Molecular Dynamics Simulation of Self-Assembly Processes of Diphenylalanine Peptide Nanotubes and Determination of Their Chirality
Abstract
1. Introduction
2. Main Methods
2.1. Model Details and Methodology for Numerical MDS Experiments
Main Software Platform Used
2.2. Model Details and Force MDS Technique for Self-Assembly of a Molecular Structure
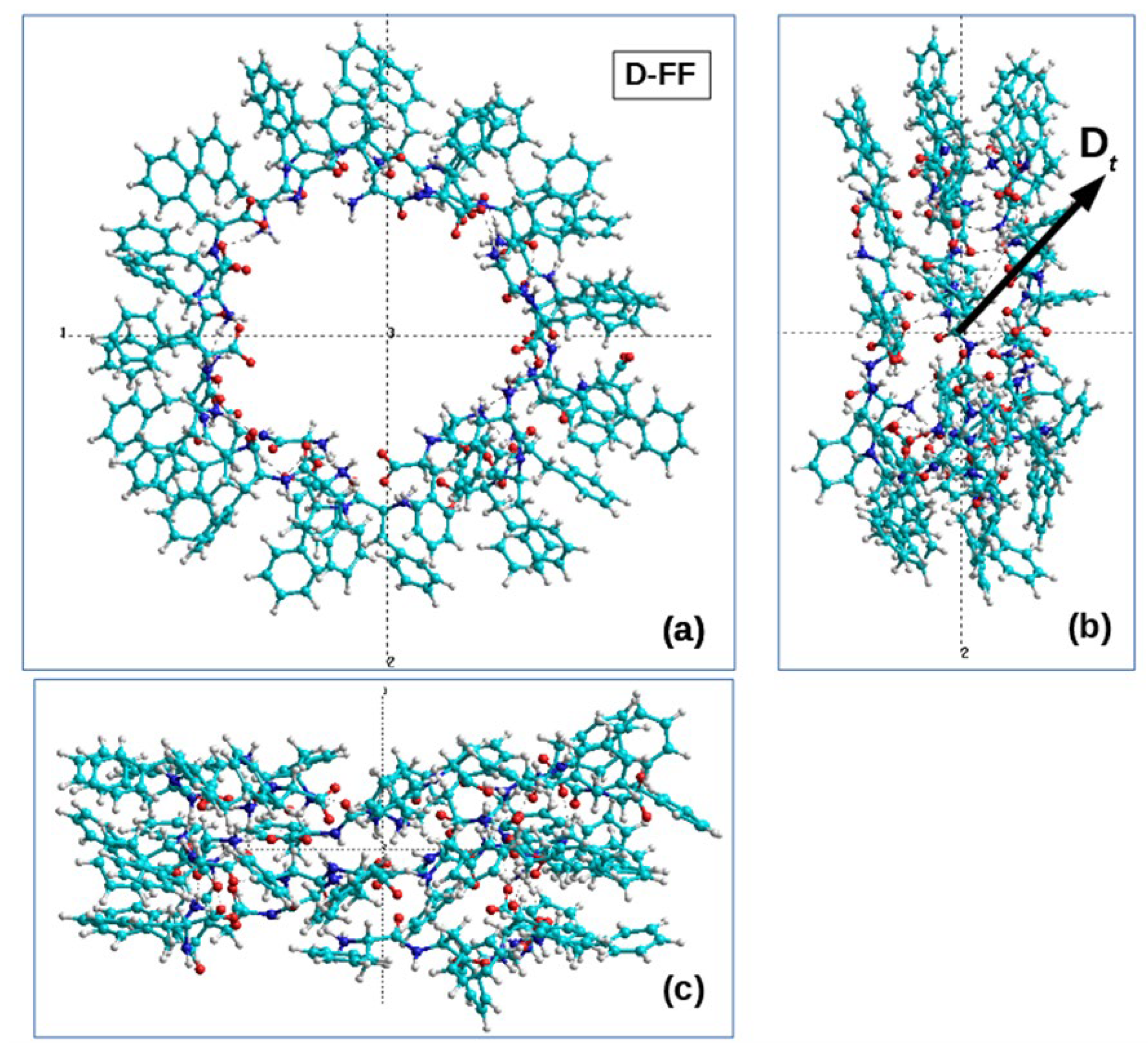
2.2.1. Initial Structure
2.2.2. The Process of Self-Assembly of the Helix
- right-handed helical nanotubes (D) from L-monomers (L-FF) and
- left-handed helical nanotubes (L) from D-monomers (D-FF).
