Classification of Amino Acids Using Hybrid Terahertz Spectrum and an Efficient Channel Attention Convolutional Neural Network
Abstract
:1. Introduction
2. Materials and Methods
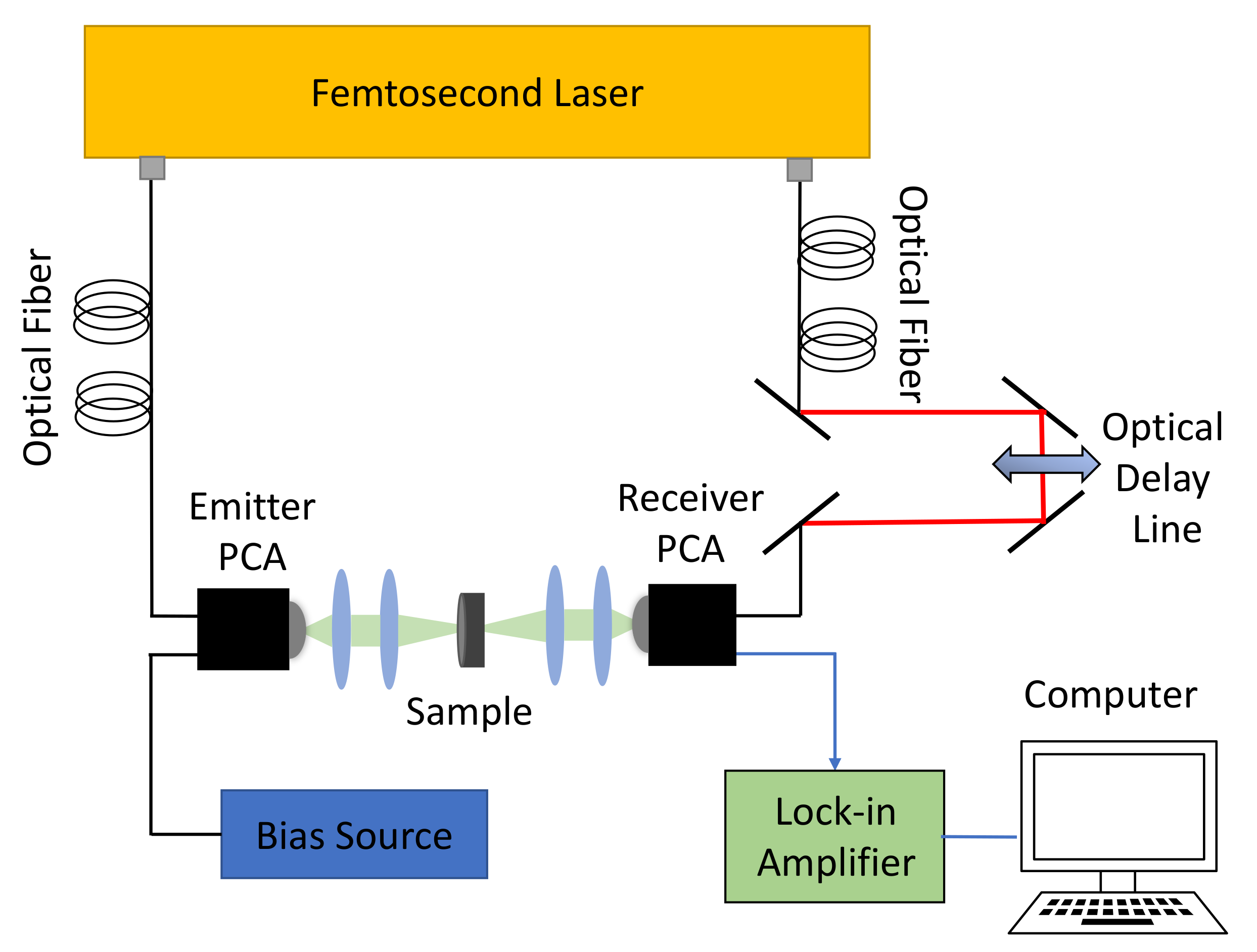
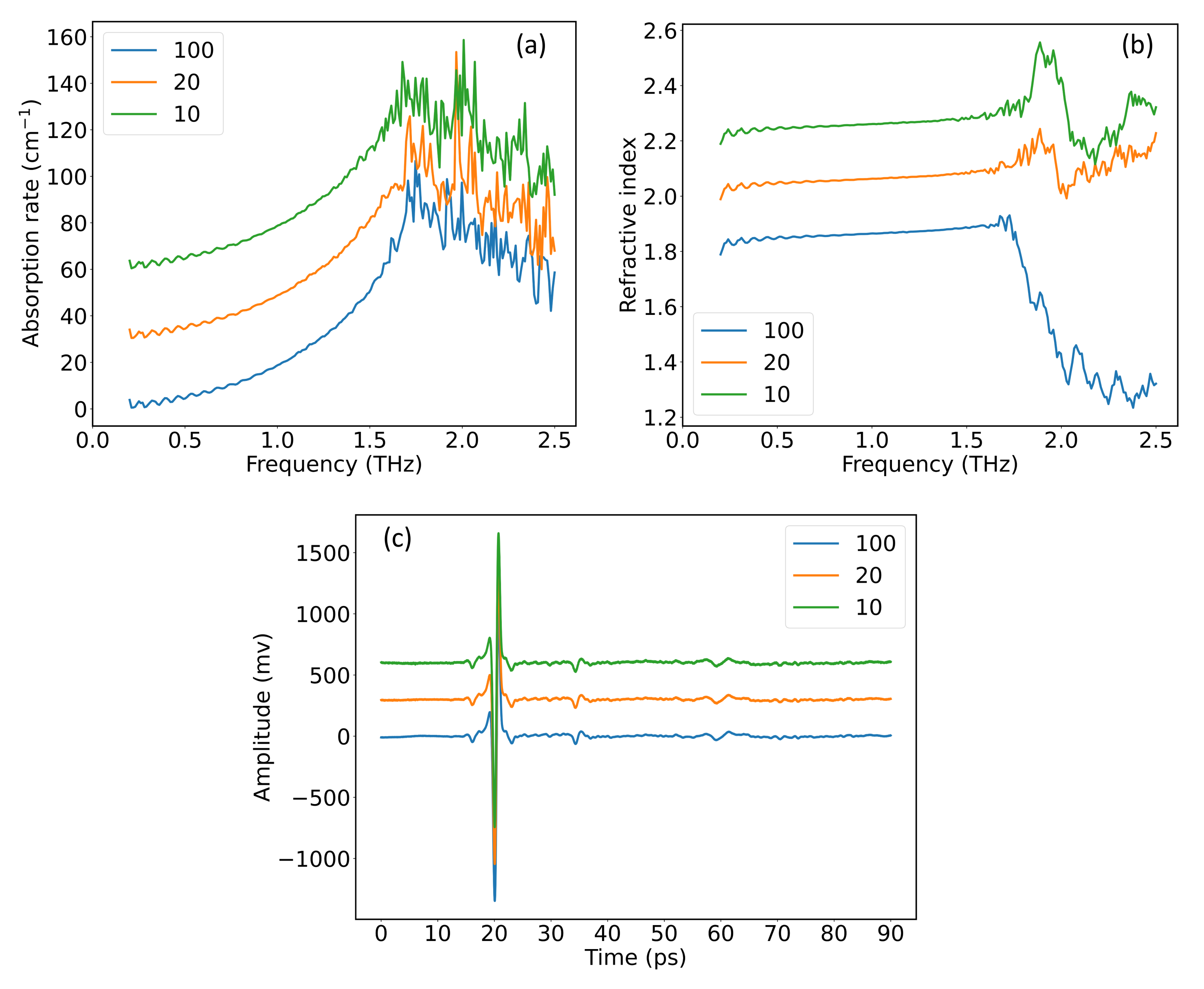
2.1. Experimental Equipment and Sample Preparation
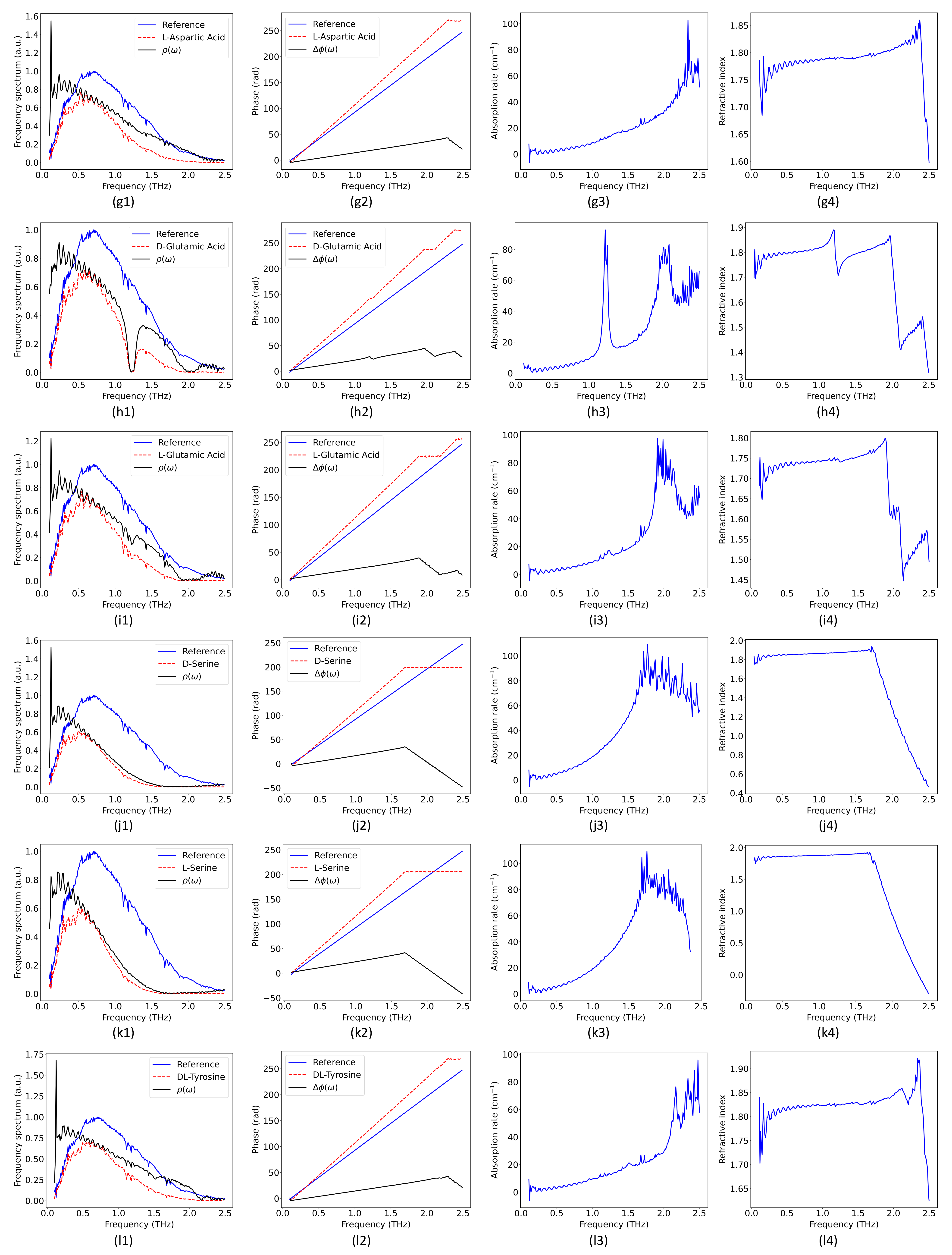
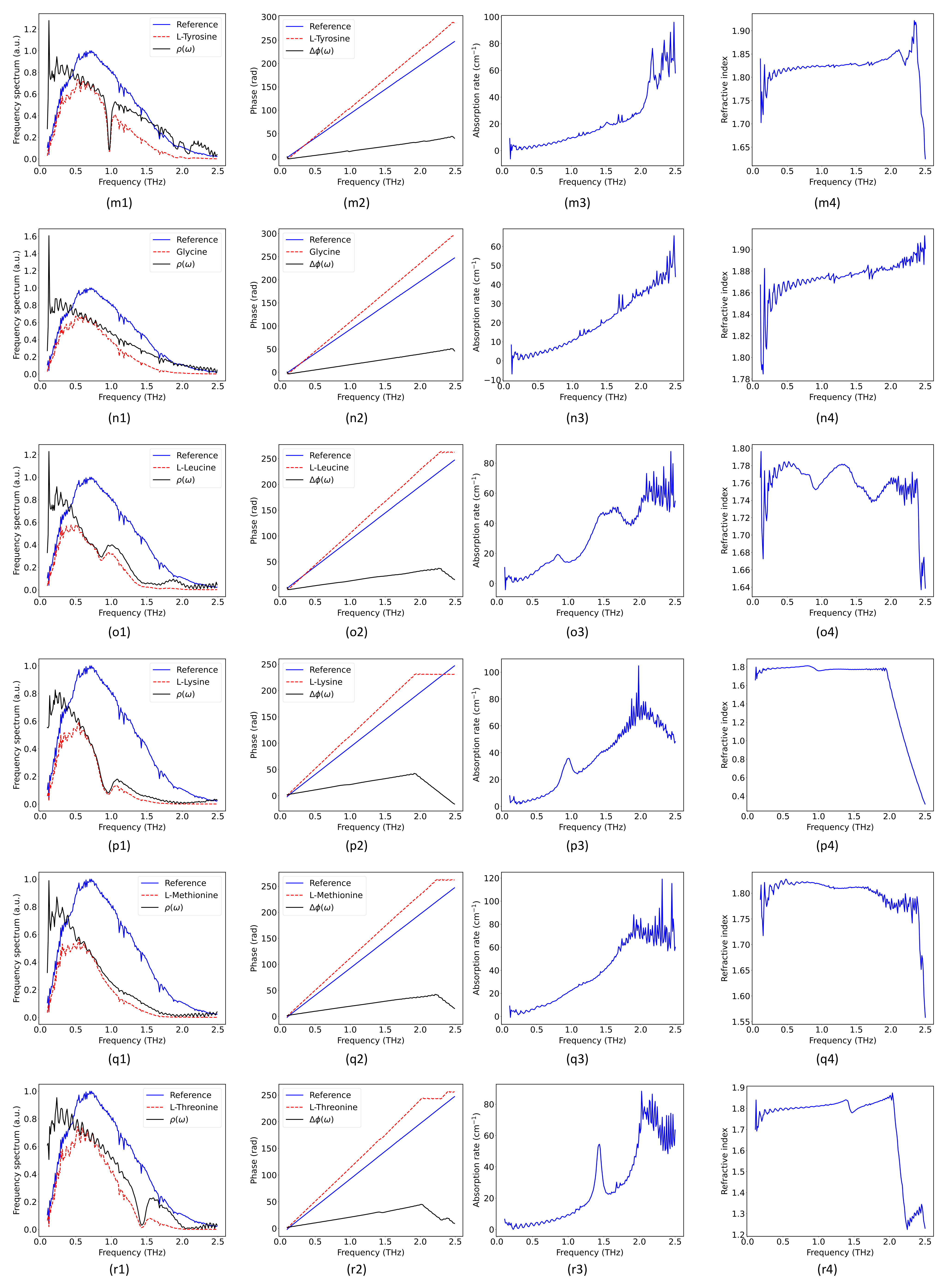
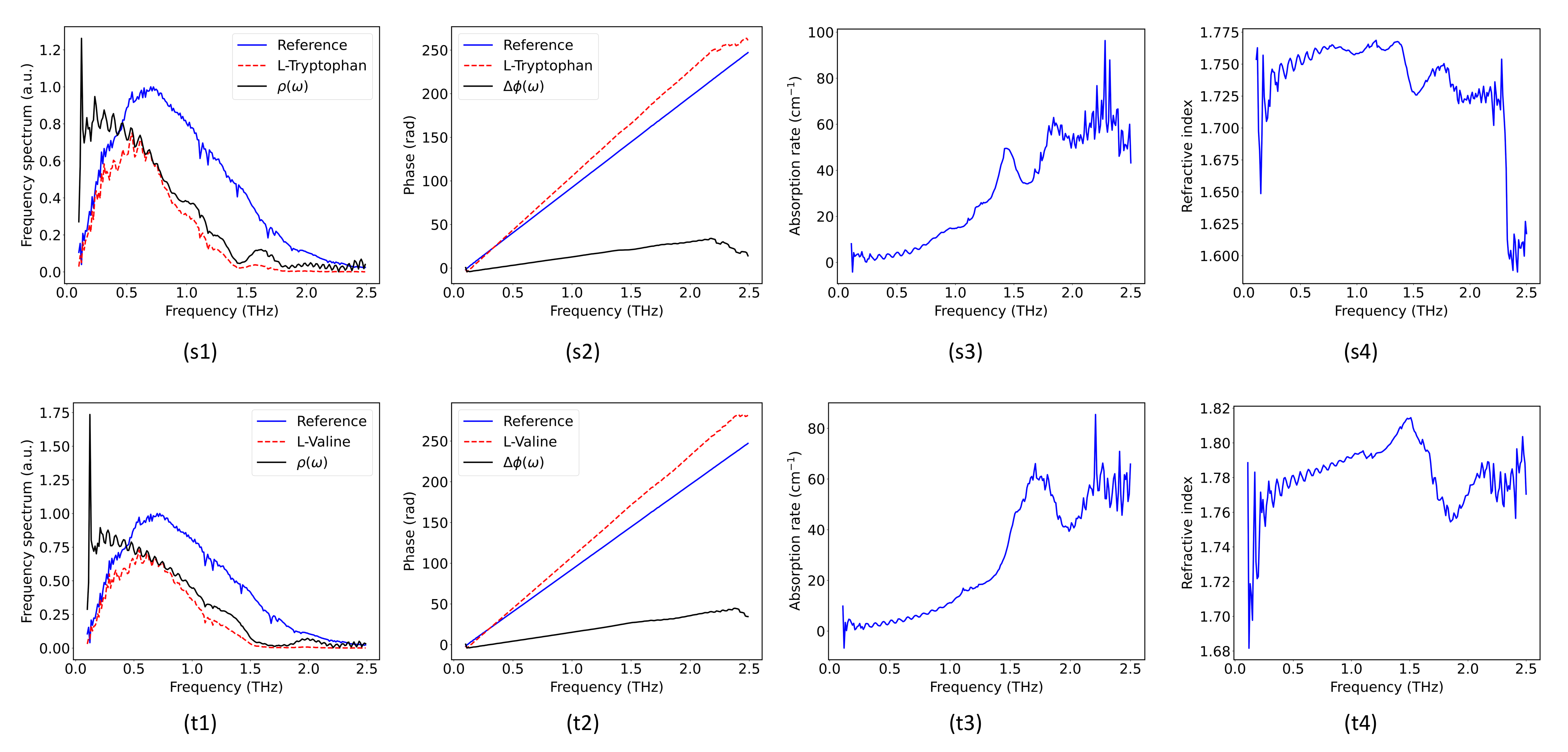
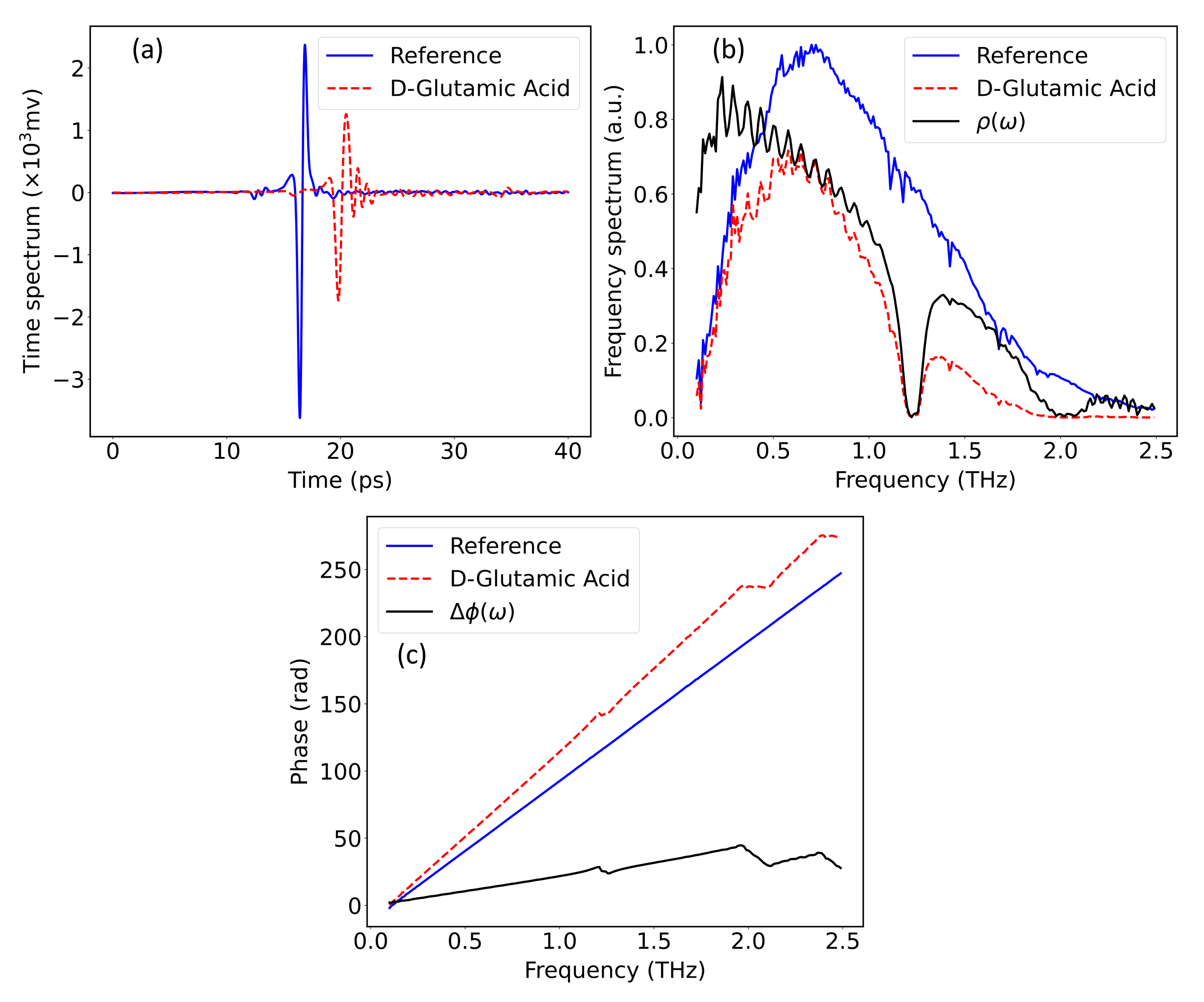
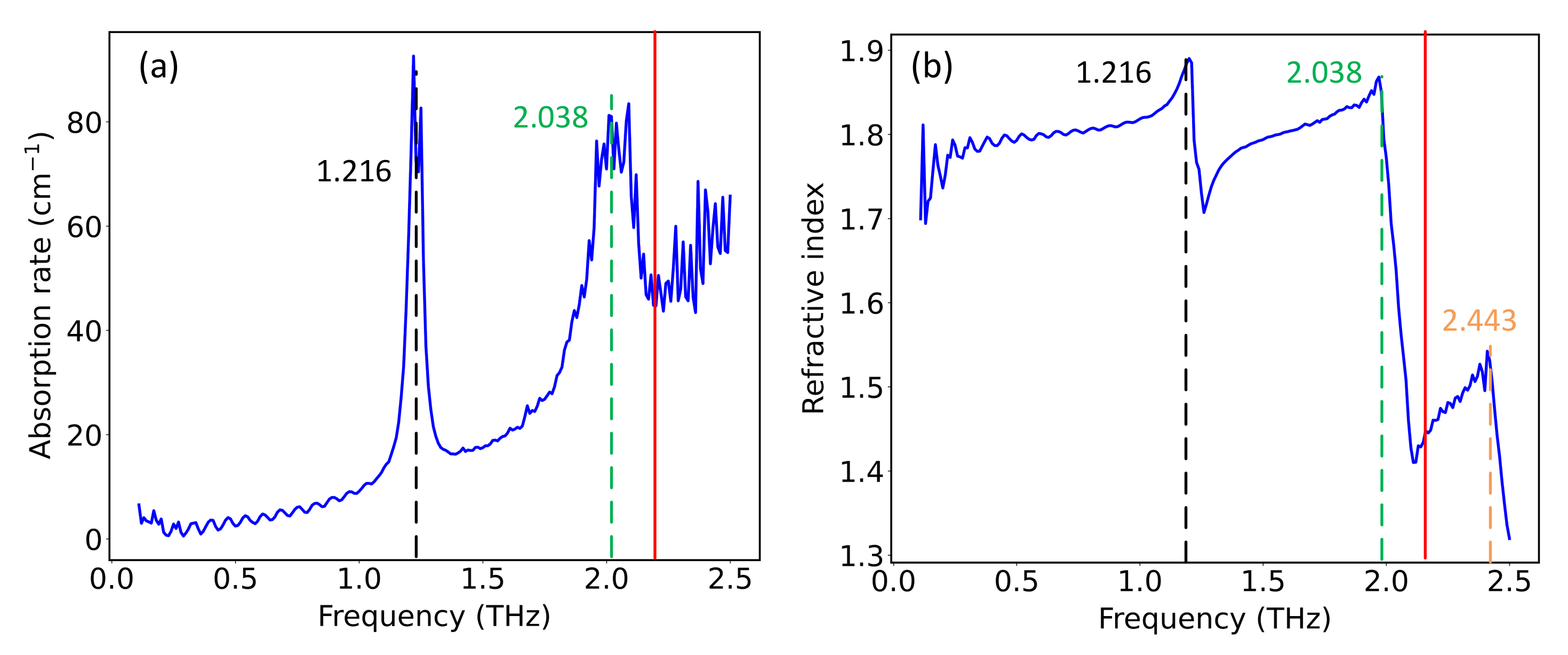
2.2. Hybrid Spectrum Combined with Absorption Rate and Refractive Index
| Amino Acid | Peaks from Literature (THz) | Measured Peaks (THz) |
|---|---|---|
| Beta-Alanine | - [27] | - |
| D-Alanine | 2.231 [27] | 2.226 |
| L-Alanine | 2.231 [27] | 2.227 |
| D-Arginine | 1.003/1.491 [27] | 0.99/1.435 |
| L-Arginine | 0.99/1.47 [28] | 1.002/1.508 |
| D-Aspartic acid | - [27] | - |
| L-Aspartic acid | - [27] | - |
| D-Glutamic acid | 1.234/2.031/2.443 [27] | 1.216/2.038/2.443 |
| L-Glutamic acid | 1.209/2.031 [27] | 1.235/1.967 |
| D-Serine | - [27] | - |
| L-Serine | - [27] | - |
| DL-Tyrosine | - [27] | - |
| L-Tyrosine | 0.951/1.929/2.083 [27] | 0.975/1.929/2.076 |
| Glycine | - [27] | - |
| L-Leucine | 0.85/1.46/1.7/2.17 [29] | 0.854/1.48/1.683/2.198 |
| L-Lysine | 0.9/2.07 [29] | 0.956/2.069 |
| L-Methionine | - [27] | - |
| L-Threonine | 1.42/2.13 [30] | 1.418/2.034 |
| L-Tryptophan | 1.44/1.851/2.289 [27] | 1.447/1.88/2.285 |
| L-Valine | 1.7/2.22 [29] | 1.678/2.236 |
2.3. Efficient Channel Attention Network
2.4. Training Details
3. Results
3.1. Metrics
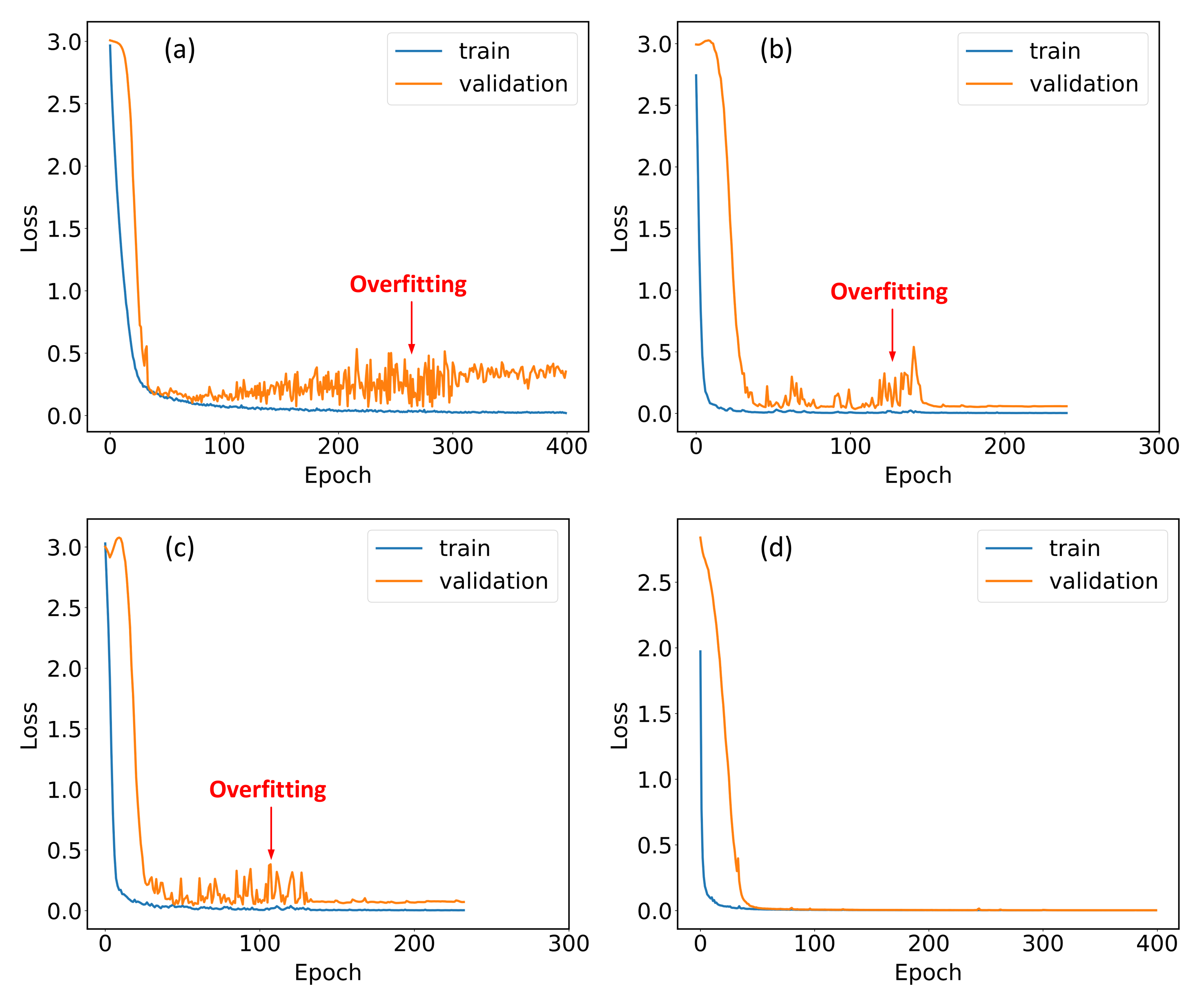
