1. Introduction
One-dimensional segmentation problems naturally appear in time series analysis across diverse application areas (often referred to as change point or jump point detection in this context). In computational biology, 1D-segmentation problems arise in the analysis of micro-array and high-throughput DNA or RNA sequencing data. Copy number variations in genomes, for example, can be detected by segmenting array-CGH or DNA coverage data into piece-wise constant segments; see Reference [
1] for a review. In epigenomics, recurrent patterns of histone modifications define genomic intervals that can be associated with functional units including promoters, enhancers, or gene bodies, see, e.g., Reference [
2] and the references therein. The identification of transcriptional units is also a segmentation problem, consisting in the distinction of expressed and non-expressed loci [
3,
4] or operons [
5] or, more generally, in the distinction of adjacent or even overlapping transcripts without the benefit of non-expressed spacers between them. The latter task is particularly relevant in organisms with “compact” genomes, such as bacteria [
5] or yeast [
6,
7], where transcribed loci are rarely separated non-expressed regions. Boundaries between transcriptional units are detectable by differences in RNA levels [
6], see, e.g., the SRG1 ncRNA in
Figure 1. A plethora of segmentation algorithms for genomic features, as well as time series data, have become available, reviewed and benchmarked, e.g., in References [
8,
9,
10].
The boundaries between segments typically are not clearly visible in individual data tracks. Such limitations can often be alleviated by aggregating multiple experiments or measurements. In the yeast RNA-seq data shown in
Figure 1, for instance, transcriptome samples at different time points of the respiratory cycle are aggregated to produce a more informative signal than just the RNA expression level at a single time point. Still, any particular choice of parameters (here the choice of the similarity measure for the temporal coverage profiles at adjacent nucleotides), produce both false positive and false negative segment boundaries.
Figure 1 suggests that an improvement could be achieved by aggregating the different segmentations to a single consensus. In a similar vein, Reference [
4] employed a simple heuristic segmentation to detect candidate loci as intervals that are scored by a statistical model for each RNA-seq experiment, followed by a problem-specific greedy heuristic to determine consensus interval boundaries for expressed ncRNA loci. The segmentation method for bacterial RNA-seq data in Reference [
5] computes optimal segmentations with different numbers
K of segments and uses a voting procedure to obtain a consensus over different values of
K. These examples beg the question whether there is a more principled way to aggregated segmentations in a single consensus segmentation.
This question also appears in a much more general context. Modern *omics studies often report their results in the form of genome browser tracks, i.e., as segmentation of the reference genome into intervals. The comparison and consolidation of such data naturally asks for a consensus or reference. This is particularly the case for annotation based on epigenomic or transcriptomic data. Here, principled, efficient methods, to compare annotations beyond quantifying overlaps would be highly desirable to avoid a complete reanalysis of the underlying raw data. In contrast to genome browser tracks, raw data typically require extensive processing and are by no means straightforward to access in all cases. Despite the obvious potential usefulness of consensus segmentations, the literature on systematic comparisons of segmentations is surprisingly sparse. Two natural ways to approach the problem have been considered:
- (i)
Focusing on the breakpoints between segments, one can treat them as signal. Significant (consensus) breakpoints are then detectable as unexpected accumulations across multiple data sets by the
C-KS algorithm [
12]. Somewhat more generally, this can be seen as clustering problem for breakpoints [
13].
- (ii)
Segmentations of linearly ordered data form partitions of an interval. (Dis)similarity measures for partitions, such as the Rand [
14], Fowlkes-Mallows [
15], Jaccard [
16], and Hubert-Arabie [
17] indices, or the Mirkin [
18], and Van Dongen [
19] metrics, are also applicable to the special case of segmentations. The
Median Partition problem, also known as
Consensus Clustering Problem, consists in finding a partition that is as close as possible to a given collection w.r.t. to one of these (dis)similarity measures [
20,
21].
Median Partition is NP-complete [
22,
23]. It is this second approach that we pursue further in this contribution.