2.2.3. MD Statistics of Nanotube Assembly
2.3. Calculation and Determination of Chirality
2.4. Calculation of Chirality from the Mixed Product of Vectors of Dipole Moments
2.5. Methods for Analyzing and Evaluating the Reliability of the Obtained MD Structures
3. Main Results and Discussions
4. Results of Comparative Visual Differential Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nepal, D.; Kang, S.; Adstedt, K.M.; Kanhaiya, K.; Bockstaller, M.R.; Brinson, L.C.; Buehler, M.J.; Coveney, P.V.; Dayal, K.; El-Awady, J.A.; et al. Hierarchically structured bioinspired nanocomposites. Nat. Mater. 2023, 22, 18–35. [Google Scholar] [CrossRef]
- Bystrov, V. Computer Simulation Nanostructures: Bioferroelectric Amino Acids. Bioferroelectricity: Peptide Nanotubes and Thymine Nucleobase; LAP LAMBERT Academic Publishing: Riga, Latvia, 2020; 137p, ISBN 978-620-2-91926-5. [Google Scholar]
- Makam, P.; Gazit, E. Minimalistic peptide supramolecular co-assembly: Expanding the conformational space for nanotechnology. Chem. Soc. Rev. 2018, 47, 3406–3420. [Google Scholar] [CrossRef]
- Scanlon, S.; Aggeli, A. Self-assembling peptide nanotubes. Nano Today 2008, 3, 22–30. [Google Scholar] [CrossRef]
- Yuan, C.; Ji, W.; Xing, R.; Li, J.; Gazit, E.; Yan, X. Hierarchically oriented organization in supramolecular peptide crystals. Nat. Rev. Chem. 2019, 3, 567–588. [Google Scholar] [CrossRef]
- Raymond, D.M.; Nilsson, B.L. Multicomponent peptide assemblies. Chem. Soc. Rev. 2018, 47, 3659–3720. [Google Scholar] [CrossRef]
- Bystrov, V.S.; Zelenovskiy, P.S.; Nuraeva, A.S.; Kopyl, S.; Zhulyabina, O.A.; Tverdislov, V. Chiral Peculiar Properties of Self-Organization of Diphenylalanine Peptide Nanotubes: Modeling of Structure and Properties. Math. Biol. Bioinform. 2019, 14, 94–125. [Google Scholar] [CrossRef]
- Kol, N.; Adler-Abramovich, L.; Barlam, D.; Shneck, R.Z.; Gazit, E.; Rousso, I. Self-Assembled Peptide Nanotubes Are Uniquely Rigid Bioinspired Supramolecular Structures. Nano Lett. 2005, 5, 1343–1346. [Google Scholar] [CrossRef] [PubMed]
- Shklovsky, J.; Beker, P.; Amdursky, N.; Gazit, E.; Rosenman, G. Bioinspired peptide nanotubes: Deposition technology and physical properties. Mater. Sci. Eng. B 2010, 169, 62–66. [Google Scholar] [CrossRef]
- Reches, M.; Gazit, E. Controlled patterning of aligned self-assembled peptide nanotubes. Nat. Nanotechnol. 2006, 1, 195–200. [Google Scholar] [CrossRef]
- Görbitz, C.H. Nanotube formation by hydrophobic dipeptides. Chem. Weinh. Bergstr. Ger. 2001, 7, 5153–5159. [Google Scholar] [CrossRef]
- Bystrov, V.S.; Paramonova, E.; Bdikin, I.; Kopyl, S.; Heredia, A.; Pullar, R.C.; Kholkin, A.L. BioFerroelectricity: Diphenylalanine Peptide Nanotubes Computational Modeling and Ferroelectric Properties at the Nanoscale. Ferroelectrics 2012, 440, 3–24. [Google Scholar] [CrossRef]
- Bystrov, V.; Sidorova, A.; Lutsenko, A.; Shpigun, D.; Malyshko, E.; Nuraeva, A.; Zelenovskiy, P.; Kopyl, S.; Kholkin, A. Modeling of Self-Assembled Peptide Nanotubes and Determination of Their Chirality Sign Based on Dipole Moment Calculations. Nanomaterials 2021, 11, 2415. [Google Scholar] [CrossRef]
- Bystrov, V.; Coutinho, J.; Zelenovskiy, P.; Nuraeva, A.; Kopyl, S.; Zhulyabina, O.; Tverdislov, V. Structures and Properties of the Self-Assembling Diphenylalanine Peptide Nanotubes Containing Water Molecules: Modeling and Data Analysis. Nanomaterials 2020, 10, 1999. [Google Scholar] [CrossRef] [PubMed]
- Bystrov, V.S.; Filippov, S.V. Molecular modelling and computational studies of peptide diphenylalanine nanotubes, containing waters: Structural and interactions analysis. J. Mol. Model. 2022, 28, 81. [Google Scholar] [CrossRef] [PubMed]
- CCDC Home|CCDC 16340 for FF—from Refs #11 (Gorbitz), CCDC 1853771—from Refs #17 (Zelenovskii). Available online: http://www.ccdc.cam.ac.uk (accessed on 6 May 2013).