3.2. The Effects of Hybrid Spectrum and ECA Module
3.3. Compare with Other ECA-Based Networks
3.4. Compare with Other Methods for Amino-Acid Classification
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

References
- Plusquellic, D.F.; Siegrist, K.; Heilweil, E.J.; Esenturk, O. Applications of Terahertz Spectroscopy in Biosystems. ChemPhysChem 2007, 8, 2412–2431. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, T.; Iwata, T.; Araki, T.; Yasui, T. Improvement of minimum paint film thickness for THz paint meters by multiple-regression analysis. Appl. Opt. 2007, 46, 7518–7526. [Google Scholar] [CrossRef] [PubMed]
- Mittleman, D.M.; Gupta, M.; Neelamani, R.; Baraniuk, R.G.; Rudd, J.V.; Koch, M. Recent advances in terahertz imaging. Appl. Phys. B 1999, 68, 1085–1094. [Google Scholar] [CrossRef]
- Wang, B.; Meng, K.; Song, T.; Li, Z. Qualitative detection of amino acids in a mixture with terahertz spectroscopic imaging. J. Opt. Soc. Am. B 2022, 39, A18–A24. [Google Scholar] [CrossRef]
- Zhang, L.; Zhong, H.; Deng, C.; Zhang, C.; Zhao, Y. Terahertz wave reference-free phase imaging for identification of explosives. Appl. Phys. Lett. 2008, 92, 091117. [Google Scholar] [CrossRef]
- Yang, X.; Zhao, X.; Yang, K.; Liu, Y.; Liu, Y.; Fu, W.; Luo, Y. Biomedical Applications of Terahertz Spectroscopy and Imaging. Trends Biotechnol. 2016, 34, 810–824. [Google Scholar] [CrossRef]
- Leitner, D.M.; Gruebele, M.; Havenith, M. Solvation dynamics of biomolecules: Modeling and terahertz experiments. HFSP J. 2008, 2, 314–323. [Google Scholar] [CrossRef] [Green Version]
