1537–1549. [Google Scholar] [CrossRef]
- Tutak, M.; Brodny, J. Forecasting methane emissions from hard coal mines including the methane drainage process. Energies 2019, 12, 3840. [Google Scholar] [CrossRef]
- Wu, Y.; Yuan, D.; Zeng, S.; Yang, L.; Dong, X.; Zhang, Q.; Xu, Y.; Liu, Z. Significant enhancement in CH4/N2 separation with amine-modified zeolite Y. Fuel 2021, 301, 121077. [Google Scholar] [CrossRef]
- Lashof, D.A.; Ahuja, D.R. Relative contributions of greenhouse gas emissions to global warming. Nature 1990, 344, 529–531. [Google Scholar] [CrossRef]
- Yin, Z.; Linga, P. Methane hydrates: A future clean energy resource. Chin. J. Chem. Eng. 2019, 27, 2026–2036. [Google Scholar] [CrossRef]
- Kapoor, R.; Ghosh, P.; Tyagi, B.; Vijay, V.K.; Vijay, V.; Thakur, I.S.; Kamyab, H.; Nguyen, D.D.; Kumar, A. Advances in biogas valorization and utilization systems: A comprehensive review. J. Clean. Prod. 2020, 273, 123052. [Google Scholar] [CrossRef]
- Gomes, V.G.; Yee, K.W.K. Pressure swing adsorption for carbon dioxide sequestration from exhaust gases. Sep. Purif. Technol. 2002, 28, 161–171. [Google Scholar] [CrossRef]
- Wiheeb, A.D.; Helwani, Z.; Kim, J.; Othman, M.R. Pressure swing adsorption technologies for carbon dioxide capture. Sep. Purif. Rev. 2016, 45, 108–121. [Google Scholar] [CrossRef]
- Mel, M.; Ibrahim MM, A.; Setyobudi, R.H. Preliminary study of biogas upgrading and purification by pressure swing adsorption. Proc. AIP Conf. 2016, 1755, 130010. [Google Scholar]
- Lin, R.B.; Xiang, S.; Zhou, W.; Chen, B. Microporous Metal-Organic Framework Materials for Gas Separation. Chem 2020, 6, 337–363. [Google Scholar] [CrossRef]
- Li, H.; Li, L.; Lin, R.B.; Zhou, W.; Zhang, Z.; Xiang, S.; Chen, B. Porous metal-organic frameworks for gas storage and separation: Status and challenges. EnergyChem 2019, 1, 100006. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; An, B.; Lin, W. Metal–organic frameworks in solid–gas phase catalysis. ACS Catal. 2018, 9, 130–146. [Google Scholar] [CrossRef]