- Zelenovskiy, P.S.; Nuraeva, A.; Kopyl, S.; Arkhipov, S.G.; Vasilev, S.G.; Bystrov, V.S.; Gruzdev, D.A.; Waliczek, M.; Svitlyk, V.; Shur, V.Y.; et al. Chirality-Dependent Growth of Self-Assembled Diphenylalanine Microtubes. Cryst. Growth Des. 2019, 19, 6414–6421. [Google Scholar] [CrossRef]
- Tverdislov, V.A. Chirality as a primary switch of hierarchical levels in molecular biological systems. Biophysics 2013, 58, 128–132. [Google Scholar] [CrossRef]
- Zelenovskiy, P.; Kornev, I.; Vasilev, S.; Kholkin, A. On the origin of the great rigidity of self-assembled diphenylalanine nano-tubes. Phys. Chem. Chem. Phys. 2016, 18, 29681–29685. [Google Scholar] [CrossRef] [PubMed]
- Tao, K.; Makam, P.; Aizen, R.; Gazit, E. Self-assembling peptide semiconductors. Science 2017, 358, eaam9756. [Google Scholar] [CrossRef] [PubMed]
- Amdursky, N.; Molotskii, M.; Aronov, D.; Adler-Abramovich, L.; Gazit, E.; Rosenman, G. Blue luminescence based on quan-tum confinement at peptide nanotubes. Nano Lett. 2009, 9, 3111–3115. [Google Scholar] [CrossRef]
- Gan, Z.; Wu, X.; Zhu, X.; Shen, J. Light-induced ferroelectricity in bioinspired self-assembled diphenylalanine nano-tubes/microtubes. Angew. Chem. Int. Ed. Engl. 2013, 52, 2055–2059. [Google Scholar] [CrossRef]
- Gan, Z.; Wu, X.; Zhang, J.; Zhu, X.; Chu, P.K. In Situ Thermal Imaging and Absolute Temperature Monitoring by Luminescent Diphenylalanine Nanotubes. Biomacromolecules 2013, 14, 2112–2116. [Google Scholar] [CrossRef]
- Nikitin, T.; Kopyl, S.; Shur, V.; Kopelevich, Y.; Kholkin, A. Low-temperature photoluminescence in self-assembled diphenylalanine microtubes. Phys. Lett. A 2016, 380, 1658–1662. [Google Scholar] [CrossRef]
- Nguyen, V.; Zhu, R.; Jenkins, K.; Yang, R. Self-assembly of diphenylalanine peptide with controlled polarization for power generation. Nat. Commun. 2016, 7, 13566. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, K.; Kelly, S.; Nguyen, V.; Wu, Y.; Yang, R. Piezoelectric diphenylalanine peptide for greatly improved flexible nano-generators. Nano Energy 2018, 51, 317–323. [Google Scholar] [CrossRef]
- Vasilev, S.; Zelenovskiy, P.; Vasileva, D.; Nuraeva, A.; Shur, V.Y.; Kholkin, A.L. Piezoelectric properties of diphenylalanine microtubes prepared from the solution. J. Phys. Chem. Solids 2016, 93, 68–72. [Google Scholar] [CrossRef]
- Bystrov, V. Photoferroelectricity in di-phenylalanine peptide nanotubes. Comput. Condens. Matter 2018, 14, 94–100. [Google Scholar] [CrossRef]
- Bystrov, V.; Paramonova, E.; Zelenovskii, P.; Kopyl, S.; Shen, H.; Lin, T.; Fridkin, V. Photoelectronic Properties of Chiral Self-Assembled Diphenylalanine Nanotubes: A Computational Study. Symmetry 2023, 15, 504. [Google Scholar] [CrossRef]
- Likhachev, I.; Bystrov, V. Assembly of a Phenylalanine Nanotube by the use of Molecular Dynamics Manipulator. Math. Biol. Bioinform. 2021, 16, 244–255. [Google Scholar] [CrossRef]
- Bystrov, V.; Likhachev, I.; Sidorova, A.; Filippov, S.; Lutsenko, A.; Shpigun, D.; Belova, E. Molecular Dynamics Simula-tion Study of the Self-Assembly of Phenylalanine Peptide Nanotubes. Nanomaterials 2022, 12, 861. [Google Scholar] [CrossRef]
- German, H.W.; Uyaver, S.; Hansmann, U.H.E. Self-Assembly of Phenylalanine-Based Molecules. J. Phys. Chem. A 2014, 119, 1609–1615. [Google Scholar] [CrossRef]
- Adler-Abramovich, L.; Vaks, L.; Carny, O.; Trudler, D.; Magno, A.; Caflisch, A.; Frenkel, D.; Gazit, E. Phenylalanine assembly into toxic fibrils suggests amyloid etiology in phenylketonuria. Nat. Chem. Biol. 2012, 8, 701–706. [Google Scholar] [CrossRef]
- HyperChem 8. Tools for Molecular Modeling. Professional Edition for Windows AC Release 8.0 USB (on CD). Gainesville, FL 32601 United States: Hypercube. Inc. 2011. Available online: http://www.hypercubeusa.com/ (accessed on 10 April 2023).
