An Operational DNA Strand Displacement Encryption Approach
Abstract
:1. Introduction
2. A Bio-Experiment-Based Encryption Approach
2.1. Encryption Framework
| Algorithm 1: A new bio-experiment-based encryption framework. |
 |
2.2. Huffman Coding and Data Transformation
2.3. SDR Modules and Seed Encoding
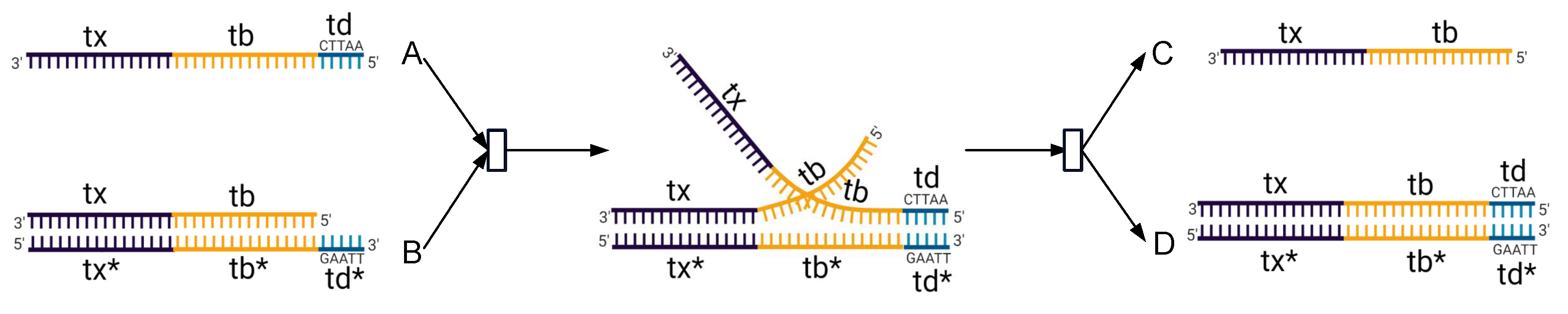
2.3.1. Degradation Reaction Module
2.3.2. Catalysis Reaction Module
2.4. Group Cyclic Shift
| Algorithm 2: groupCS(, , ℓ), a procedure that extends a DNA sequence of length to a new one of length at least ℓ. |
 |
2.5. The BioEN Algorithm
| Algorithm 3: BioEN (a DNA-strand-displacement-based encryption algorithm). |
 |
3. Validation of Feasibility
3.1. Experimental Setup
3.2. Tools and Data
3.3. Experimentation Procedure
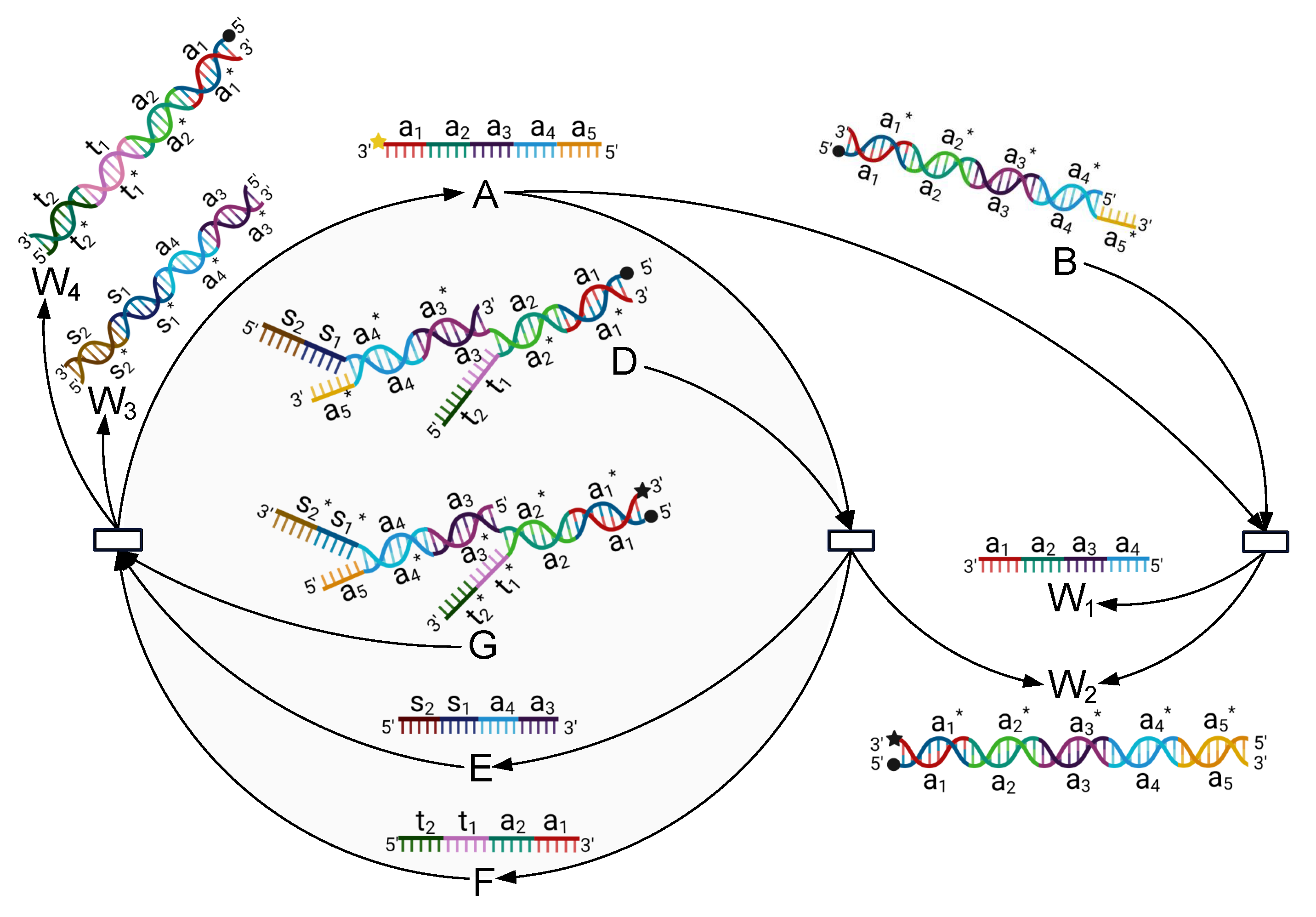
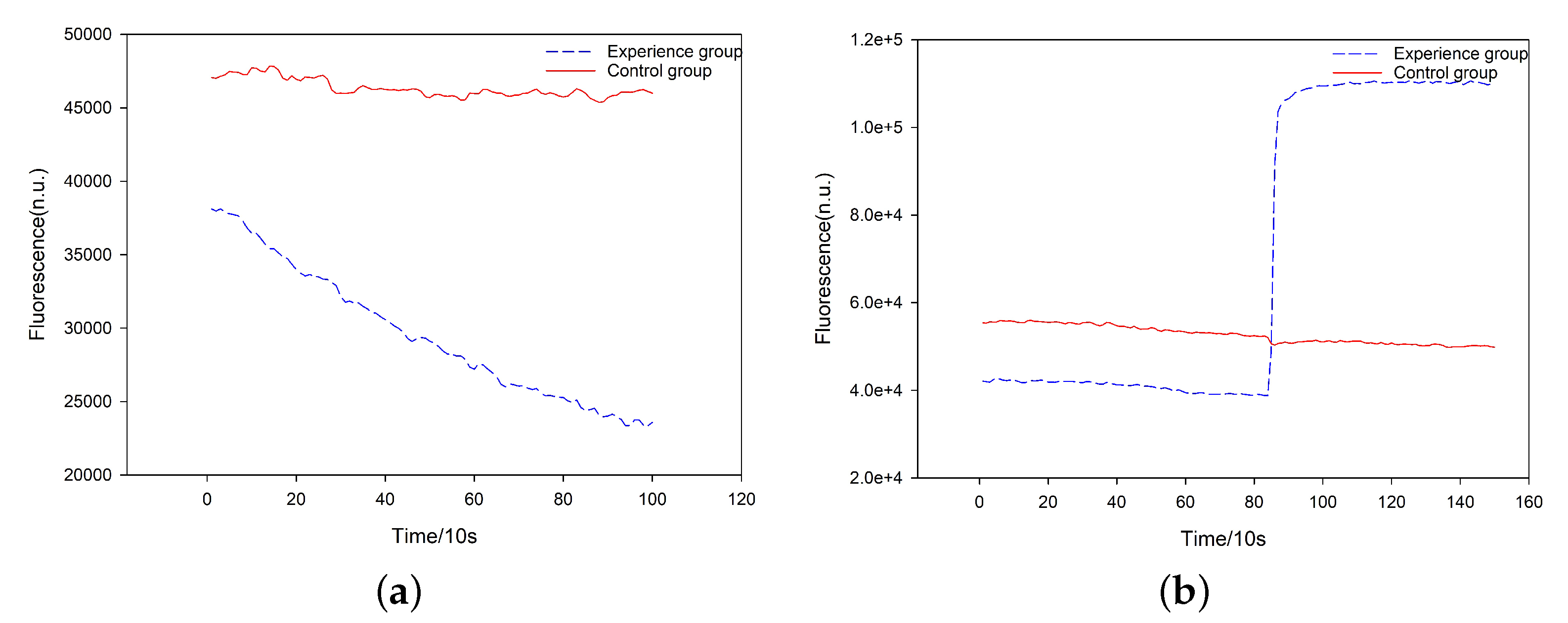
3.3.1. Simulation Experiment of the DR-Module
3.3.2. Biological Experiments of the DR-Module
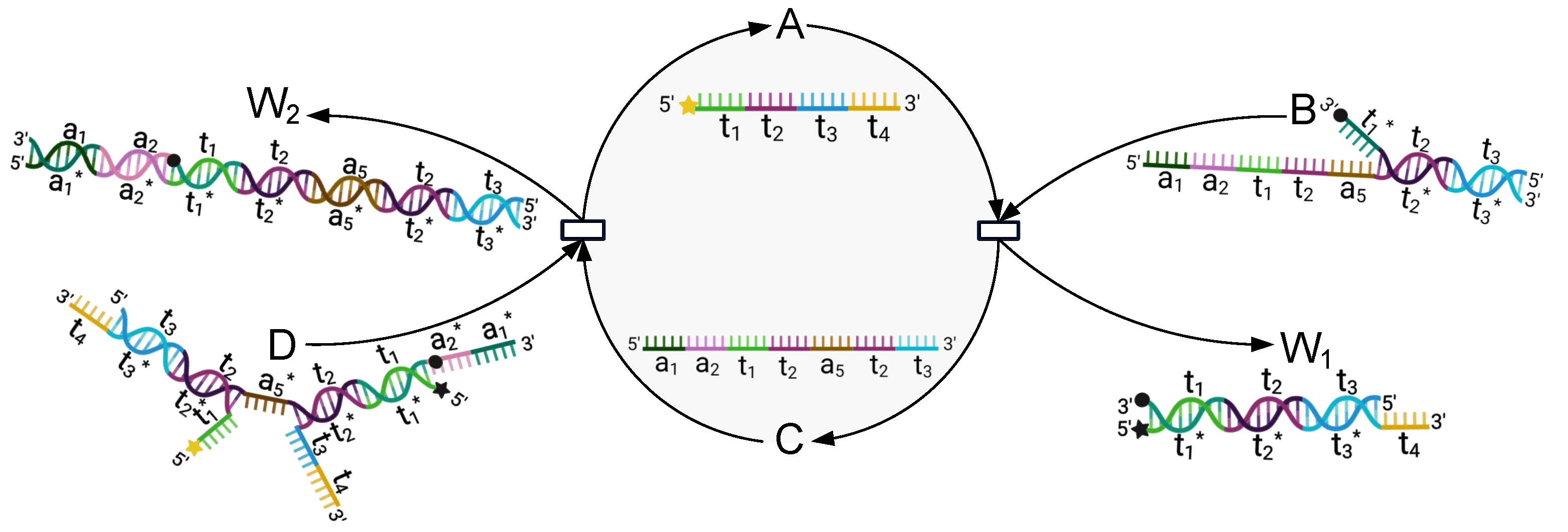
3.3.3. Simulation Experiment of the CR-Module
3.3.4. Biological Experiments of CR-Module
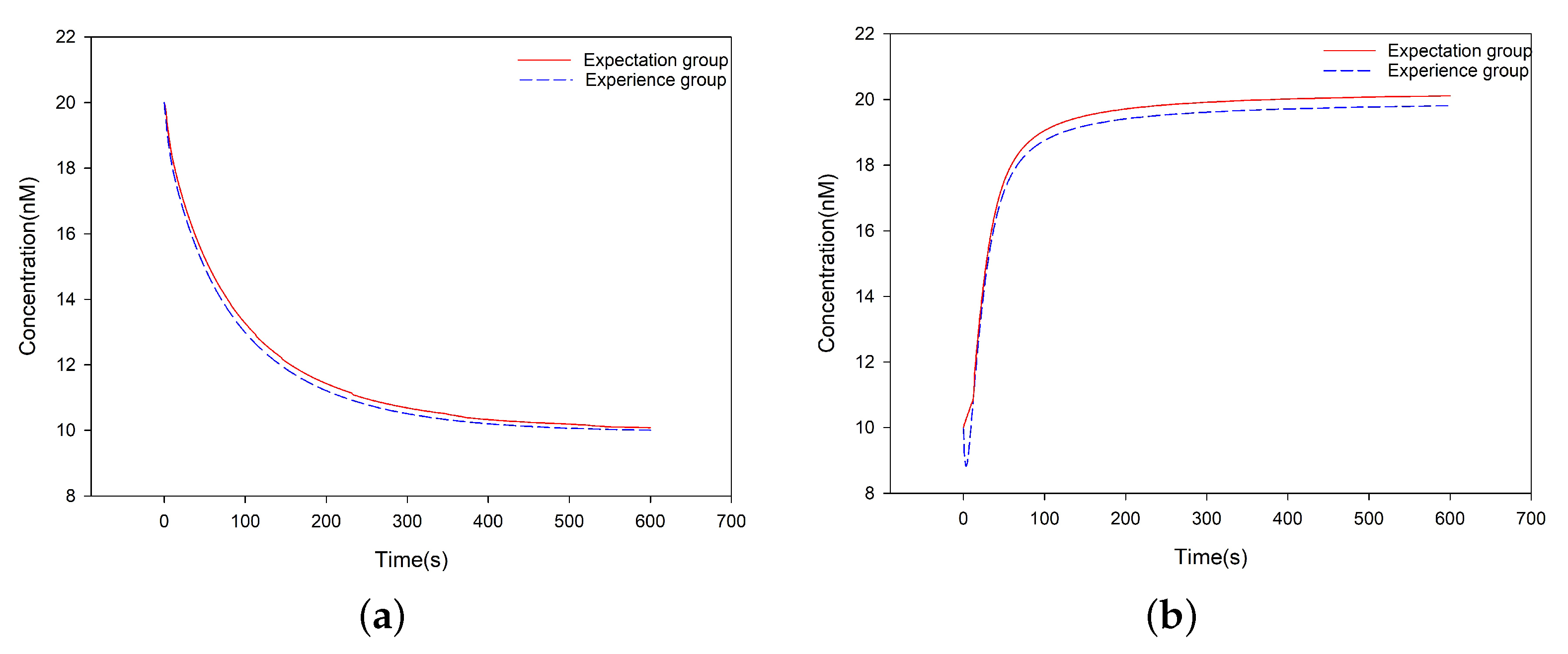
3.4. Experimental Results
4. Security Analysis
