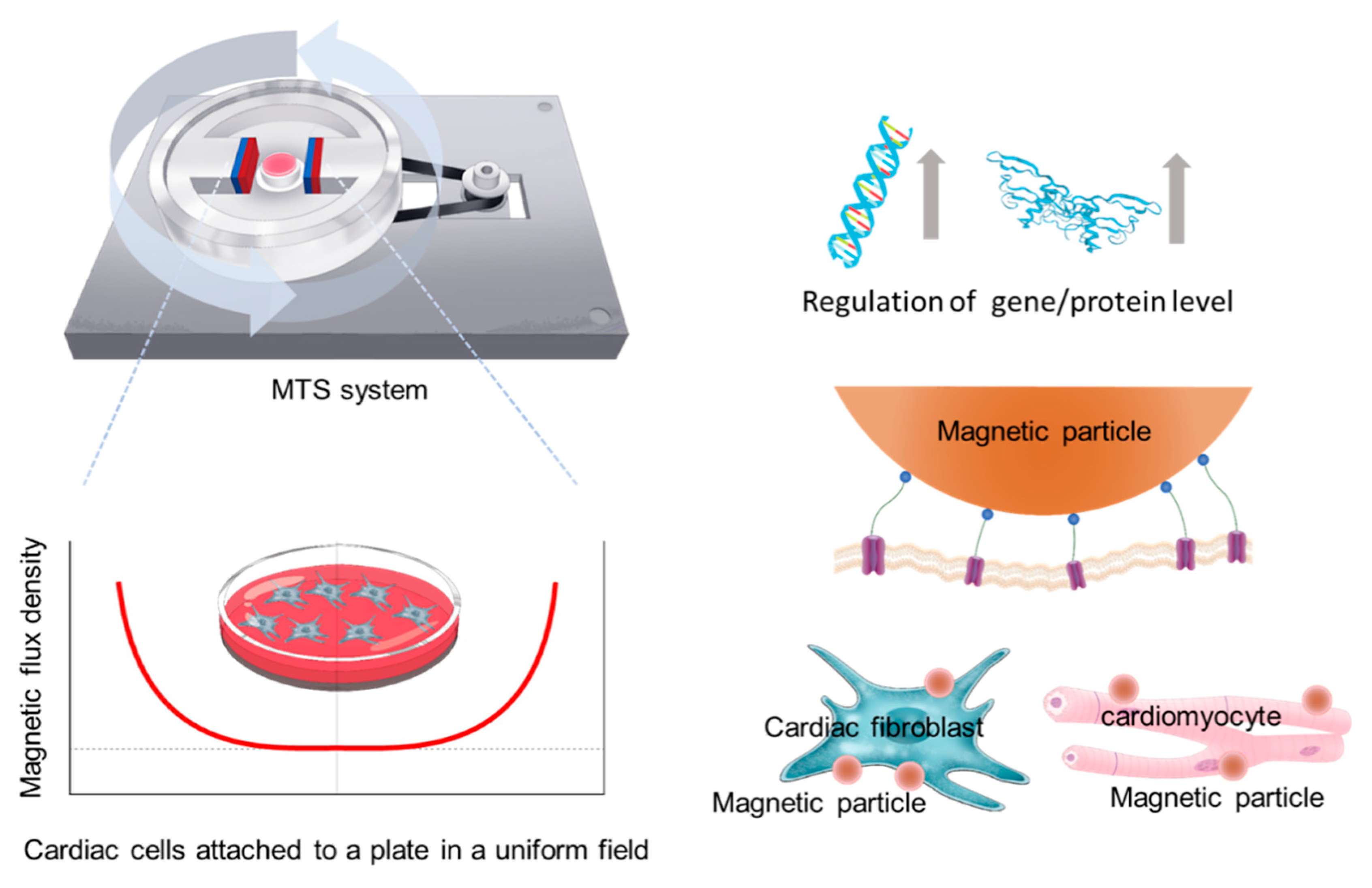
Development of Magnetic Torque Stimulation (MTS) Utilizing Rotating Uniform Magnetic Field for Mechanical Activation of Cardiac Cells
Abstract
:1. Introduction
2. Materials and Methods
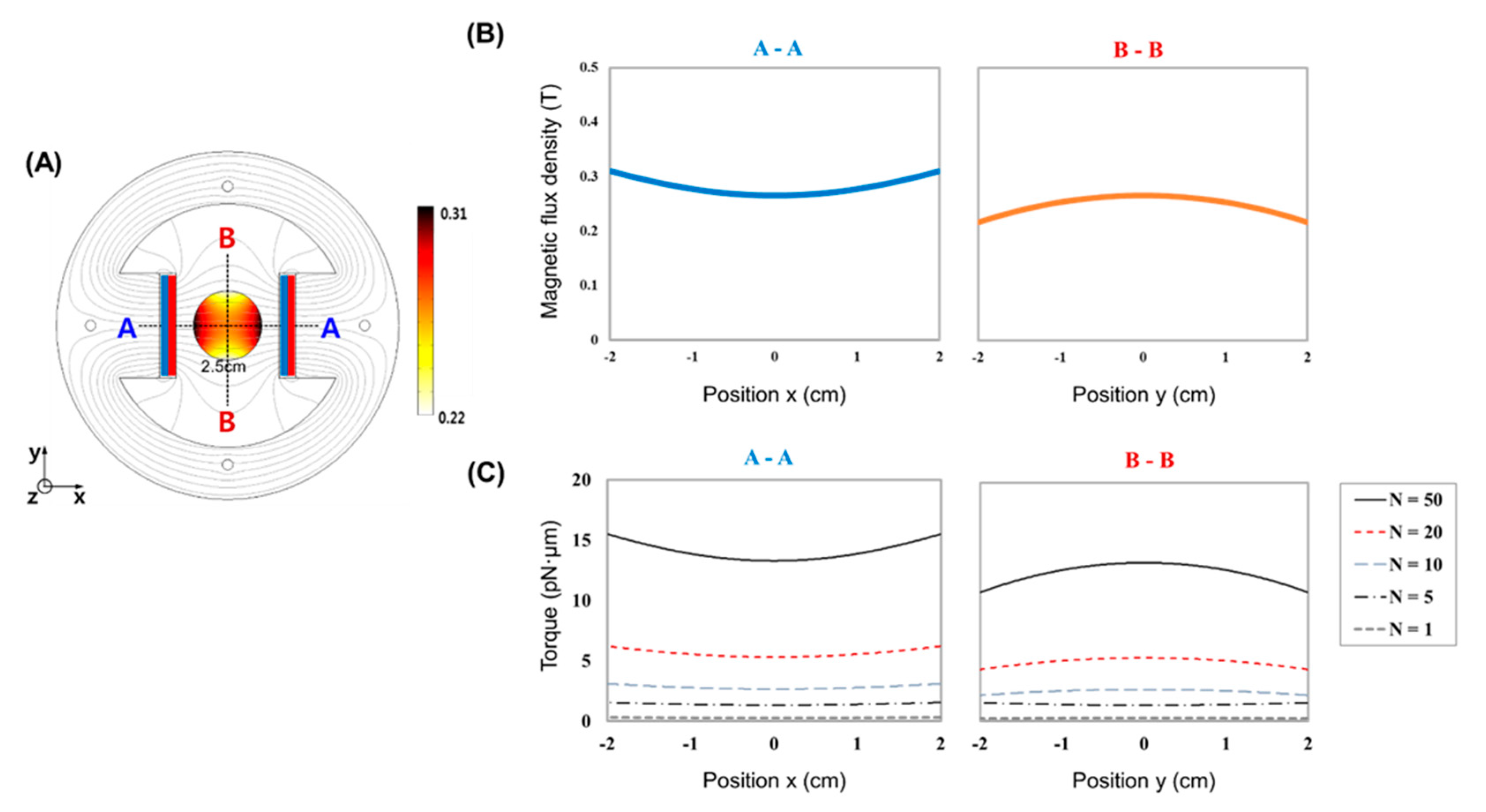
2.1. MTS Assembly
2.2. Cell Culture
2.3. WGA-Coated MPs and MTS Stimulation
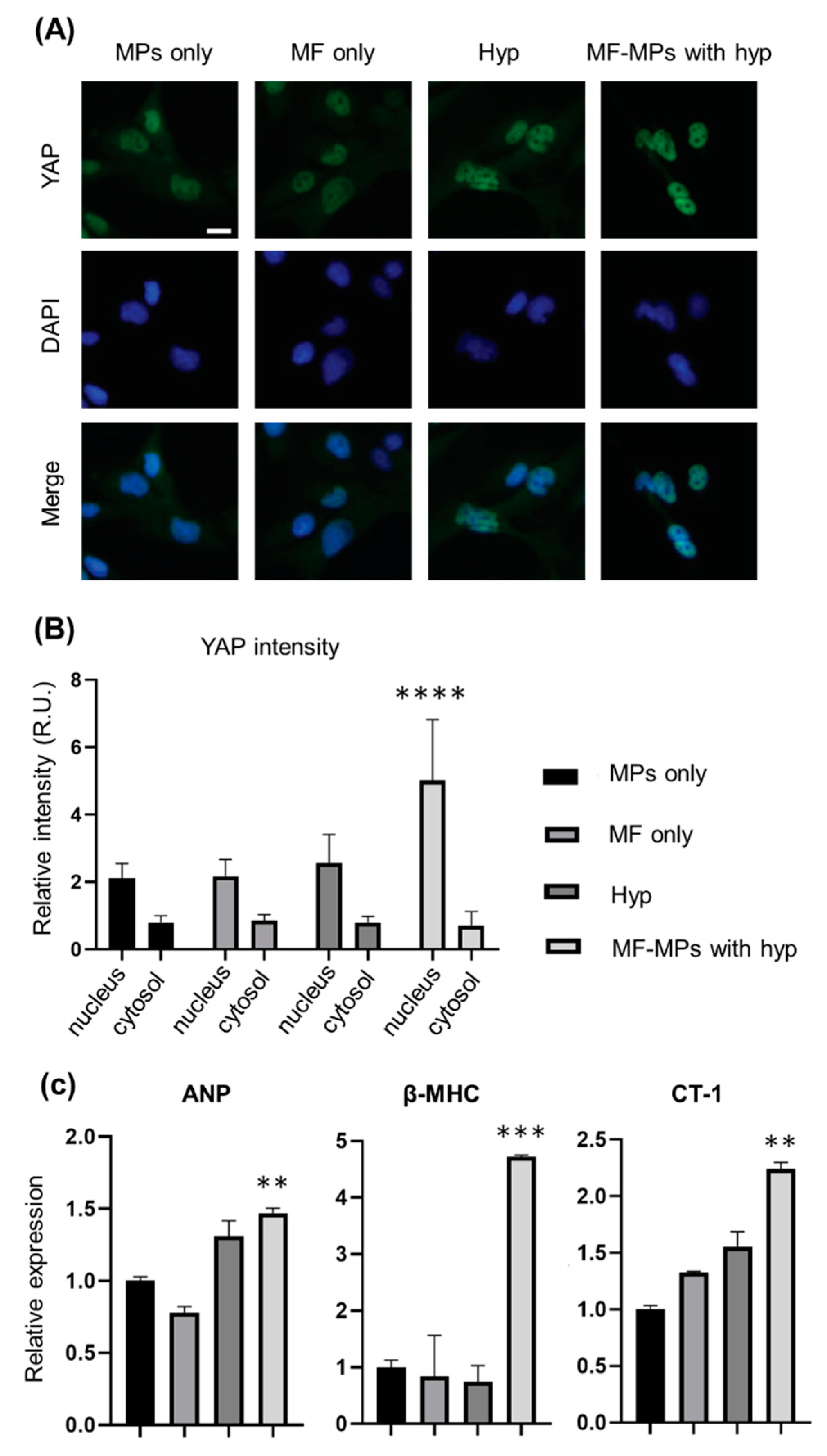
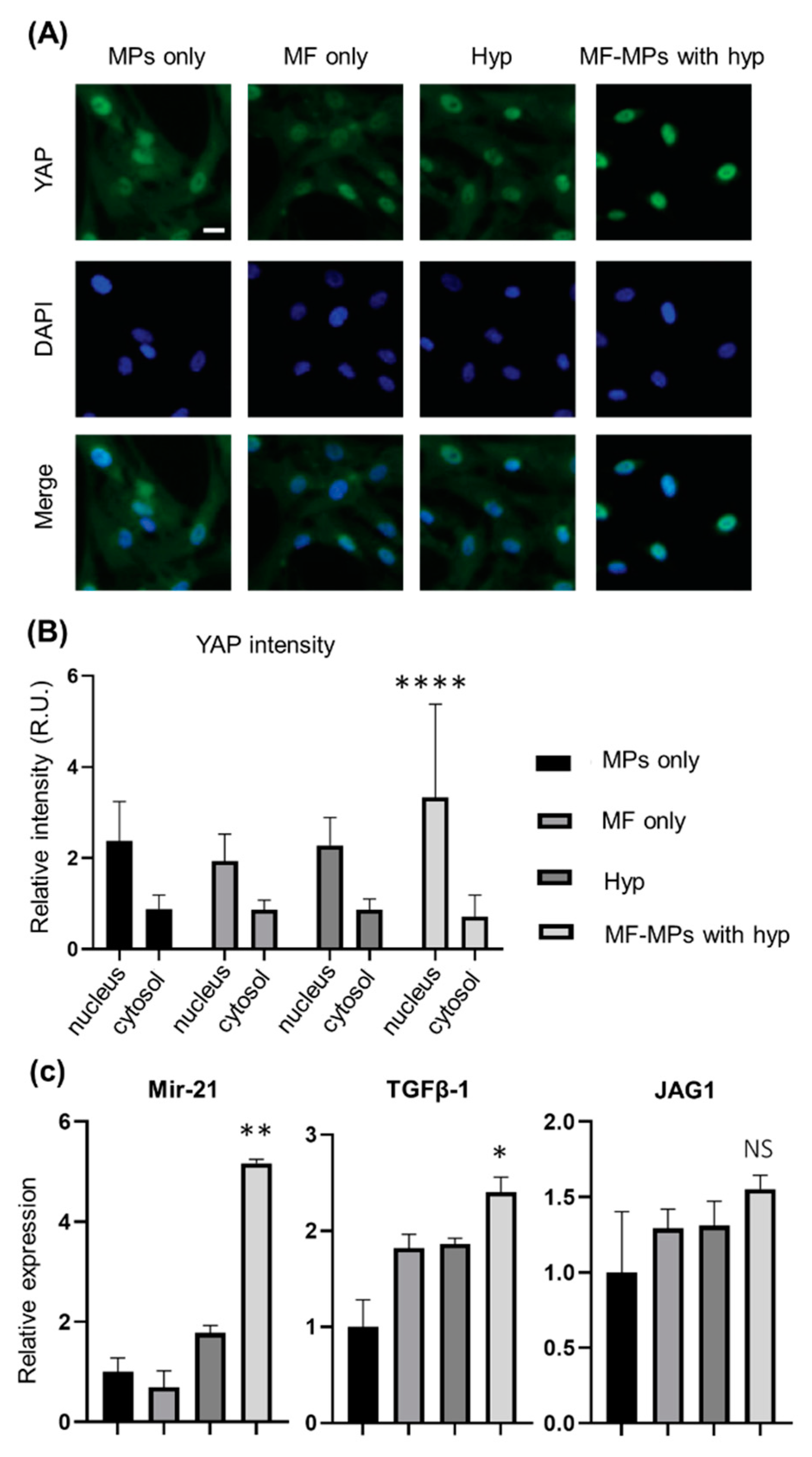
2.4. Immunofluorescence Assay
2.5. RNA Isolation and RT-PCR Analysis
2.6. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Happe, C.L.; Engler, A.J. Mechanical Forces Reshape Differentiation Cues That Guide Cardiomyogenesis. Circ. Res. 2016, 118, 296–310. [Google Scholar] [CrossRef] [Green Version]
- Janmey, P.A.; Miller, R.T. Mechanisms of mechanical signaling in development and disease. J. Cell Sci. 2011, 124, 9–18. [Google Scholar] [CrossRef] [Green Version]
- Ladoux, B.; Mege, R.M. Mechanobiology of collective cell behaviours. Nat. Rev. Mol. Cell Biol. 2017, 18, 743–757. [Google Scholar] [CrossRef]
- Lepore, D.; De Santis, R.; Pagliara, M.M.; Gloria, A.; Oliviero, O.; Nucci, C.; Improta, G.; Triassi, M.; Ambrosio, L. Effect of topical antiinflammatory drugs on mechanical behavior of rabbit cornea. J. Appl. Biomater. Funct. Mater. 2017, 15, e142–e148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martino, F.; Perestrelo, A.R.; Vinarsky, V.; Pagliari, S.; Forte, G. Cellular Mechanotransduction: From Tension to Function. Front. Physiol. 2018, 9, 824. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.S. Mechanotransduction—A field pulling together? J. Cell Sci. 2008, 121, 3285–3292. [Google Scholar] [CrossRef] [Green Version]