- Cai, M.; Chen, G.; Qin, L.; Qu, C.; Dong, X.; Ni, J.; Yin, X. Metal organic frameworks as drug targeting delivery vehicles in the treatment of cancer. Pharmaceutics 2020, 12, 232. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.L.; Jiang, X.L.; Xu, H.; Zhao, B. Applications of MOF as luminescent sensors for environmental pollutants. Small 2021, 17, 2005327. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.Q.; Yin, X.B. Metal–organic frameworks with multiple luminescence emissions: Designs and applications. Acc. Chem. Res. 2020, 53, 485–495. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Huang, H.; Zhang, X.; Zhao, H.; Li, F.; Gu, Y. Designed metal-organic frameworks with potential for multi-component hydrocarbon separation. Coord. Chem. Rev. 2023, 484, 215111. [Google Scholar] [CrossRef]
- Shi, Z.; Yang, W.; Deng, X.; Cai, C.; Yan, Y.; Liang, H.; Liu, Z.; Qiao, Z. Machine-learning-assisted high-throughput computational screening of high performance metal–organic frameworks. Mol. Syst. Des. Eng. 2020, 5, 725–742. [Google Scholar] [CrossRef]
- Anbia, M.; Hoseini, V.; Sheykhi, S. Sorption of methane, hydrogen and carbon dioxide on metal-organic framework, iron terephthalate (MOF-235). J. Ind. Eng. Chem. 2012, 18, 1149–1152. [Google Scholar] [CrossRef]
- Niu, Z.; Cui, X.; Pham, T.; Lan, P.C.; Xing, H.; Forrest, K.A.; Wojtas, L.; Space, B.; Ma, S. A metal–organic framework based methane Nano-trap for the capture of coal-mine methane. Angew. Chem. Int. Ed. 2019, 58, 10138–10141. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Gao, Q.; Li, Y.N.; Yang, L.; Gao, Q.; Li, Y.N.; Liu, Q.N.; Liu, X.T.; Wang, S.; Chen, L.Z. Efficient purification of CH4 from ternary mixtures by a microporous heterometal-organic framework. Sep. Purif. Technol. 2024, 335, 126235. [Google Scholar] [CrossRef]