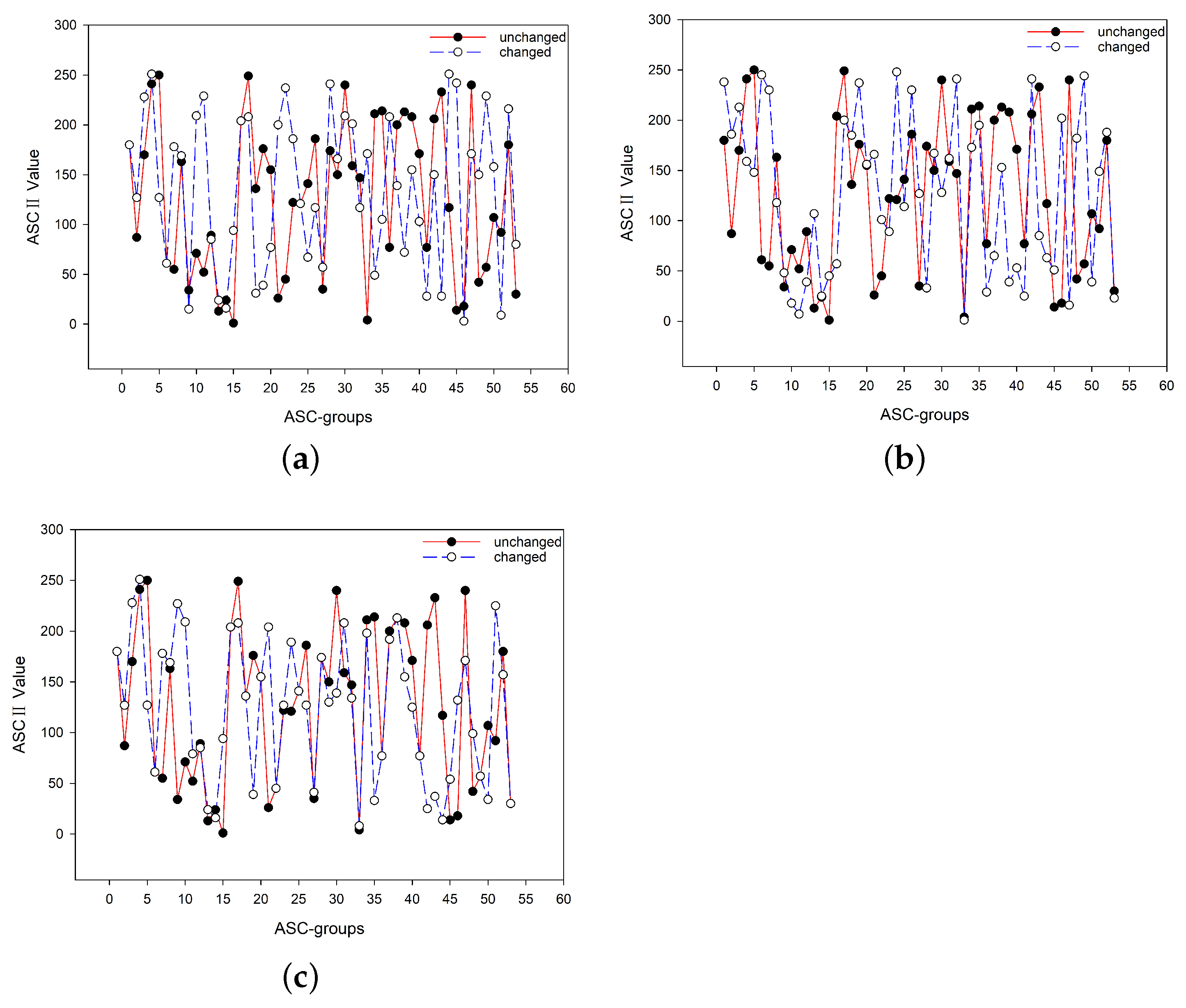
4.1. Key Sensitivity
4.1.1. Change One Base
4.1.2. Change Experiment Conditions
4.1.3. Change One Element in the Process of Extending the Key
4.2. Key Space Analysis
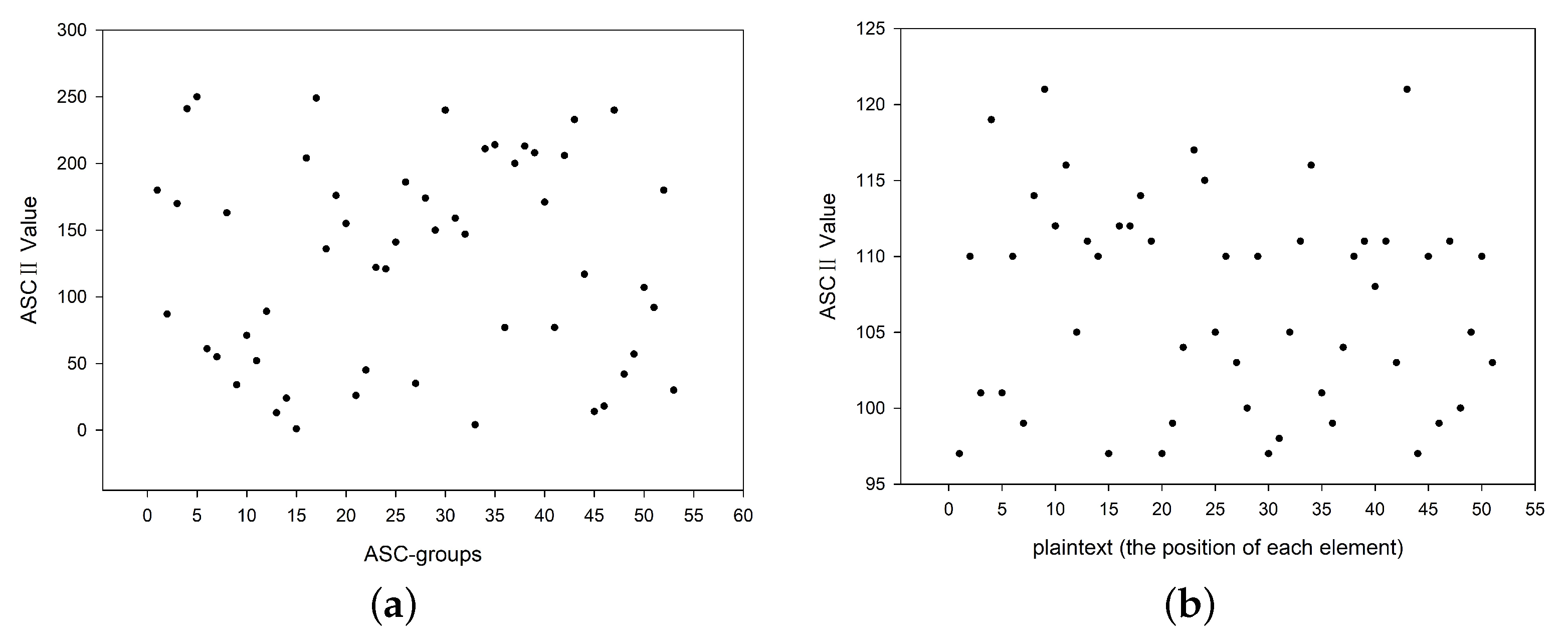
4.3. Statistic Characteristic
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mali, K.; Chakraborty, S.; Roy, M. A study on statistical analysis and security evaluation parameters in image encryption. Int. J. Sci. Res. Dev. 2015, 3, 339–343. [Google Scholar]
- Liang, Z.; Qin, Q.; Zhou, C.; Wang, N.; Xu, Y.; Zhou, W. Medical image encryption algorithm based on a new five-dimensional three-leaf chaotic system and genetic operation. PLoS ONE 2021, 16, e0260014. [Google Scholar] [CrossRef] [PubMed]
- Gehani, A.; LaBean, T.; Reif, J. DNA-based cryptography. In Aspects of Molecular Computing; Springer: Berlin/Heidelberg, Germany, 2003; pp. 167–188. [Google Scholar]
- Roy, M.; Mali, K.; Chatterjee, S.; Chakraborty, S.; Debnath, R.; Sen, S. A study on the applications of the biomedical image encryption methods for secured computer aided diagnostics. In Proceedings of the 2019 Amity International Conference on Artificial Intelligence (AICAI), Dubai, United Arab Emirates, 4–6 February 2019; pp. 881–886. [Google Scholar]