- Trepat, X.; Wasserman, M.R.; Angelini, T.E.; Millet, E.; Weitz, D.A.; Butler, J.P.; Fredberg, J.J. Physical forces during collective cell migration. Nat. Phys. 2009, 5, 426–430. [Google Scholar] [CrossRef] [Green Version]
- Rief, M.; Oesterhelt, F.; Heymann, B.; Gaub, H.E. Single molecule force spectroscopy on polysaccharides by atomic force microscopy. Science 1997, 275, 1295–1297. [Google Scholar] [CrossRef] [Green Version]
- Wen, J.D.; Lancaster, L.; Hodges, C.; Zeri, A.C.; Yoshimura, S.H.; Noller, H.F.; Bustamante, C.; Tinoco, I. Following translation by single ribosomes one codon at a time. Nature 2008, 452, 598–603. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Weitemier, A.Z.; Zeng, X.; He, L.; Wang, X.; Tao, Y.; Huang, A.J.Y.; Hashimotodani, Y.; Kano, M.; Iwasaki, H.; et al. Near-infrared deep brain stimulation via upconversion nanoparticle-mediated optogenetics. Science 2018, 359, 679–684. [Google Scholar] [CrossRef] [Green Version]
- Grossman, N.; Bono, D.; Dedic, N.; Kodandaramaiah, S.B.; Rudenko, A.; Suk, H.J.; Cassara, A.M.; Neufeld, E.; Kuster, N.; Tsai, L.H.; et al. Noninvasive Deep Brain Stimulation via Temporally Interfering Electric Fields. Cell 2017, 169, 1029–1041.e16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kringelbach, M.L.; Jenkinson, N.; Owen, S.L.; Aziz, T.Z. Translational principles of deep brain stimulation. Nat. Rev. Neurosci. 2007, 8, 623–635. [Google Scholar] [CrossRef] [PubMed]
- Yizhar, O.; Fenno, L.E.; Davidson, T.J.; Mogri, M.; Deisseroth, K. Optogenetics in neural systems. Neuron 2011, 71, 9–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stanley, S.A.; Gagner, J.E.; Damanpour, S.; Yoshida, M.; Dordick, J.S.; Friedman, J.M. Radio-wave heating of iron oxide nanoparticles can regulate plasma glucose in mice. Science 2012, 336, 604–608. [Google Scholar] [CrossRef] [Green Version]