- Lemak, A.S.; Balabaev, N.K. A Comparison Between Collisional Dynamics and Brownian Dynamics. Mol. Simul. 1995, 15, 223–231. [Google Scholar] [CrossRef]
- Lemak, A.S.; Balabaev, N.K. Molecular dynamics simulation of a polymer chain in solution by collisional dynamics method. J. Comput. Chem. 1996, 17, 1685–1695. [Google Scholar] [CrossRef]
- Filippov, S.V.; Bystrov, V.S. A Visual Differential Analysis of Structural Features of Internal Cavities in Two Chiral Forms of Diphenylalanine Nanotubes. Biophysics 2020, 65, 374–380. [Google Scholar] [CrossRef]
- Bystrov, V.S.; Filippov, S.V. Computer modeling and numerical studies of peptide nanotubes based on diphenylalanine. Keldysh Inst. Prepr. 2021, 78, 1–54. [Google Scholar] [CrossRef]
- Wang, J.M.; Cieplak, P.; Kollman, P. How Well Does a Restrained Electrostatic Potential (RESP) Model Perform in Calculat-ing Conformational Energies of Organic and Biological Molecules? J. Comput. Chem. 1999, 21, 1049. [Google Scholar] [CrossRef]
- Glyakina, A.V.; Likhachev, I.V.; Balabaev, N.K.; Galzitskaya, O.V. Comparative mechanical unfolding studies of spectrin domains R15, R16 and R17. J. Struct. Biol. 2018, 201, 162–170. [Google Scholar] [CrossRef] [PubMed]
- Balabayev, N.K.; Likhachev, I. Trajectory Analyzer of Molecular Dynamics. Math. Biol. Bioinform. 2007, 2, 120–129. [Google Scholar] [CrossRef]
- Likhachev, I.V.; Balabaev, N.; Galzitskaya, O.V. Available Instruments for Analyzing Molecular Dynamics Trajectories. Open Biochem. J. 2016, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- PyMOL. Pymol.Org. Available online: https://pymol.org/2/ (accessed on 11 November 2022).