- Wang, X.; Teng, L. A one-time one-key encryption algorithm based on the ergodicity of chaos. Chin. Phys. B 2012, 21, 020504. [Google Scholar] [CrossRef]
- Mokhtar, M.A.; Gobran, S.N.; El-Badawy, E.S.A. Colored image encryption algorithm using DNA code and chaos theory. In Proceedings of the 2014 International Conference on Computer and Communication Engineering, Tianjin, China, 27–28 March 2014; pp. 12–15. [Google Scholar]
- Yang, J.; Ma, J.; Liu, S.; Zhang, C. A molecular cryptography model based on structures of DNA self-assembly. Chin. Sci. Bull. 2014, 59, 1192–1198. [Google Scholar] [CrossRef]
- Liu, H.; Wang, X. Image encryption using DNA complementary rule and chaotic maps. Appl. Soft Comput. 2012, 12, 1457–1466. [Google Scholar] [CrossRef]
- Rehman, A.; Liao, X.; Kulsoom, A.; Abbas, S. Selective encryption for gray images based on chaos and DNA complementary rules. Multimed. Tools Appl. 2015, 74, 4655–4677. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, J.; Fan, J.; Gong, L. Image encryption algorithm based on chaotic system and dynamic S-boxes composed of DNA sequences. Multimed. Tools Appl. 2016, 75, 4363–4382. [Google Scholar] [CrossRef]