- Stanley, S.A.; Sauer, J.; Kane, R.S.; Dordick, J.S.; Friedman, J.M. Remote regulation of glucose homeostasis in mice using genetically encoded nanoparticles. Nat. Med. 2015, 21, 92–98. [Google Scholar] [CrossRef] [Green Version]
- Wheeler, M.A.; Smith, C.J.; Ottolini, M.; Barker, B.S.; Purohit, A.M.; Grippo, R.M.; Gaykema, R.P.; Spano, A.J.; Beenhakker, M.P.; Kucenas, S.; et al. Genetically targeted magnetic control of the nervous system. Nat. Neurosci. 2016, 19, 756–761. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.; Romero, G.; Christiansen, M.G.; Mohr, A.; Anikeeva, P. Wireless magnetothermal deep brain stimulation. Science 2015, 347, 1477–1480. [Google Scholar] [CrossRef] [Green Version]
- Nimpf, S.; Keays, D.A. Is magnetogenetics the new optogenetics? EMBO J. 2017, 36, 1643–1646. [Google Scholar] [CrossRef]
- Marmugi, L.; Renzoni, F. Optical Magnetic Induction Tomography of the Heart. Sci. Rep. 2016, 6, 23962. [Google Scholar] [CrossRef] [Green Version]
- Karbassi, E.; Fenix, A.; Marchiano, S.; Muraoka, N.; Nakamura, K.; Yang, X.; Murry, C.E. Cardiomyocyte maturation: Advances in knowledge and implications for regenerative medicine. Nat. Rev. Cardiol. 2020, 17, 341–359. [Google Scholar] [CrossRef]
- Du, V.; Luciani, N.; Richard, S.; Mary, G.; Gay, C.; Mazuel, F.; Reffay, M.; Menasche, P.; Agbulut, O.; Wilhelm, C. A 3D magnetic tissue stretcher for remote mechanical control of embryonic stem cell differentiation. Nat. Commun. 2017, 8, 400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, S.; Zhang, X.; Zhan, C.; Wu, J.; Xu, J.; Cheung, J. Measuring single cardiac myocyte contractile force via moving a magnetic bead. Biophys. J. 2005, 88, 1489–1495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, T.H.; Kang, S.; Park, S.; Choi, J.S.; Kim, P.K.; Cheon, J. A magnetic resonance tuning sensor for the MRI detection of biological targets. Nat. Protoc. 