- Tverdislov, V.A.; Sidorova, A.E.; Bagrova, O.E.; Belova, E.V.; Bystrov, V.S.; Levashova, N.T.; Lutsenko, A.O.; Semenova, E.V.; Shpigun, D.K. Chirality as a Symmetric Basis of Self-Organization of Biomacromolecules. Biophysics 2022, 67, 673–691. [Google Scholar] [CrossRef]
- Sidorova, A.; Bystrov, V.; Lutsenko, A.; Shpigun, D.; Belova, E.; Likhachev, I. Quantitative Assessment of Chirality of Protein Secondary Structures and Phenylalanine Peptide Nanotubes. Nanomaterials 2021, 11, 3299. [Google Scholar] [CrossRef]
- Filippov, S.V. Blender software platform as an environment for modeling objects and processes of science disciplines. Keldysh Inst. Prepr. 2018, 230, 1–42. [Google Scholar] [CrossRef]
- Filippov, S.V. Method for the identification of atoms of macromolecules visualized in 3D-editors. Keldysh Inst. Prepr. 2019, 97, 1–10. [Google Scholar] [CrossRef]
- Blender Foundation. blender.org—Home of the Blender Project—Free and Open 3D Creation Software [Electronic Resource]. Available online: http://www.blender.org (accessed on 10 April 2023).
- G’MIC—GREYC’s Magic for Image Computing: A Full-Featured Open-Source Framework for Image Processing—Main [Electronic Resource]. Available online: https://gmic.eu/ (accessed on 10 April 2023).
- Reches, M.; Gazit, E. Casting Metal Nanowires Within Discrete Self-Assembled Peptide Nanotubes. Science 2003, 300, 625–627. [Google Scholar] [CrossRef] [PubMed]
- Adler-Abramovich, L.; Gazit, E. The physical properties of supramolecular peptide assemblies: From building block association to technological applications. Chem. Soc. Rev. 2014, 43, 6881–6893, Correction in Chem. Soc. Rev. 2014, 43, 7236. [Google Scholar] [CrossRef]
- Tamamis, P.; Adler-Abramovich, L.; Reches, M.; Marshall, K.; Sikorski, P.; Serpell, L.; Gazit, E.; Archontis, G. Self-Assembly of Phenylalanine Oligopeptides: Insights from Experiments and Simulations. Biophys. J. 2009, 96, 5020–5029. [Google Scholar] [CrossRef]