- Mulu, E.; M’Arimi, M.M.; Ramkat, R.C.; Mecha, A.C. Potential of wood ash in purification of biogas. Energy Sustain. Dev. 2021, 65, 45–52. [Google Scholar] [CrossRef]
- Wang, Y.; Dong, Q.; Cao, H.; Ji, W.; Duan, J.; Jing, S.; Jin, W. Finely Tuned Porous Coordination Polymers To Boost Methane Separation Efficiency. Chem. Eur. J. 2019, 25, 8790–8796. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Xu, E.L.; Li, M. Purification of coal mine methane on carbon molecular sieve by vacuum pressure swing adsorption. Sep. Sci. Technol. 2016, 51, 909–916. [Google Scholar] [CrossRef]
- Tu, S.; Yu, L.; Lin, D.; Chen, Y.; Wu, Y.; Zhou, X.; Li, Z.; Xia, Q. Robust nickel-based metal–organic framework for highly efficient methane purification and capture. ACS Appl. Mater. Interfaces 2022, 14, 4242–4250. [Google Scholar] [CrossRef] [PubMed]
- Kang, Z.; Xue, M.; Fan, L.; Huang, L.; Guo, L.; Wei, G.; Chen, B.; Qiu, S. Highly selective sieving of small gas molecules by using an ultra-microporous metal—Organic framework membrane. Energy Environ. Sci. 2014, 7, 4053–4060. [Google Scholar] [CrossRef]
- Chung, Y.G.; Camp, J.; Haranczyk, M.; Sikora, B.J.; Bury, W.; Krungleviciute, V.; Yildirim, T.; Sholl, D.S.; Snurr, R.Q. Computation-ready, experimental metal—Organic frameworks: A tool to enable high-throughput screening of nanoporous crystals. Chem. Mater. 2014, 26, 6185–6192. [Google Scholar] [CrossRef]
- Willems, T.F.; Rycroft, C.H.; Kazi, M.; Meza, J.C.; Haranczyk, M. Algorithms and tools for high-throughput geometry-based analysis of crystalline porous materials. Microporous Mesoporous Mater. 2012, 149, 134–141. [Google Scholar] [CrossRef]