2018, 13, 2664–2684. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.W.; Jeong, H.K.; Southard, K.M.; Jun, Y.W.; Cheon, J. Magnetic Nanotweezers for Interrogating Biological Processes in Space and Time. Acc. Chem. Res. 2018, 51, 839–849. [Google Scholar] [CrossRef]
- Russo, T.; Peluso, V.; Gloria, A.; Oliviero, O.; Rinaldi, L.; Improta, G.; De Santis, R.; D’Anto, V. Combination Design of Time-Dependent Magnetic Field and Magnetic Nanocomposites to Guide Cell Behavior. Nanomaterials 2020, 10, 577. [Google Scholar] [CrossRef] [Green Version]
- Vecchione, R.; Quagliariello, V.; Giustetto, P.; Calabria, D.; Sathya, A.; Marotta, R.; Profeta, M.; Nitti, S.; Silvestri, N.; Pellegrino, T.; et al. Oil/water nano-emulsion loaded with cobalt ferrite oxide nanocubes for photo-acoustic and magnetic resonance dual imaging in cancer: In vitro and preclinical studies. Nanomedicine 2017, 13, 275–286. [Google Scholar] [CrossRef]
- Iaccarino, G.; Profeta, M.; Vecchione, R.; Netti, P.A. Matrix metalloproteinase-cleavable nanocapsules for tumor-activated drug release. Acta Biomater. 2019, 89, 265–278. [Google Scholar] [CrossRef]
- Herum, K.M.; Choppe, J.; Kumar, A.; Engler, A.J.; McCulloch, A.D. Mechanical regulation of cardiac fibroblast profibrotic phenotypes. Mol. Biol. Cell 2017, 28, 1871–1882. [Google Scholar] [CrossRef]
- MacKenna, D.; Summerour, S.R.; Villarreal, F.J. Role of mechanical factors in modulating cardiac fibroblast function and extracellular matrix synthesis. Cardiovasc. Res. 2000, 46, 257–263. [Google Scholar] [CrossRef] [Green Version]