- Serita, K.; Matsuda, E.; Okada, K.; Murakami, H.; Kawayama, I.; Tonouchi, M. Invited Article: Terahertz microfluidic chips sensitivity-enhanced with a few arrays of meta-atoms. APL Photonics 2018, 3, 051603. [Google Scholar] [CrossRef] [Green Version]
- Vang, Z.P.; Reyes, A.; Sonstrom, R.E.; Holdren, M.S.; Sloane, S.E.; Alansari, I.Y.; Neill, J.L.; Pate, B.H.; Clark, J.R. Copper-Catalyzed Transfer Hydrodeuteration of Aryl Alkenes with Quantitative Isotopomer Purity Analysis by Molecular Rotational Resonance Spectroscopy. J. Am. Chem. Soc. 2021, 143, 7707–7718. [Google Scholar] [CrossRef]
- Liu, H.; Fan, Y.X.; Li, L.; Chen, H.G.; Wang, P.F.; Tao, Z.Y. Self-adaptive terahertz spectroscopy from atmospheric vapor based on Hilbert-Huang transform. Opt. Express 2018, 26, 27279–27293. [Google Scholar] [CrossRef]
- Huang, Y.; Sun, P.; Zhang, Z.; Jin, C. Numerical method based on transfer function for eliminating water vapor noise from terahertz spectra. Appl. Opt. 2017, 56, 5698–5704. [Google Scholar] [CrossRef]
- Naftaly, M.; Miles, R. A method for removing etalon oscillations from THz time-domain spectra. Opt. Commun. 2007, 280, 291–295. [Google Scholar] [CrossRef]
- King, M.D.; Buchanan, W.D.; Korter, T.M. Identification and quantification of polymorphism in the pharmaceutical compound diclofenac acid by terahertz spectroscopy and solid-state density functional theory. Anal. Chem. 2011, 8310, 3786–3792. [Google Scholar] [CrossRef]
- Burnett, A.D.; Fan, W.; Upadhya, P.C.; Cunningham, J.E.; Hargreaves, M.D.; Munshi, T.; Edwards, H.G.M.; Linfield, E.H.; Davies, A.G. Broadband terahertz time-domain spectroscopy of drugs-of-abuse and the use of principal component analysis. Analyst 2009, 134, 1658–1668. [Google Scholar] [CrossRef] [PubMed]