The
Segmentation Aggregation problem [
24] is the specialization to partitions of a linearly ordered set, such as a time series or genomic sequence: Given a set of
m segmentations
,
, ...,
on an interval, and a distance function
D between segmentations, the task is to find a segmentation
that minimizes the distance sum
We assume that
is a dissimilarity, i.e., that (i)
, and (ii)
if . In most cases,
will be a metric. However, neither symmetry nor the triangle inequality are necessary. It is shown in Reference [
24] that
Segmentation Aggregation, in contrast to
Median Partition in the general case, can be solved exactly by dynamic programming for several interesting distance measures, including the disagreement or Mirkin metric and the information distance. Mailă [
25] showed, using an axiomatic approach based on certain additivity conditions in the lattice of partitions, that the “variation of information” [
26], i.e., the information distance [
24] serves as the essentially unique natural distance between partitions. Nevertheless we consider here a much broader class of dissimilarity measures. Despite its appealing features, namely the almost complete absence of model assumptions and the fact that no detailed knowledge on the provenance of the input segmentations is required,
Segmentation Aggregation has rarely been used in practical data analysis. Here, we demonstrate that it can be a useful and efficient approach.
A given set of input data is frequently subject to biases, such as an uneven phylogenetic distribution of taxa in comparative genomics or unbalanced distributions of samples between treatment groups. For the purpose of consensus formation, it is usually desirable to retain all data. As a remedy for sampling biases, a plethora of weighting schemes have been proposed to correct for biases by giving larger weights to underrepresented and smaller weights to over-represented data; see Reference [
27] for a comparison of different approaches. In the context of segmentations of genomic features or time series, the confidence in individual segmentations
may be different, e.g., due to different noise levels in individual data tracks. It may also be desirable to treat biological replicates different from technical replicates. Naturally, such differences can be expressed by introducing segmentation-specific weights
. We shall see below that such weights can be introduced in
Segmentation Aggregation in a straightforward manner.
In this contribution, we investigate the weighted version of the Segmentation Aggregation problem with the aim of getting insights into the properties of consensus segmentations. In particular, we generalize previous results of the positioning of consensus break points and we derive an upper bound on length of consensus segments for a large class of distance functions. We then consider two very different applications of consensus segmentations: the identification of transcriptional units, using yeast transcriptomes as a show-case example, and the segmentation of microbial growth curves. We close with a brief discussion of several open problems both regarding the theory behind consensus segmentations and their practical applications.
2. Theory
2.1. Dynamic Programming Algorithm
The
Weighted Segmentation Aggregation problem is a moderate generalization of the unweighted version considered in Reference [
24]. Given a set
of input segmentations and corresponding weights
that quantify the relative importance of the contributing segmentations
, the task is to minimize the objective function
i.e., the weighted total dissimilarity of the unknown consensus segmentation
. Without loosing generality we may assume that
.
Following Reference [
24], we consider a distance measure
D that can be expressed in terms of the
common refinement of two segmentations.
consists of all intersections of the segments of
and
. The common refinement is also known as the
union segmentation since its set of segment boundaries is exactly the union of the boundaries of
and
. In particular, therefore,
. Now, define the
potential of a segmentation
as
where
is a potential function evaluating the individual segments. This gives rise to a class of distances between segmentations defined by
Substituting for
D in Equation (
2) yields
where the middle term depends only on the input. It is, therefore, a constant that can be dropped for the purpose of optimization. Making use of the fact that the weights are normalized, we obtain the objective function
Now, we explicitly consider
as a sequence of intervals
A. Using the additivity of the potential
E, we obtain
The additive form of Equation (
7) as a sum of the contributions
for the consensus segments
makes it possible to minimize
by dynamic programming [
24]. To this end, consider the subset
of segmentations that have a segment boundary at
k, i.e. , position
k is the endpoint of a segment. For a given segmentation
, denote by
the sum of the contributions
with
. Write
for the minimal value of
. Since
k is a segment boundary, the last segment
A before
k is necessarily of the form
, where
denotes the segment boundary immediately preceding
k. Using this notation, we can compute
Thus, we obtain a simple dynamic programming recursion that has the same form for the weighted and unweighted consensus segmentation; also see [
24]. The weights appear only in the scoring function
. We note, furthermore, that the recursion (
8) is the same as for segmentation problems in general [
28]. It appears, e.g., in Reference [
29] for financial time series, in Reference [
30] in context of text segmentation, in Reference [
31] for the analysis of array CGH data, and in References [
5,
7,
32] for the identification of transcripts in tiling array and RNA-seq data. It is discussed in the setting of very general similarity measures in Reference [
11]. As we shall see below, the effort to compute
is dominated by the effort to compute the score
.
Before we proceed, we briefly consider a general condition on the form of the potential function
. Denote by
the discrete segmentation in which every interval is a single point and by
the indiscrete segmentation consisting of a single interval. A function
is
subadditive if
for every
A and every subdivision
of
A. This inequality is strict for at least one interval if and only if
. Comparing
and
, we observe that, in this case,
, violating that
D is a proper distance function. For the limiting case of an additive potential,
for all intervals and their subdivisions, we obtain
for any two segmentations
and
. Thus, only potentials that satisfy
for at least some
are of interest. A function is
superadditive if
for all
. One easily checks that
D is a metric whenever
is superadditive. This condition is not necessary, however. For example, the negentropy defined in Equation (
17) below, is not superadditive.