- Stoppel, W.L.; Kaplan, D.L.; Black, L.D., 3rd. Electrical and mechanical stimulation of cardiac cells and tissue constructs. Adv. Drug Deliv. Rev. 2016, 96, 135–155. [Google Scholar] [CrossRef] [Green Version]
- Saucerman, J.J.; Tan, P.M.; Buchholz, K.S.; McCulloch, A.D.; Omens, J.H. Mechanical regulation of gene expression in cardiac myocytes and fibroblasts. Nat. Rev. Cardiol. 2019, 16, 361–378. [Google Scholar] [CrossRef] [PubMed]
- Curtis, M.W.; Russell, B. Micromechanical regulation in cardiac myocytes and fibroblasts: Implications for tissue remodeling. Pflügers Arch. Eur. J. Physiol. 2011, 462, 105–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LaBarge, W.; Mattappally, S.; Kannappan, R.; Fast, V.G.; Pretorius, D.; Berry, J.L.; Zhang, J. Maturation of three-dimensional, hiPSC-derived cardiomyocyte spheroids utilizing cyclic, uniaxial stretch and electrical stimulation. PLoS ONE 2019, 14, e0219442. [Google Scholar] [CrossRef]
- Tallawi, M.; Rai, R.; Boccaccini, A.R.; Aifantis, K.E. Effect of substrate mechanics on cardiomyocyte maturation and growth. Tissue Eng. Part B Rev. 2015, 21, 157–165. [Google Scholar] [CrossRef] [Green Version]
- Colombo, M.; Carregal-Romero, S.; Casula, M.F.; Gutierrez, L.; Morales, M.P.; Bohm, I.B.; Heverhagen, J.T.; Prosperi, D.; Parak, W.J. Biological applications of magnetic nanoparticles. Chem. Soc. Rev. 2012, 41, 4306–4334. [Google Scholar] [CrossRef]
- Bastos, J.P.A.; Sadowski, N. Electromagnetic Modeling by Finite Element Methods; Marcel Dekker: New York, NY, USA, 2003; 490p. [Google Scholar]
- Shevkoplyas, S.S.; Siegel, A.C.; Westervelt, R.M.; Prentiss, M.G.; Whitesides, G.M. The force acting on a superparamagnetic bead due to an applied magnetic field. Lab Chip 2007, 7, 1294–1302. [Google Scholar] [CrossRef]