- Wentao, Z.; Yuewen, L.; Pingping, Z.; Xianming, X. Recognition of transgenic soybean oil based on terahertz time-domain spectroscopy and PCA-SVM. Infrared Laser Eng. 2017, 46, 1125004. [Google Scholar] [CrossRef]
- Pohl, A.; Deßmann, N.; Dutzi, K.; Hübers, H.W. Identification of Unknown Substances by Terahertz Spectroscopy and Multivariate Data Analysis. J. Infrared 2016, 37, 175–188. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Wang, Q.; Li, L. PLS model investigation of thiabendazole based on THz spectrum. J. Quant. Spectrosc. Radiat. Transf. 2013, 117, 7–14. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, Z.; Zhu, R.; Xiang, Y.; Yang, Y.; Harrington, P.B. Application of terahertz time-domain spectroscopy combined with chemometrics to quantitative analysis of imidacloprid in rice samples. J. Quant. Spectrosc. Radiat. Transf. 2015, 167, 1–9. [Google Scholar] [CrossRef]
- Li, Z. Wavelength Selection for Quantitative Analysis in Terahertz Spectroscopy Using a Genetic Algorithm. IEEE Trans. Terahertz Sci. Technol. 2016, 6, 658–663. [Google Scholar] [CrossRef]
- Li, J.Y.; Sun, P.; Zou, Y.; Liu, W.; Wang, W.A. Comparison and Analysis of Iterative and Genetic Algorithms Used to Extract the Optical Parameters of Glucose Polycrystalline. Spectrosc. Spectr. Anal. 2016, 36, 3875. [Google Scholar] [CrossRef]
- Li, Z.; Guan, A.; Ge, H.; Lian, F. Wavelength selection of amino acid THz absorption spectra for quantitative analysis by a self-adaptive genetic algorithm and comparison with mwPLS. Microchem. J. 2017, 132, 185–189. [Google Scholar] [CrossRef]