- Dubbeldam, D.; Calero, S.; Ellis, D.E.; Snurr, R.Q. RASPA: Molecular simulation software for adsorption and diffusion in flexible nanoporous materials. Mol. Simul. 2016, 42, 81–101. [Google Scholar] [CrossRef]
- Rappé, A.K.; Casewit, C.J.; Colwell, K.S.; Goddard, W.A., III; Skiff, W.M. UFF, a full periodic table force field for molecular mechanics and molecular dynamics simulations. J. Am. Chem. Soc. 1992, 114, 10024–10035. [Google Scholar] [CrossRef]
- Potoff, J.J.; Siepmann, J.I. Vapor–liquid equilibria of mixtures containing alkanes, carbon dioxide, and nitrogen. AIChE J. 2001, 47, 1676–1682. [Google Scholar] [CrossRef]
- Chokbunpiam, T.; Chanajaree, R.; Caro, J.; Janke, W.; Remsungnen, T.; Hannongbua, S.; Fritzsche, S. Separation of nitrogen dioxide from the gas mixture with nitrogen by use of ZIF materials; computer simulation studies. Comput. Mater. Sci. 2019, 168, 246–252. [Google Scholar] [CrossRef]
- Li, J.R.; Kuppler, R.J.; Zhou, H.C. Selective gas adsorption and separation in metal–organic frameworks. Chem. Soc. Rev. 2009, 38, 1477–1504. [Google Scholar] [CrossRef] [PubMed]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Natekin, A.; Knoll, A. Gradient boosting machines, a tutorial. Front. Neurorobot. 2013, 7, 21. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.; Luo, S.; Zhao, G.; Wu, H. Predicting hard rock pillar stability using GBDT, XGBoost, and LightGBM algorithms. Mathematics 2020, 8, 765. [Google Scholar] [CrossRef]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.Y. Lightgbm: A highly efficient gradient boosting decision tree. Adv. Neural Inf. Process. Syst. 2017, 3149–3157, 3149–3157. [Google Scholar]
- Bergstra, J.; Komer, B.; Eliasmith, C.; Yamins, D.; Cox, D.D. Hyperopt: A python library for model selection and hyperparameter optimization. Comput. Sci. Discov. 2015, 8, 014008. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Lee, S.I. A unified approach to interpreting model predictions. Adv. Neural Inf. Process. Syst. 2017, 4768–4777, 4768–4777. [Google Scholar]