- Mosconi, F.; Allemand, J.F.; Croquette, V. Soft magnetic tweezers: A proof of principle. Rev. Sci. Instrum. 2011, 82, 034302. [Google Scholar] [CrossRef] [Green Version]
- Rivas-Pardo, J.A.; Eckels, E.C.; Popa, I.; Kosuri, P.; Linke, W.A.; Fernandez, J.M. Work Done by Titin Protein Folding Assists Muscle Contraction. Cell Rep. 2016, 14, 1339–1347. [Google Scholar] [CrossRef] [Green Version]
- Paluch, E.K.; Nelson, C.M.; Biais, N.; Fabry, B.; Moeller, J.; Pruitt, B.L.; Wollnik, C.; Kudryasheva, G.; Rehfeldt, F.; Federle, W. Mechanotransduction: Use the force(s). BMC Biol. 2015, 13, 47. [Google Scholar] [CrossRef] [Green Version]
- Granegger, M.; Choi, Y.; Locher, B.; Aigner, P.; Hubmann, E.J.; Lemme, F.; Cesarovic, N.; Hubler, M.; Schweiger, M. Comparative analysis of cardiac mechano-energetics in isolated hearts supported by pulsatile or rotary blood pumps. Sci. Rep. 2019, 9, 20058. [Google Scholar] [CrossRef] [Green Version]
- Saini, H.; Navaei, A.; Van Putten, A.; Nikkhah, M. 3D cardiac microtissues encapsulated with the co-culture of cardiomyocytes and cardiac fibroblasts. Adv. Healthc. Mater. 2015, 4, 1961–1971. [Google Scholar] [CrossRef] [PubMed]
- Lyon, R.C.; Zanella, F.; Omens, J.H.; Sheikh, F. Mechanotransduction in cardiac hypertrophy and failure. Circ. Res. 2015, 116, 1462–1476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Li, X.; Jing, X.; Li, M.; Ren, Y.; Chen, J.; Yang, C.; Wu, H.; Guo, F. Hypoxia promotes maintenance of the chondrogenic phenotype in rat growth plate chondrocytes through the HIF-1alpha/YAP signaling pathway. Int. J. Mol. Med. 2018, 42, 3181–3192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Jiang, C.; Hong, H.; Liu, J.; Qiu, L.; Huang, Y.; Ye, L. Effects of hypoxia on cardiomyocyte proliferation and association with stage of development. Biomed. Pharmacother. 2019, 118, 109391. [Google Scholar] [CrossRef] [PubMed]