- Hu, Q.f.; Cai, J. Research of Terahertz Time-Domain Spectral Identification Based on Deep Learning. Spectrosc. Spectr. Anal. 2021, 41, 94–99. [Google Scholar]
- Li, T.; Xu, Y.; Luo, J.; He, J.; Lin, S. A Method of Amino Acid Terahertz Spectrum Recognition Based on the Convolutional Neural Network and Bidirectional Gated Recurrent Network Model. Sci. Program. 2021, 2021, 2097257. [Google Scholar] [CrossRef]
- Wang, Q.; Wu, B.; Zhu, P.; Li, P.; Zuo, W.; Hu, Q. ECA-Net: Efficient Channel Attention for Deep Convolutional Neural Networks. In Proceedings of the The IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 14–19 June 2020. [Google Scholar]
- Yamamoto, K.; Masui, A.; Ishida, H. Kramers–Kronig analysis of infrared reflection spectra with perpendicular polarization. Appl. Opt. 1994, 33, 6285–6293. [Google Scholar] [CrossRef]
- Chen, Y.; Huang, S.; Pickwell-MacPherson, E. Frequency-wavelet domain deconvolution for terahertz reflection imaging and spectroscopy. Opt. Express 2010, 18, 1177–1190. [Google Scholar] [CrossRef]
- THz Database. Available online: http://www.thzdb.org/index.php?s=0 (accessed on 4 May 2022).
- Wang, W.N.; Li, Y.B.; Yue, W.W. Vibrational spectrum of histidine and arginine in THz range. Acta Phys. Sin. 2007, 56, 781–785. [Google Scholar] [CrossRef]
- Yu, Q.; Jiang, L.; Zhang, L. Research Progress of THz Spectroscopy of Amino Acids. Infrared Technol. 2018, 40, 220–226. [Google Scholar]
- Li, W.; Yan, F.; Wang, Z.C. Quantum Chemical Identification of Terahertz Absorption Peaks of Threonine with Different Molecular Configurations. Spectrosc. Spectr. Anal. 2020, 40, 2054–2058. [Google Scholar]