- Wu, J.; Liao, X.; Yang, B. Image encryption using 2D Hénon-Sine map and DNA approach. Signal Process. 2018, 153, 11–23. [Google Scholar] [CrossRef]
- Zhang, J.; Huo, D. Image encryption algorithm based on quantum chaotic map and DNA coding. Multimed. Tools Appl. 2019, 78, 15605–15621. [Google Scholar] [CrossRef]
- Zheng, J.; Liu, L. Novel image encryption by combining dynamic DNA sequence encryption and the improved 2D logistic sine map. IET Image Process. 2020, 14, 2310–2320. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, M. A new image encryption algorithm based on ladder transformation and DNA coding. Multimed. Tools Appl. 2021, 80, 13339–13365. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, Y.; Zhao, Y. A novel image encryption scheme based on 2-D logistic map and DNA sequence operations. Nonlinear Dyn. 2015, 82, 1269–1280. [Google Scholar] [CrossRef]
- Wei, D.; Jiang, M. A fast image encryption algorithm based on parallel compressive sensing and DNA sequence. Optik 2021, 238, 166748. [Google Scholar] [CrossRef]
- Popli, M. DNA Cryptography: A Novel Approach for Data Security Using Genetic Algorithm. Int. J. Adv. Res. Comput. Sci. Manag. Stud. 2018, 6, 53–63. [Google Scholar]
- Ravichandran, D.; Murthy, B.; Balasubramanian, V.; Fathima, S.; Amirtharajan, R. An efficient medical image encryption using hybrid DNA computing and chaos in transform domain. Med. Biol. Eng. Comput. 2021, 59, 589–605. [Google Scholar] [CrossRef]
- Zhu, E.; Chen, C.; Rao, Y.; Xiong, W. Biochemical Logic Circuits Based on DNA Combinatorial Displacement. IEEE Access 2020, 8, 34096–34103. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Z.; Sun, J. Three-variable chaotic oscillatory system based on DNA strand displacement and its coupling combination synchronization. IEEE Trans. Nanobioscience 2020, 19, 434–445. [Google Scholar] [CrossRef]
- Zou, C.; Wei, X.; Zhang, Q.; Liu, C.; Liu, Y. Solution of equations based on analog DNA strand displacement circuits. IEEE Trans. Nanobioscience 2019, 18, 191–204. [Google Scholar] [CrossRef]
- Wang, F.; Lv, H.; Li, Q.; Li, J.; Zhang, X.; Shi, J.; Wang, L.; Fan, C. Implementing digital computing with DNA-based switching circuits. Nat. Commun. 