- Sadoshima, J.; Izumo, S. Mechanical stretch rapidly activates multiple signal transduction pathways in cardiac myocytes: Potential involvement of an autocrine/paracrine mechanism. EMBO J. 1993, 12, 1681–1692. [Google Scholar] [CrossRef] [PubMed]
- Komuro, I.; Katoh, Y.; Kaida, T.; Shibazaki, Y.; Kurabayashi, M.; Hoh, E.; Takaku, F.; Yazaki, Y. Mechanical loading stimulates cell hypertrophy and specific gene expression in cultured rat cardiac myocytes. Possible role of protein kinase C activation. J. Biol. Chem. 1991, 266, 1265–1268. [Google Scholar] [PubMed]
- Robador, P.A.; San Jose, G.; Rodriguez, C.; Guadall, A.; Moreno, M.U.; Beaumont, J.; Fortuno, A.; Diez, J.; Martinez-Gonzalez, J.; Zalba, G. HIF-1-mediated up-regulation of cardiotrophin-1 is involved in the survival response of cardiomyocytes to hypoxia. Cardiovasc. Res. 2011, 92, 247–255. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Liew, O.W.; Richards, A.M.; Chen, Y.T. Overview of MicroRNAs in Cardiac Hypertrophy, Fibrosis, and Apoptosis. Int. J. Mol. Sci. 2016, 17, 749. [Google Scholar] [CrossRef]
- Song, J.; Hu, B.; Qu, H.; Bi, C.; Huang, X.; Zhang, M. Mechanical stretch modulates microRNA 21 expression, participating in proliferation and apoptosis in cultured human aortic smooth muscle cells. PLoS ONE 2012, 7, e47657. [Google Scholar] [CrossRef]
- Kunita, A.; Morita, S.; Irisa, T.U.; Goto, A.; Niki, T.; Takai, D.; Nakajima, J.; Fukayama, M. MicroRNA-21 in cancer-associated fibroblasts supports lung adenocarcinoma progression. Sci. Rep. 2018, 8, 8838. [Google Scholar] [CrossRef]