- Tsinalis, O.; Matthews, P.M.; Guo, Y.; Zafeiriou, S. Automatic sleep stage scoring with single-channel EEG using convolutional neural networks. arXiv 2016, arXiv:1610.01683. [Google Scholar]
- Liu, C.; Belkin, M. Accelerating SGD with momentum for over-parameterized learning. arXiv 2018, arXiv:1810.13395. [Google Scholar]
- Du, W.; Rao, N.; Dong, C.; Wang, Y.; Hu, D.; Zhu, L.; Zeng, B.; Gan, T. Automatic classification of esophageal disease in gastroscopic images using an efficient channel attention deep dense convolutional neural network. Biomed. Opt. Express 2021, 12, 3066–3081. [Google Scholar] [CrossRef] [PubMed]
- Lecun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef] [Green Version]
- Ferguson, B.; Zhang, X.C. Materials for terahertz science and technology. Nat. Mater. 2002, 1, 26–33. [Google Scholar] [CrossRef] [PubMed]


| 20 | Beta-Ala | D-Ala | L-Ala | D-Arg | L-Arg | D-Asp | L-Asp | D-Glu | L-Glu | D-Ser | L-Ser | DL-Tyr | L-Tyr | Gly | L-Leu | L-Tys | L-Met | L-Thr | L-Try | L-Val | Acc |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a | 99.2 | 100 | 99.8 | 99.6 | 100 | 99.6 | 100 | 100 | 99.8 | 80.8 | 97.8 | 99.4 | 100 | 99.8 | 100 | 99.8 | 100 | 99.6 | 100 | 99.8 | 98.7 |
| b | 100 | 99.6 | 100 | 90.6 | 99.3 | 100 | 99.6 | 100 | 99.8 | 54.9 | 95.4 | 100 | 99.8 | 100 | 100 | 99.8 | 91.3 | 100 | 99.8 | 99.1 | 96.5 |
| c | 99.8 | 100 | 99.8 | 100 | 99.8 | 99.8 | 100 | 100 | 99.8 | 98.0 | 98.3 | 99.1 | 99.6 | 100 | 100 | 99.8 | 100 | 100 | 100 | 99.8 | 99.7 |
| d | 98.9 | 98.7 | 99.8 | 84.1 | 100 | 51.9 | 49.4 | 100 | 97.8 | 99.6 | 98.1 | 97.6 | 100 | 100 | 98.5 | 99.6 | 99.8 | 98.1 | 100 | 100 | 88.6 |