- Rissanou, A.N.; Georgilis, E.; Kasotakis, E.; Mitraki, A.; Harmandaris, V. Effect of Solvent on the Self-Assembly of Dialanine and Diphenylalanine Peptides. J. Phys. Chem. B 2013, 117, 3962–3975. [Google Scholar] [CrossRef]
- Guo, C.; Luo, Y.; Zhou, R.; Wei, G. Probing the Self-Assembly Mechanism of Diphenylalanine-Based Peptide Nanovesicles and Nanotubes. ACS Nano 2012, 6, 3907–3918. [Google Scholar] [CrossRef]
- Xiong, Q.; Jiang, Y.; Cai, X.; Yang, F.; Li, Z.; Han, W. Conformation Dependence of Diphenylalanine Self-Assembly Structures and Dynamics: Insights from Hybrid-Resolution Simulations. ACS Nano 2019, 13, 4455–4468. [Google Scholar] [CrossRef]
- Bellotto, O.; D’andrea, P.; Marchesan, S. Nanotubes and water-channels from self-assembling dipeptides. J. Mater. Chem. B 2023. [Google Scholar] [CrossRef]
- Bellotto, O.; Pierri, G.; Rozhin, P.; Polentarutti, M.; Kralj, S.; D’Andrea, P.; Tedesco, C.; Marchesan, S. Dipeptide self-assembly into water-channels and gel biomaterial. Org. Biomol. Chem. 2022, 20, 6211–6218. [Google Scholar] [CrossRef] [PubMed]
- Bystrov, V.S.; Bdikin, I.K.; Singh, B. Piezoelectric and ferroelectric properties of various amino acids and tubular dipeptide nanostructures: Molecular modeling. Nanomater. Sci. Eng. 2020, 2, 11–24. [Google Scholar] [CrossRef]
- Bystrov, V.S.; Paramonova, E.V.; Yurkova, D.O.; Ledeneva, O.R.; Belova, E.V. Photoelectronic properties of peptide nanotubes based on various amino acids. In Proceedings of the VII Cogress of Biophysics of Russia, Krasnodar, Russia, 17–23 April 2023; Kuban State Technological University: Krasnodar, Russia, 2023; Volume 1, pp. 128–129. [Google Scholar]
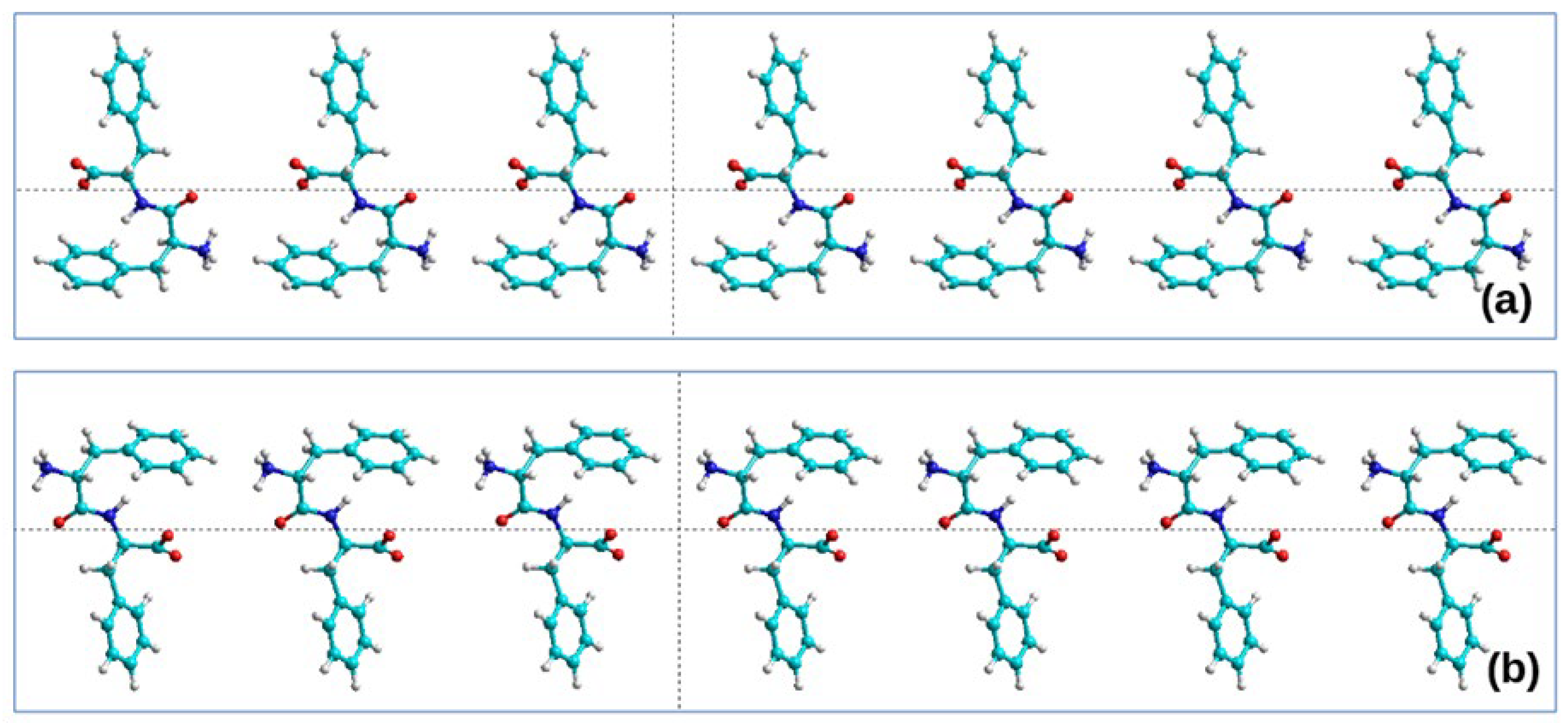