2.2. Efficient Computation of the Segment Scores
The direct evaluation of
according to its definition, Equation (
8), for given
i and
k, requires
operations because this entails the summation over
segments for each of the
m input segmentations. This results in an impractical total effort of
compared to the quadratic cost of the dynamic programming recursion itself. It is of considerable practical interest, therefore, to find a more efficient way of computing the scoring function. The key idea is to consider, for a given position
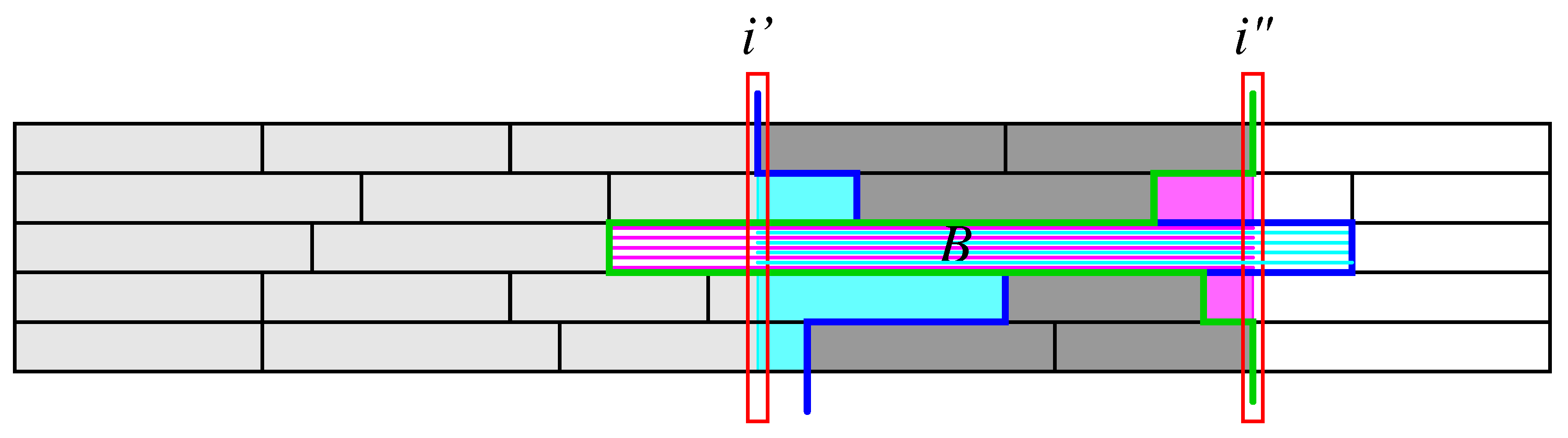
i, two slightly different partial sums:
For a given boundary
i in
, the first term sums all intervals in
that do not extend beyond
i, while the second sum also includes those that begin before or at
i and extend beyond
i. Thus,
captures all segments of the
that are contained within
with one important exception: Segments, such as
B in
Figure 2, that contain
contribute to
but not to
. Such overlapping segments will be taken care of in a correction term discussed below. Using the notation
and
we, furthermore, define terms
where each sum contains at most a single term, namely the interval
that extends across
i. Note that intervals that begin or end in position
i do not contribute to
or
, respectively. The correction terms correspond to the parts of segments that are non-trivially intersected by
, shown in magenta and cyan, resp., in
Figure 2. Thus
covers exactly all the intervals contributing to
– with the exception of segments
that begin at
and end at
as mentioned above. For such segments, instead of the contributions for
and
, a single contribution for the interval
has to be used. In addition, the contribution for
B that is erroneously subtracted with
, needs to be restored. Collecting these contributions, we obtain the following correction term for intervals that span across the interval
of interest:
For the interval
, the correction term
defined in Equation (
11) can be understood as follows: the first term accounts for the correct contribution of
, the second term compensates for the error introduced by
, and the remaining two terms remove the superfluous contributions introduced by
and
. We summarize this derivation in the following form:
Theorem 1. The potential-dependent segment scores defined in Equation (7) can be expressed as The only term that depends on both
and
k is the correction of long input intervals
. The restricted sum over the
in Equation (
11) contains at most one segment for each input segmentation and, thus, can be evaluated in
time for a given interval. Furthermore, the sum is certainly empty whenever
is larger than the largest segment in any of the
; this can be used to speed up the evaluation from
to
if the segment lengths in the input are bounded by a constant, except for the short intervals.