2020, 11, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Liu, Y.; Zhu, E.; Zhang, Q.; Wei, X.; Wang, B. Cross-Inhibitor: A time-sensitive molecular circuit based on DNA strand displacement. Nucleic Acids Res. 2020, 48, 10691–10701. [Google Scholar] [CrossRef]
- Wang, R.; Wang, S.; Xu, X.; Jiang, W.; Zhang, N. MNAzyme probes mediated DNA logic platform for microRNAs logic detection and cancer cell identification. Anal. Chim. Acta 2021, 1149, 338213. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Yu, H.; Tian, J.; Xiao, B. Nonenzymatic DNA-Based Fluorescence Biosensor Combining Carbon Dots and Graphene Oxide with Target-Induced DNA Strand Displacement for microRNA Detection. Nanomaterials 2021, 11, 2608. [Google Scholar] [CrossRef] [PubMed]
- Okumura, S.; Nixon Hapsianto, B.; Lobato-Dauzier, N.; Ohno, Y.; Benner, S.; Torii, Y.; Tanabe, Y.; Takada, K.; Baccouche, A.; Shinohara, M.; et al. Morphological Manipulation of DNA Gel Microbeads with Biomolecular Stimuli. Nanomaterials 2021, 11, 293. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Yin, Z.; Wang, L.; Cui, J.; Yang, J.; Wang, R. Solving 0–1 Integer Programming Problem Based on DNA Strand Displacement Reaction Network. ACS Synth. Biol. 2021, 10, 2318–2330. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Xiao, B.; Zheng, X.; Zhou, C. An image encryption algorithm based on chaos system and DNA strand displacement model. In Proceedings of the 2nd International Conference on Artificial Intelligence: Techniques and Applications, DEStech Transactions on Computer Science and Engineering, Settat, Morocco, 28–30 June 2017; pp. 102–107. [Google Scholar]
- Zou, C.; Wei, X.; Zhang, Q.; Zhou, C.; Zhou, S. Encryption Algorithm Based on DNA Strand Displacement and DNA Sequence Operation. IEEE Trans. Nanobioscience 2021, 20, 223–234. [Google Scholar] [CrossRef] [PubMed]
- Ailenberg, M.; Rotstein, O.D. An improved Huffman coding method for archiving text, images, and music characters in DNA. Biotechniques 2009, 47, 747–754. [Google Scholar] [CrossRef]
- Jäntschi, L. Formulas, Algorithms and Examples for Binomial Distributed Data Confidence Interval Calculation: Excess Risk, Relative Risk and Odds Ratio. Mathematics 2021, 9, 2506. [Google Scholar] [CrossRef]
- Zhu, E.; Jiang, F.; Liu, C.; Xu, J. Partition independent set and reduction based approach for partition coloring problem. IEEE Trans. Cybern. 2020, in press. [Google Scholar] [CrossRef]