| e | 99.6 | 96.5 | 89.9 | 99.1 | 99.4 | 91.7 | 97.2 | 99.8 | 99.8 | 99.4 | 99.6 | 98.9 | 98.3 | 98.7 | 100 | 99.8 | 98.9 | 98.2 | 97.4 | 98.5 | 93.1 |
| f | 99.6 | 94.3 | 92.3 | 99.3 | 99.1 | 93.2 | 79.7 | 99.6 | 99.6 | 99.8 | 98.9 | 98.7 | 97.8 | 99.6 | 100 | 99.8 | 98.5 | 98.2 | 97.4 | 98.7 | 92.2 |
| g | 75.9 | 90.2 | 83.3 | 95.9 | 94.5 | 87.5 | 84.3 | 98.2 | 97.6 | 53.2 | 79.2 | 91.9 | 96.9 | 82.0 | 94.7 | 89.9 | 90.0 | 96.9 | 93.7 | 91.0 | 87.4 |
| h | 99.8 | 99.8 | 99.4 | 100 | 100 | 98.5 | 99.8 | 100 | 100 | 99.3 | 99.3 | 99.1 | 100 | 99.1 | 100 | 100 | 100 | 100 | 99.6 | 99.8 | 99.7 |
| a | 98.5 | 99.4 | 98.0 | 99.3 | 99.5 | 99.1 | 99.2 | 99.4 | 99.4 | 65.7 | 96.3 | 98.7 | 99.8 | 98.9 | 99.9 | 99.2 | 100 | 98.9 | 98.0 | 98.9 | 97.3 |
| b | 100 | 98.6 | 98.3 | 77.1 | 94.5 | 99.9 | 98.1 | 100 | 99.6 | 57.1 | 95.5 | 99.2 | 99.6 | 100 | 100 | 100 | 77.2 | 99.9 | 96.1 | 97.3 | 93.6 |
| c | 99.6 | 99.6 | 99.0 | 99.4 | 99.2 | 99.6 | 99.7 | 100 | 99.4 | 92.2 | 96.5 | 97.8 | 99.4 | 99.8 | 99.6 | 99.9 | 100 | 100 | 98.4 | 98.4 | 98.9 |
| d | 88.3 | 97.3 | 97.2 | 63.7 | 99.2 | 54.8 | 67.0 | 100 | 61.5 | 88.7 | 52.0 | 64.2 | 99.9 | 99.4 | 80.3 | 96.4 | 98.8 | 67.2 | 99.1 | 99.5 | 73.0 |
| e | 96.0 | 93.0 | 74.4 | 93.9 | 99.2 | 78.5 | 74.5 | 99.2 | 98.8 | 98.9 | 97.8 | 92.2 | 96.8 | 96.0 | 99.2 | 98.3 | 94.9 | 97.9 | 95.8 | 97.1 | 88.7 |
| f | 90.6 | 88.2 | 82.8 | 97.2 | 98.5 | 61.9 | 34.7 | 98.6 | 97.5 | 95.7 | 60.5 | 90.6 | 96.0 | 97.8 | 98.5 | 96.3 | 93.9 | 95.1 | 95.4 | 96.2 | 83.5 |
| g | 59.7 | 79.5 | 71.3 | 87.6 | 88.6 | 78.8 | 68.9 | 89.9 | 88.5 | 48.8 | 61.3 | 79.3 | 92.7 | 68.2 | 81.4 | 67.9 | 82.9 | 90.4 | 83.9 | 83.7 | 76.2 |