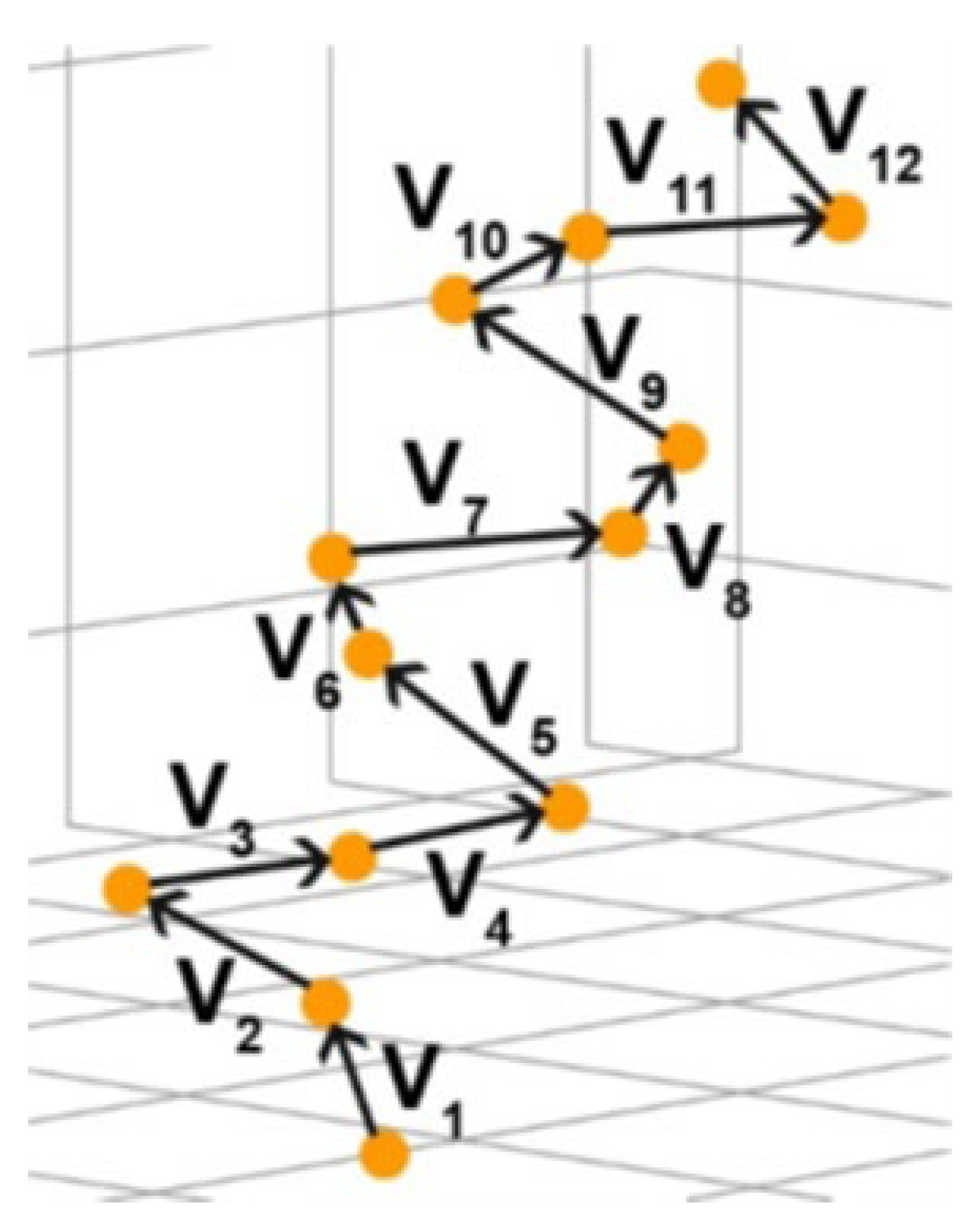
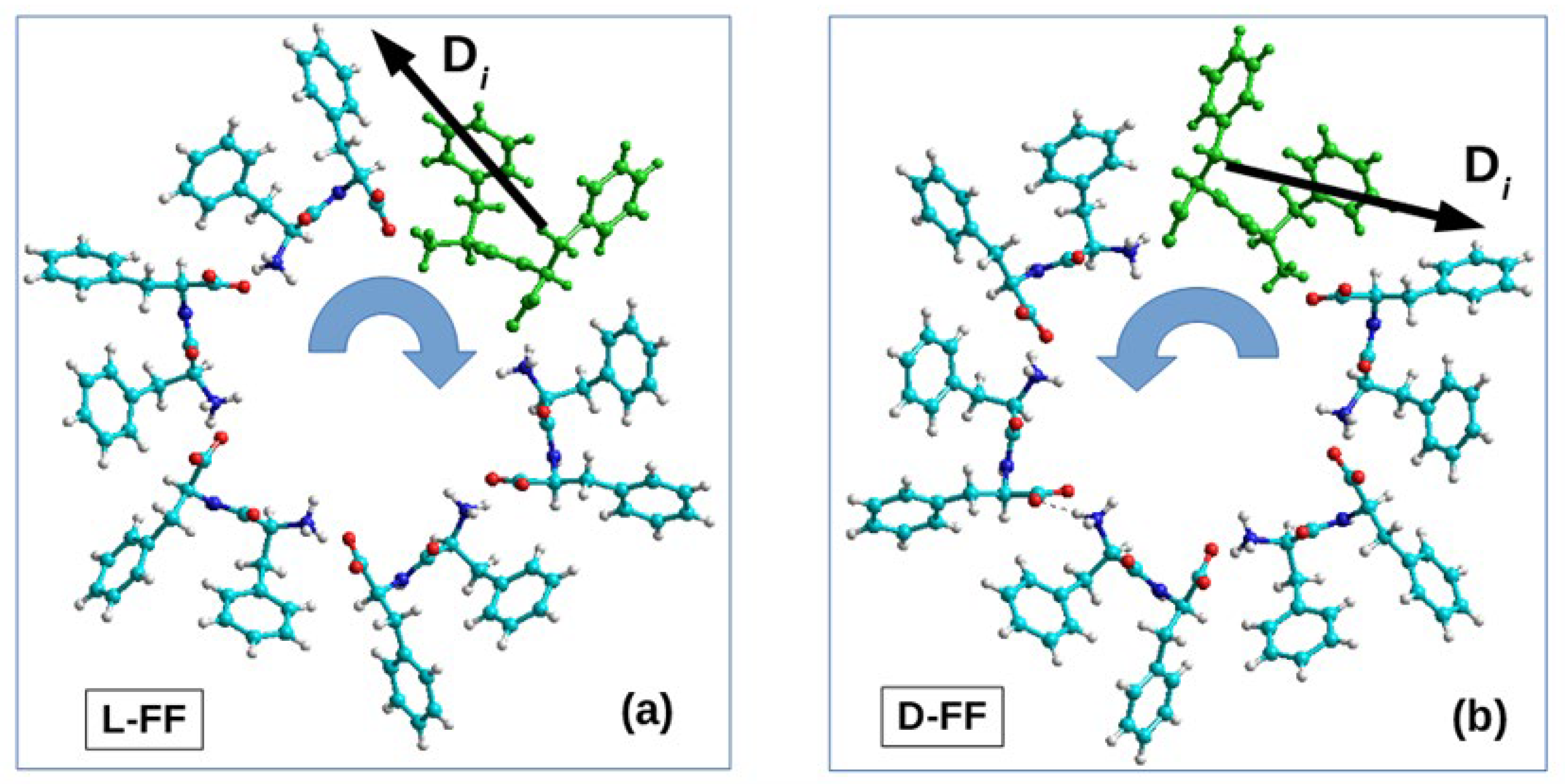
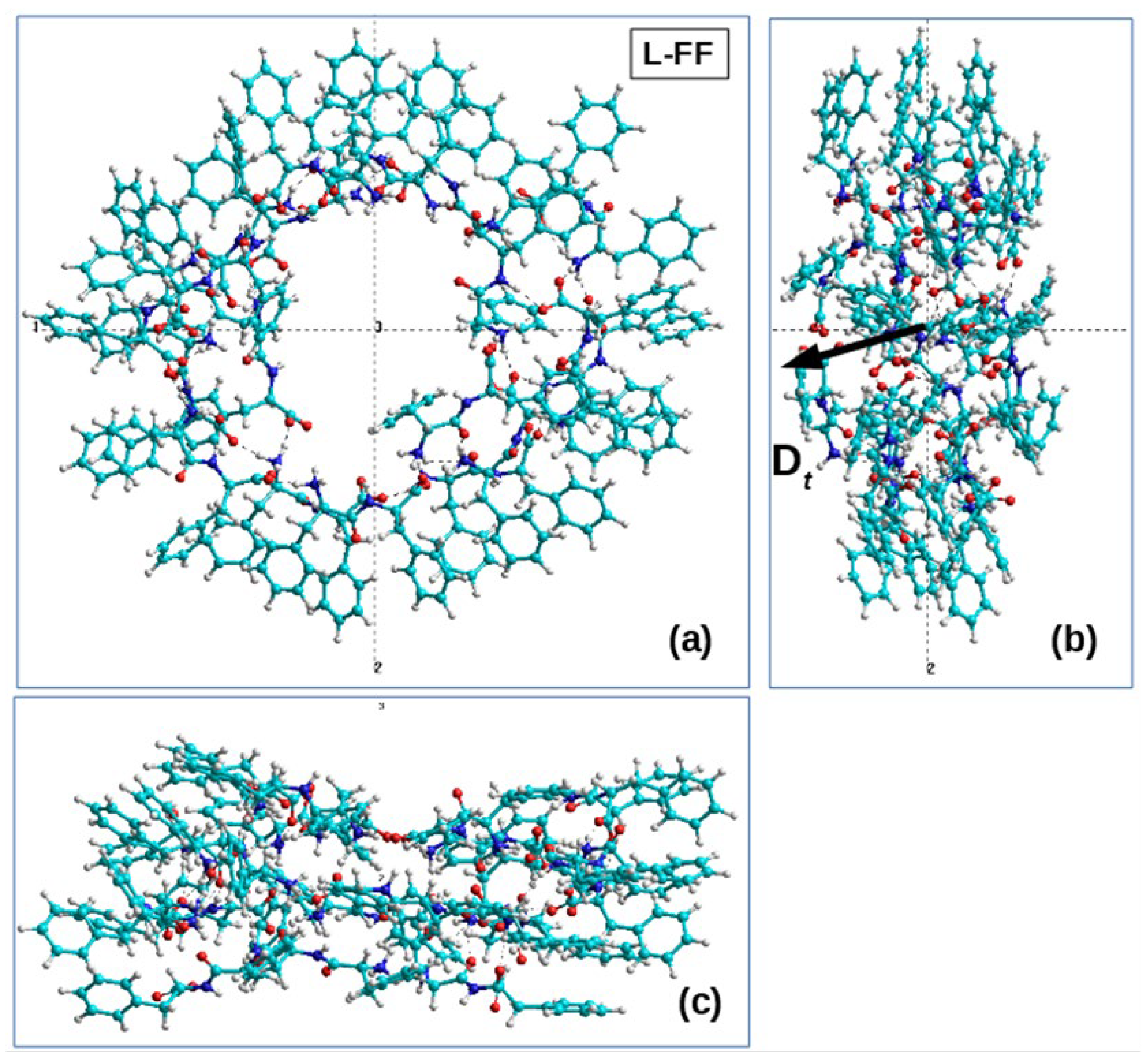
| Run Number | L-Monomer | D-Monomer | ||||
|---|---|---|---|---|---|---|
| Number of Left Helices | Number of Right Helices | Value According to the Formulas (1)–(3) | Number of Left Helices | Number of Right Helices | Value According to the Formulas (1)–(3) | |
| 1 | 1 | −0.006 | 0.8 | 0.2 | −0.046 | |
| 2 | 1 | 0.033 | 1 | −0.098 | ||
| 3 | 1 | 0.009 | 0.2 | 0.8 | −0.031 | |
| 4 | 1 | 0.043 | 0.8 | 0.2 | −0.045 | |
| 5 | 1 | 0.005 | 1 | −0.001 | ||
| 6 | 0.5 | 0.5 | −0.032 | 0.5 | 0.5 | −0.007 |
| 7 | 1 | 0.018 | 1 | −0.030 | ||
| 8 | 1 | −0.027 | 1 | −0.031 | ||
| 9 | 1 | −0.038 | 1 | −0.023 | ||
| 10 | 0.5 | 0.5 | 0.042 | 1 | −0.002 | |
| 11 | 1 | 0.008 | 0.5 | 0.5 | 0.050 | |
| 12 | 1 | 0.016 | 1 | −0.027 | ||
| 13 | 0.5 | 0.5 | 0.062 | 0.5 | 0.5 | 0.004 |
| 14 | 1 | 0.001 | 0.5 | 0.5 | 0.003 | |
| 15 | 1 | 0.039 | 0.5 | 0.5 | 0.018 | |
| 16 | 1 | 0.031 | 1 | −0.003 | ||
| 17 | 0.5 | 0.5 | 0.040 | 1 | 0.004 | |
| 18 | 1 | 0.003 | 0.5 | 0.5 | −0.028 | |
| 19 | 1 | −0.004 | 1 | −0.043 | ||
| 20 | 0.5 | 0.5 | 0.015 | 0.8 | 0.2 | −0.063 |
| 21 | 1 | 0.073 | 0.5 | 0.5 | 0.003 | |
| 22 | 1 | 0.050 | 0.2 | 0.8 | −0.054 | |
| 23 | 1 | 0.059 | 0.5 | 0.5 | −0.030 | |
| 24 | 1 | 0.054 | 0.5 | 0.5 | −0.029 | |
| 25 | 1 | 0.048 | 1 | −0.022 | ||
| 26 | 1 | 0.015 | 1 | −0.011 | ||
| 27 | 0.5 | 0.5 | 0.044 | 0.5 | 0.5 | −0.039 |
| 28 | 0.5 | 0.5 | 0.086 | 0.5 | 0.5 | 0.012 |
| 29 | 0.5 | 0.5 | 0.003 | 0.8 | 0.2 | −0.012 |
| 30 | 1 | −0.021 | 0.5 | 0.5 | −0.011 | |
| 31 | 1 | 0.043 | 0.5 | 0.5 | −0.007 | |
| 32 | 1 | 0.034 | 0.8 | 0.2 | −0.016 | |
| Total expert opinion | 7 (22%) | 25 (78%) | 21.9 (68%) | 10.1 (32%) | ||