- Bolboacǎ, S.D.; Roşca, D.D.; Jäntschi, L. Structure-Activity Relationships from Natural Evolution. MATCH Commun. Math. Comput. Chem. 2014, 71, 149–172. [Google Scholar]
- Lakin, M.R.; Youssef, S.; Polo, F.; Emmott, S.; Phillips, A. Visual DSD: A design and analysis tool for DNA strand displacement systems. Bioinformatics 2011, 27, 3211–3213. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, E.; Luo, X.; Liu, C.; Chen, C. An Operational DNA Strand Displacement Encryption Approach. Nanomaterials 2022, 12, 877. https://doi.org/10.3390/nano12050877
Zhu E, Luo X, Liu C, Chen C. An Operational DNA Strand Displacement Encryption Approach. Nanomaterials. 2022; 12(5):877. https://doi.org/10.3390/nano12050877
Chicago/Turabian StyleZhu, Enqiang, Xianhang Luo, Chanjuan Liu, and Congzhou Chen. 2022. "An Operational DNA Strand Displacement Encryption Approach" Nanomaterials 12, no. 5: 877. https://doi.org/10.3390/nano12050877
APA StyleZhu, E., Luo, X., Liu, C., & Chen, C. (2022). An Operational DNA Strand Displacement Encryption Approach. Nanomaterials, 12(5), 877. https://doi.org/10.3390/nano12050877