Lemma 1. The arrays of correction terms , , , and can be computed in total time.
Proof. The values of
and
can be computed iteratively: we obtain
by adding the contribution
to
whenever
for the segmentation
. Similarly,
is obtained by adding
to
if
. For each
i, therefore, we require
operations. The sums in Equation (
10) comprise at most one segment of
for every
q. All terms can be computed in constant time using auxiliary arrays that return, for each
i and
q, the values of
and
for
and
. These auxiliary arrays in turn can obviously be constructed in
time for the breakpoint list of the input segmentations. □
It, therefore, makes sense to precompute the arrays , , , and .
Corollary 1. The score can be computed in time with preprocessing cost to compute the arrays , , , and .
It is worth noting, finally, that there is nothing to be gained by storing the score values since each entry is used only once in the recursion.
2.3. Boundaries of Consensus Segments
For a function g on , we define the local curvature at x as . A function g is (strictly) convex at x if . This condition immediately implies that x is not a local maximum of g since at least one of or is larger than . Correspondingly, g is (strictly) concave in x if , whence x is not a local minimum.
Definition 1. The potential is boundedly convex
if satisfied for all intervals For boundedly convex , the curvature in non-increasing as the intervals become larger. In particular, suppose depends only on the length of the interval and is a smooth function, then and for all implies is boundedly convex.
Theorem 2. Let be a set of segmentations with union segmentation and suppose is boundedly convex. Then, the consensus is refined by the union segmentation .
Proof. Following References [
24,
33], we assume, for contradiction, that the optimal consensus
has a segment boundary
that is not contained in the union segmentation
. We aim to show that moving
to some close-by position
x will reduce the cost
. We focus on a fixed input segmentation
and denote by
. Denote by
and
q the first boundaries to the left and to the right of
in
. Analogously,
and
are the first boundary to the left and to the right of
in
, respectively. Thus,
contains the two segments
and
. Since every segment of
is a subset of a unique segment of
, we have
.
We proceed by evaluating how
varies when the boundary
is perturbed. Let
x be the perturbed boundary position. Since only
and
depends on
x and all boundaries except
are fixed, it suffices to focus on the intervals
and
, respectively. Collecting all constant terms in
, we obtain
Since is boundedly convex, we have and , whence . Thus, is concave at x for every and, thus, also for any non-negative contribution to the linear combination of input segmentations. Thus, as a function of the moving boundary x cannot have a minimum in the interior of the interval , contradicting the assumption that is a boundary in the optimal consensus . □
Theorem 2 establishes a very useful property: All segment boundaries of the consensus are contained in the union segmentation. This property was observed for disagreement distance and information distance (see below) in the unweighted setting [
24,
33]. Here, we show it holds for a broader class of distance functions and arbitrary weighting schemes. The techniques used in the proof of Theorem 2 do not seem to generalize to potentials with increasing curvature. Numerical data, however, indicate that the union segmentation refines the consensus for a much larger class of potential functions.
From an algorithmic point of view, it implies that it suffices to compute the
for those values of
k where segment boundaries are in the union segmentation of the inputs
. Correspondingly, we need to store the auxiliary variables only for the intervals of the union segmentation, instead of each
i. That is, the recursion (
8) reduced to
where
denotes the
i-th segment boundary in the union segmentation
.
Recursion (
15) also speeds up the computation of the scoring function
, which now is also needed only for the segment boundaries. First, note that we still obtain
from
by adding the contributions
for the intervals ending at the boundary
to
since by definition
and
are consecutive breakpoints. Analogously,
is obtained by adding
for all blocks beginning at
to
. The terms
and
remain the same. The correction term
could be stored for all pairs of the boundaries in
. Alternatively, it suffices to store the
m boundaries at which the intervals crossing
start and to keep track of the correct correction term directly in recursion (
15). Equation (
15), thus, can be evaluated in
, where
s is the number of breakpoints in the union segmentation.
2.4. Length Bounds on Consensus Segments
It is reasonable to expect that a consensus segmentation cannot be a lot coarser than the individual input segmentations. To see that this is indeed the case, we start with a technical observation.