| Total according to Formulas (1)–(3) | 6 (19%) | 26 (81%) | 25 (78%) | 7 (22%) | ||
| Initial Chirality Type of the FF Molecule | L-FF | D-FF |
|---|---|---|
| Computed data for 6FF PNT coil from [14,15] | 1.219 | −0.674 |
| Computed data for 6FF PNT coil in present work | 1.357 | −1.347 |
| Computed data for 19FF PNT obtained by MD method in present work | 0.7771 | −1.2302 |
| Chirality PNT sign | Positive | Negative |
| Chirality PNT symbol | D | L |
| Experimental D-FF NT | 9.08 |
| Experimental L-FF NT | 9.17 |
| Molecular dynamics D-FF NT | 10.89 |
| Molecular dynamics L-FF NT | 10.87 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bystrov, V.; Likhachev, I.; Filippov, S.; Paramonova, E. Molecular Dynamics Simulation of Self-Assembly Processes of Diphenylalanine Peptide Nanotubes and Determination of Their Chirality. Nanomaterials 2023, 13, 1905. https://doi.org/10.3390/nano13131905
Bystrov V, Likhachev I, Filippov S, Paramonova E. Molecular Dynamics Simulation of Self-Assembly Processes of Diphenylalanine Peptide Nanotubes and Determination of Their Chirality. Nanomaterials. 2023; 13(13):1905. https://doi.org/10.3390/nano13131905
Chicago/Turabian StyleBystrov, Vladimir, Ilya Likhachev, Sergey Filippov, and Ekaterina Paramonova. 2023. "Molecular Dynamics Simulation of Self-Assembly Processes of Diphenylalanine Peptide Nanotubes and Determination of Their Chirality" Nanomaterials 13, no. 13: 1905. https://doi.org/10.3390/nano13131905
APA StyleBystrov, V., Likhachev, I., Filippov, S., & Paramonova, E. (2023). Molecular Dynamics Simulation of Self-Assembly Processes of Diphenylalanine Peptide Nanotubes and Determination of Their Chirality. Nanomaterials, 13(13), 1905. https://doi.org/10.3390/nano13131905