- Lundberg, S.M.; Erion, G.; Chen, H.; DeGrave, A.; Prutkin, J.M.; Nair, B.; Lee, S.I. From local explanations to global understanding with explainable AI for trees. Nat. Mach. Intell. 2020, 2, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Vassallo, A.; Wu, J. Toward the inverse design of MOF membranes for efficient D2/H2 separation by combination of physics-based and data-driven modeling. J. Membr. Sci. 2020, 598, 117675. [Google Scholar] [CrossRef]
- Qiao, Z.; Peng, C.; Zhou, J.; Jiang, J. High-throughput computational screening of 137953 metal–organic frameworks for membrane separation of a CO2/N2/CH4 mixture. J. Mater. Chem. A 2016, 4, 15904–15912. [Google Scholar] [CrossRef]
- Yu, S.; Bo, J.; Meijun, Q. Molecular dynamic simulation of self-and transport diffusion for CO2/CH4/N2 in low-rank coal vitrinite. Energy Fuels 2018, 32, 3085–3096. [Google Scholar] [CrossRef]
- Li, L.; Zhang, T.; Duan, Y.; Wei, Y.; Dong, C.; Ding, L.; Qiao, Z.; Wang, H. Selective gas diffusion in two-dimensional MXene lamellar membranes: Insights from molecular dynamics simulations. J. Mater. Chem. A 2018, 6, 11734–11742. [Google Scholar] [CrossRef]
- Tang, H.; Jiang, J. In silico screening and design strategies of ethane-selective metal–organic frameworks for ethane/ethylene separation. AIChE J. 2021, 67, e17025. [Google Scholar] [CrossRef]
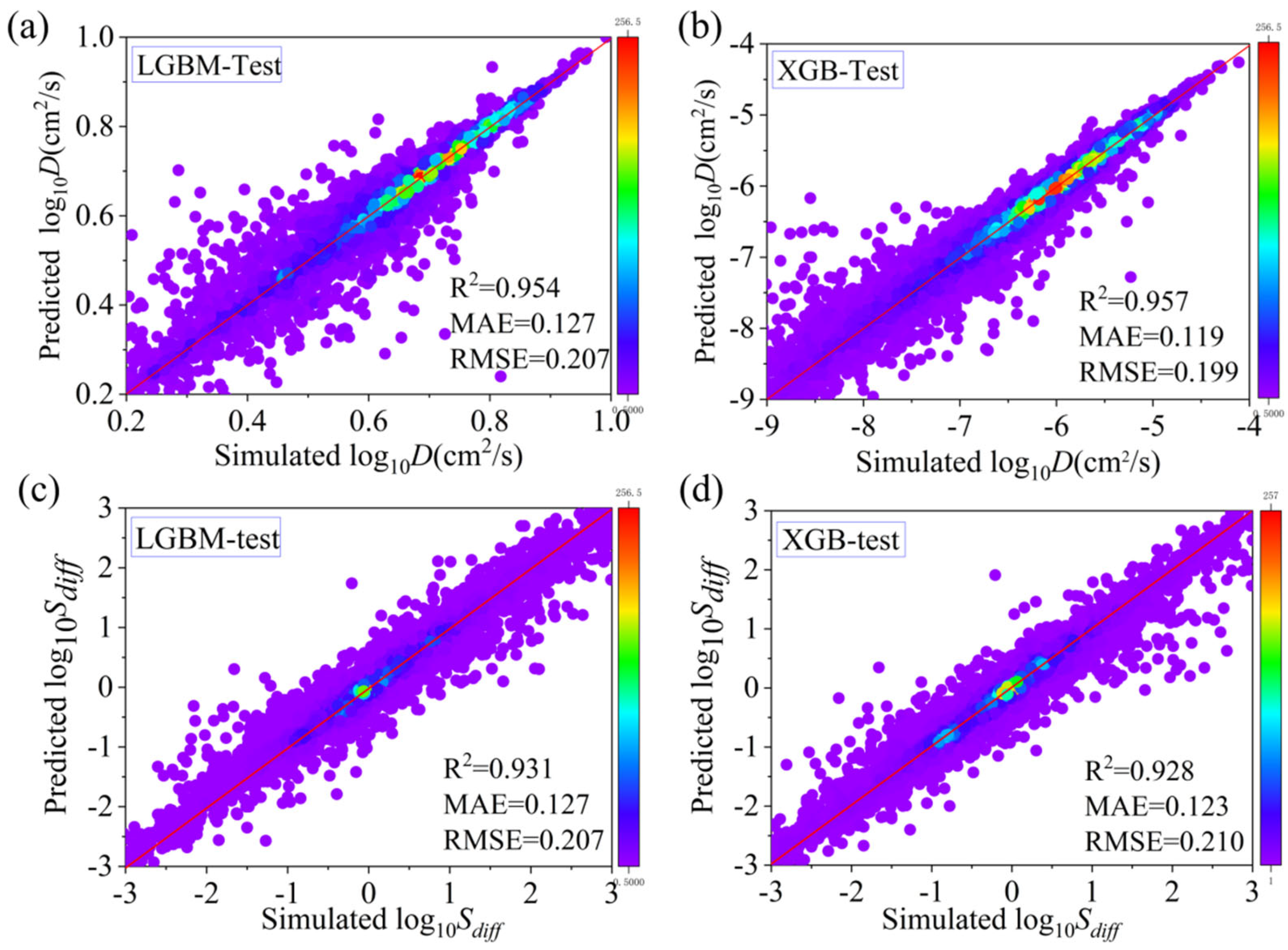

| Indicators | Algorithm | R2 | MAE | RMSE |
|---|---|---|---|---|
| D | RF | 0.934 | 0.151 | 0.247 |
| LGBM | 0.954 | 0.127 | 0.207 | |
| XGB | 0.957 | 0.119 | 0.199 | |
| GBRT | 0.947 | 0.129 | 0.222 | |
| S | RF | 0.887 | 0.155 | 0.264 |
| LGBM | 0.931 | 0.127 | 0.207 | |
| XGB | 0.928 | 0.123 | 0.210 | |
| GBRT | 0.907 | 0.135 | 0.240 |