| h | 99.8 | 99.3 | 98.1 | 100 | 100 | 97.9 | 99.6 | 100 | 100 | 98.1 | 99.0 | 98.3 | 100 | 98.0 | 100 | 100 | 100 | 100 | 99.3 | 99.2 | 99.3 |
| ours | 99.5 | 100 | 98.6 | 100 | 99.4 | 99.7 | 99.7 | 100 | 99.6 | 90.3 | 99.1 | 99.5 | 99.8 | 99.7 | 99.9 | 100 | 100 | 99.8 | 99.6 | 99.7 | 99.2 |
| Depth | #.Params | FLOPs | Training Time | Test Rate | |
|---|---|---|---|---|---|
| ECA-DCNN [33] | 125 | 7.33 M | 421.42 M | 21.60 min | 943.93 fps |
| ECA-Resnet50 [24] | 67 | 24.14 M | 588.23 M | 11.56 min | 1134.35 fps |
| ECA-Resnet101 [24] | 134 | 43.21 M | 675.45 M | 19.74 min | 675.45 fps |
| ECA network (ours) | 4 | 4.72 M | 64.61 M | 3.67 min | 3782.46 fps |
| CNN-BiGRU [23] | - | 3.23 M | 6.31 M | 169.63 min | 84.00 fps |
| CNN [22] | - | 0.93 M | 103.07 M | 11.4 min | 1887.13 fps |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, B.; Qin, X.; Meng, K.; Zhu, L.; Li, Z. Classification of Amino Acids Using Hybrid Terahertz Spectrum and an Efficient Channel Attention Convolutional Neural Network. Nanomaterials 2022, 12, 2114. https://doi.org/10.3390/nano12122114
Wang B, Qin X, Meng K, Zhu L, Li Z. Classification of Amino Acids Using Hybrid Terahertz Spectrum and an Efficient Channel Attention Convolutional Neural Network. Nanomaterials. 2022; 12(12):2114. https://doi.org/10.3390/nano12122114
Chicago/Turabian StyleWang, Bo, Xiaoling Qin, Kun Meng, Liguo Zhu, and Zeren Li. 2022. "Classification of Amino Acids Using Hybrid Terahertz Spectrum and an Efficient Channel Attention Convolutional Neural Network" Nanomaterials 12, no. 12: 2114. https://doi.org/10.3390/nano12122114