- Xiong, X.; Liu, Y.; Mei, Y.; Peng, J.; Wang, Z.; Kong, B.; Zhong, P.; Xiong, L.; Quan, D.; Li, Q.; et al. Novel Protective Role of Myeloid Differentiation 1 in Pathological Cardiac Remodelling. Sci. Rep. 2017, 7, 41857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, R.L.; Nithiyanantham, S.; Huang, C.Y.; Pai, P.Y.; Chang, T.T.; Hu, L.C.; Chen, R.J.; VijayaPadma, V.; Kuo, W.W.; Huang, C.Y. Synergistic cardiac pathological hypertrophy induced by high-salt diet in IGF-IIRalpha cardiac-specific transgenic rats. PLoS ONE 2019, 14, e0216285. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, M.; Kim, J.; Shin, H.; Kim, Y.; Jang, H.; Park, Y.; Kim, S.-J. Development of Magnetic Torque Stimulation (MTS) Utilizing Rotating Uniform Magnetic Field for Mechanical Activation of Cardiac Cells. Nanomaterials 2020, 10, 1684. https://doi.org/10.3390/nano10091684
Song M, Kim J, Shin H, Kim Y, Jang H, Park Y, Kim S-J. Development of Magnetic Torque Stimulation (MTS) Utilizing Rotating Uniform Magnetic Field for Mechanical Activation of Cardiac Cells. Nanomaterials. 2020; 10(9):1684. https://doi.org/10.3390/nano10091684
Chicago/Turabian StyleSong, Myeongjin, Jongseong Kim, Hyundo Shin, Yekwang Kim, Hwanseok Jang, Yongdoo Park, and Seung-Jong Kim. 2020. "Development of Magnetic Torque Stimulation (MTS) Utilizing Rotating Uniform Magnetic Field for Mechanical Activation of Cardiac Cells" Nanomaterials 10, no. 9: 1684. https://doi.org/10.3390/nano10091684
APA StyleSong, M., Kim, J., Shin, H., Kim, Y., Jang, H., Park, Y., & Kim, S.-J. (2020). Development of Magnetic Torque Stimulation (MTS) Utilizing Rotating Uniform Magnetic Field for Mechanical Activation of Cardiac Cells. Nanomaterials, 10(9), 1684. https://doi.org/10.3390/nano10091684