| Gas Mixture i/j | CSD Cord | LCD [Å] | ϕ | PLD [Å] | ρ [kg/m3] | Di [cm2/s] | Dj [cm2/s] | Sdiff(i/j) |
|---|---|---|---|---|---|---|---|---|
| He/CH4 | ELUQIM04 | 2.92 | 0.04 | 2.44 | 1764.65 | 9.03 × 10−6 | 1.47 × 10−10 | 61,406.36 |
| ELUQIM05 | 2.90 | 0.04 | 2.43 | 1773.83 | 8.42 × 10−6 | 1.76 × 10−10 | 47,815.36 | |
| ELUQIM06 | 2.89 | 0.04 | 2.41 | 1779.43 | 9.74 × 10−6 | 8.11 × 10−10 | 12,015.02 | |
| H2/CH4 | ELUQIM05 | 2.90 | 0.04 | 2.43 | 1773.83 | 7.08 × 10−6 | 1.76 × 10−10 | 40,173.07 |
| ELUQIM04 | 2.92 | 0.04 | 2.44 | 1764.65 | 4.14 × 10−6 | 1.47 × 10−10 | 28,148.27 | |
| FAPYEA04 | 2.47 | 0.00 | 2.40 | 1583.54 | 2.83 × 10−6 | 3.01 × 10−10 | 9397.11 | |
| CO2/CH4 | XEKDUO | 2.98 | 0.02 | 2.75 | 1903.55 | 1.70 × 10−7 | 6.92 × 10−11 | 2463.01 |
| ELUQIM05 | 2.90 | 0.04 | 2.43 | 1773.83 | 3.91 × 10−7 | 1.76 × 10−10 | 2220.35 | |
| HIQPEE | 3.84 | 0.15 | 3.12 | 1440.14 | 1.24 × 10−6 | 5.69 × 10−10 | 2185.18 | |
| O2/CH4 | FAPYEA04 | 2.47 | 0.00 | 2.40 | 1583.54 | 2.92 × 10−7 | 3.01 × 10−10 | 968.16 |
| ELUQIM05 | 2.90 | 0.04 | 2.43 | 1773.83 | 1.58 × 10−7 | 1.76 × 10−10 | 898.78 | |
| GUXQAS | 2.79 | 0.02 | 2.52 | 1598.28 | 1.16 × 10−7 | 1.30 × 10−10 | 893.53 | |
| H2S/CH4 | GUXQAS | 2.79 | 0.02 | 2.52 | 1598.28 | 1.70 × 10−9 | 1.30 × 10−10 | 13.04 |
| RUPZIM | 3.48 | 0.11 | 3.25 | 1549.49 | 1.51 × 10−7 | 1.39 × 10−8 | 10.87 | |
| GUXPUL | 2.79 | 0.02 | 2.58 | 1595.02 | 1.18 × 10−9 | 1.10 × 10−10 | 10.70 | |
| N2/CH4 | FAPYEA04 | 2.47 | 0.00 | 2.40 | 1583.54 | 1.11 × 10−7 | 3.01 × 10−10 | 369.05 |
| PARFOF | 2.77 | 0.05 | 2.46 | 1541.02 | 4.78 × 10−7 | 2.16 × 10−9 | 221.18 | |
| HIWXER01 | 3.29 | 0.13 | 2.76 | 2533.00 | 1.80 × 10−7 | 1.27 × 10−9 | 141.34 |
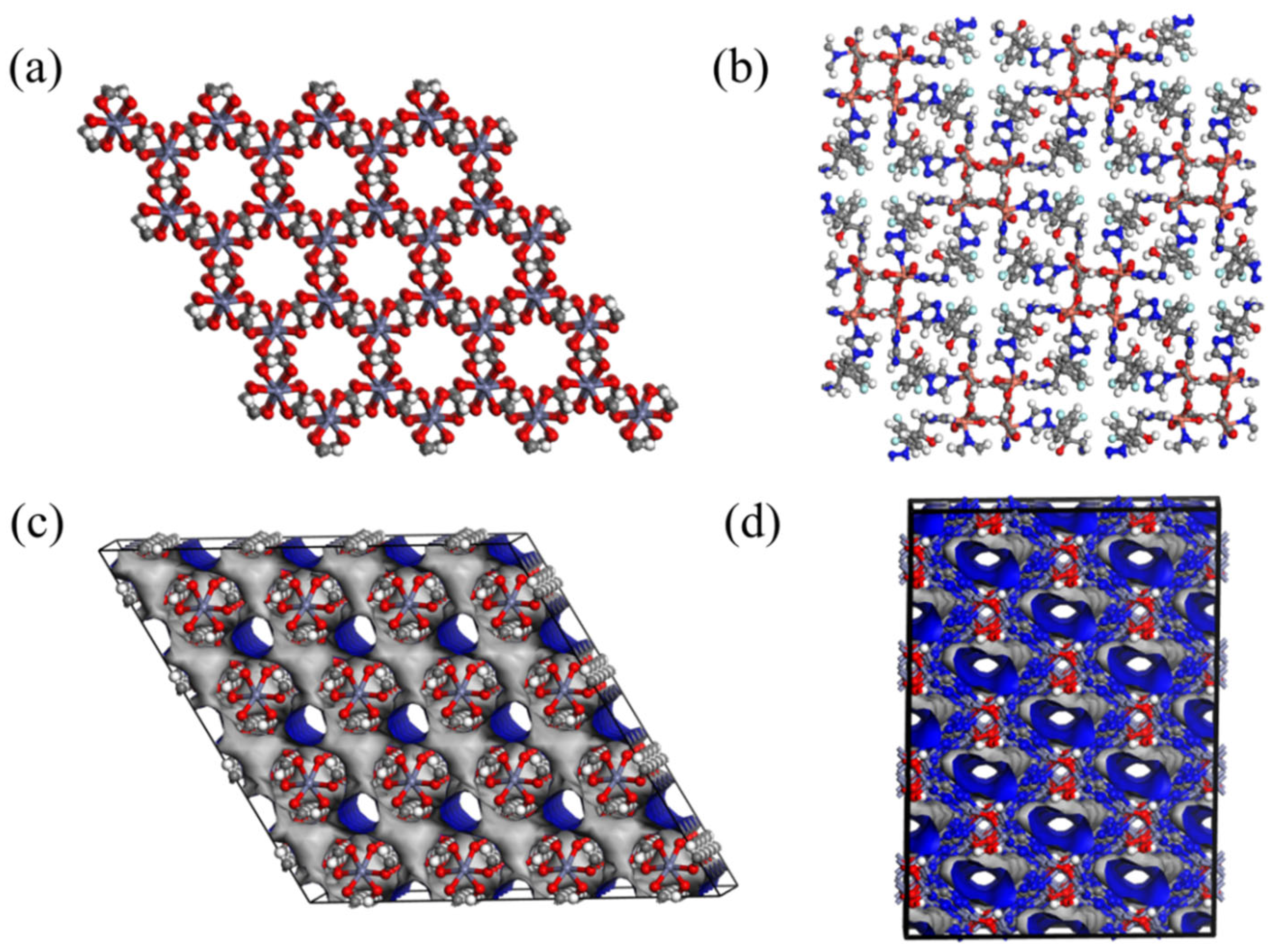
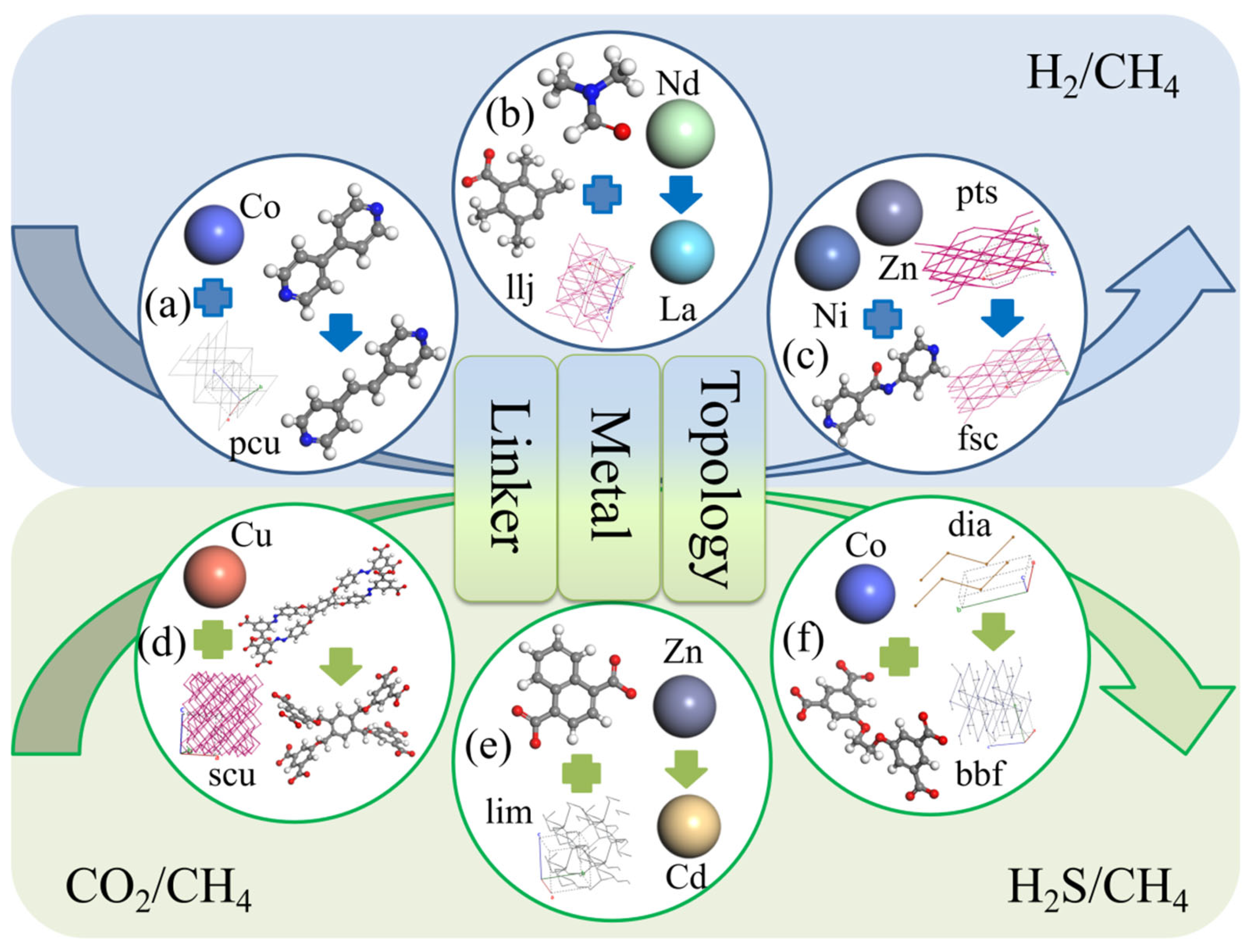
| Gas Mixture (i/j) | NO. | CSD Cord | Metal Center | Organic Links | Top Structure | Sdiff(i/j) |
|---|---|---|---|---|---|---|
| H2/CH4 | a | GIRDUI | Co | MGFJDEHFNMWYBD | pcu | 7.93 |
| GIRGUL | Co | MTAVBTGOXNGCJR | pcu | 643.48 | ||
| b | CIMTAV | La | BVKZGUZCCUSVTD | llj | 4.58 | |
| CIMTEZ | Nd | BVKZGUZCCUSVTD | llj | 12.97 | ||
| c | EBELUU | NiZn | JEVCWSUVFOYBFI | fsc | 5.36 | |
| EBEMEF | NiZn | JEVCWSUVFOYBFI | pts | 147.93 | ||
| CO2/CH4 | d | CAJQEL | Cu | GRYHAGOZZMMYAO | scu | 0.31 |
| CAJQIP | Cu | DUKMDOUQAIDJRW | scu | 2.24 | ||
| e | HEBTEP | Zn | ABMFBCRYHDZLRD | lim | 21.93 | |
| SETFUT | Cd | ABMFBCRYHDZLRD | lim | 1.22 | ||
| H2S/CH4 | f | ISIKIF | Co | GEBVRXNOWAYDCP | dia | 16.07 |
| ISIKOL | Co | GEBVRXNOWAYDCP | bbf | 0.97 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guan, Y.; Huang, X.; Xu, F.; Wang, W.; Li, H.; Gong, L.; Zhao, Y.; Guo, S.; Liang, H.; Qiao, Z. Data-Driven and Machine Learning to Screen Metal–Organic Frameworks for the Efficient Separation of Methane. Nanomaterials 2024, 14, 1074. https://doi.org/10.3390/nano14131074
Guan Y, Huang X, Xu F, Wang W, Li H, Gong L, Zhao Y, Guo S, Liang H, Qiao Z. Data-Driven and Machine Learning to Screen Metal–Organic Frameworks for the Efficient Separation of Methane. Nanomaterials. 2024; 14(13):1074. https://doi.org/10.3390/nano14131074
Chicago/Turabian StyleGuan, Yafang, Xiaoshan Huang, Fangyi Xu, Wenfei Wang, Huilin Li, Lingtao Gong, Yue Zhao, Shuya Guo, Hong Liang, and Zhiwei Qiao. 2024. "Data-Driven and Machine Learning to Screen Metal–Organic Frameworks for the Efficient Separation of Methane" Nanomaterials 14, no. 13: 1074. https://doi.org/10.3390/nano14131074
APA StyleGuan, Y., Huang, X., Xu, F., Wang, W., Li, H., Gong, L., Zhao, Y., Guo, S., Liang, H., & Qiao, Z. (2024). Data-Driven and Machine Learning to Screen Metal–Organic Frameworks for the Efficient Separation of Methane. Nanomaterials, 14(13), 1074. https://doi.org/10.3390/nano14131074