Lemma 2. Consider intervals , and . Then if for every there is with such that Proof. Equation (
7) implicitly defined
as the term in parentheses, which in turn is the
-weighted sum of contributions for each
. Consider
with
. The contribution
of
to
is
Now, consider an alternative segmentation in which
A is subdivided into
at some position
x inside
B. Then,
A contributes
The terms corresponding to the segments
that intersect
A are the same as before since either
or
, depending on whether
comes before or after
B in
. Thus, we have
if and only if Equation (
16) is satisfied. Since
,
, and
are convex linear combinations of the
,
, and
, respectively, it is sufficient for
that
holds for all
. □
In other words, if A satisfies the condition of Lemma 2, then strictly decreases when A is subdivided into and . Thus, we conclude:
Corollary 2. An interval A satisfying the conditions specified in Lemma 2 cannot appear in a consensus segmentation.
Our goal is now to show that sufficiently long intervals
A always satisfy the conditions of Lemma 2 and, thus, can never be part of the consensus segmentation. Here, we need that
is superadditive, i.e.,
for all
and
. This is the case particularly for the polynomial potentials. It fails for the negentropy potential, Equation (
17), however, because this function is not monotonically increasing with the segment length
.
Theorem 3. Let be a superadditive potential. Let B be the longest segment in the input segmentations and denote by the length of the shortest interval A such that , where and . Then every segment of the consensus segmentation is shorter than .
Proof. If
is superadditive, the l.h.s. of Equation (
16) is maximal if
(for even
) or
for odd
, i.e., we assume that
x is located in the middle of
A. In order to ensure that segments containing
x are completely contained in
A we need
. If this condition is satisfied, Equation (
16) applies. We obtain a sufficient condition by replacing the r.h.s. with the maximal possible contribution of the subdivided interval
B. By superadditivity, this term monotonically increases with the size of
B. The assumption that
x equally divides
A fixed the l.h.s. of the inequality. Since
is strictly superadditive
is strictly monotonically increasing with
, thus, there is a unique smallest value
of
unless
for all
A, in which case no bound
exists. □
Corollary 3. The consensus segmentation with superadditive potential for m input segmentations with length bound as specified in Theorem 3 can be computed in time.
Proof. We observe that for each
k, only values of
j between
and
appear in Equation (
8) since longer segments by Theorem 3 cannot be part of an optimal consensus segmentation. The corollary now follows immediately from Corollary 1. □
The length bound on consensus segments, thus, leads to a reduction of the computational efforts. Although in Theorem 3 may be inconvenient to compute for some choices of the potential , we shall see below that a simple, uniform bound can be obtained for an interesting class of potentials.
2.5. Special Potential Functions
Let us now consider plausible distance functions. The
disagreement distance between segmentation was introduced in Reference [
24] using the potential
. A natural generalization is
for
. We note that a linear potential
, i.e.,
, yields a constant value of
because the sum of all segment lengths adds up to
n; thus,
is independent of the segmentation
.
Recall that the entropy of a discrete distribution is defined as
. Given a segmentation
, we consider the probabilities
of randomly picking a point from a segment, i.e.,
is the relative length of a segment
, where
n denotes the total length of the segmented genome or time series. The
information distance is the symmetrized conditional entropy, which can also be computed as
[
24,
34]. It corresponds to the potential function
given by the negative of the entropy (negentropy) contribution of the interval
A.
It has been shown in References [
24,
33] that the union segmentation
refines the unweighted consensus segmentation for both the disagreement distance and the information distance. This result generalizes to the weighted case and the
-disagreement distances with
.
Corollary 4. The consensus segmentation is refined by the union segmentation for the disagreement distance, its α generalization with , as well as the information distance.
Proof. It suffices to show that the potentials are boundedly convex. For the disagreement distance, we have , and we have and ; for , we have and for . For the negentropy, , we have and , where . The scaling by obviously does not affect the signs. □
Is does not seem possible to generalize the result to potentials that grow faster than quadratically.
Let us finally consider the consequence of Theorem 3. Reusing the convexity results above we can replace by where . A short computation then shows that the inequality in Theorem 3 is satisfied for . Since , we have
Corollary 5. The consensus segmentation of a collection of segmentations with respect to the α-disagreement potentials contains no segment longer than twice the length of the longest input segment.
This allows us immediately to limit the range of the indices in recursion (
8) to
.
2.6. Generalization: Symmetrized Boundary Mover’s Distance
Equation (
7) highlights the fact that the cost function
measures, for each segment
, how well
A conforms to the input segmentations. As noted above, the additive structure of Equation (
7) is sufficient to enable minimization by dynamic programming for arbitrary choices of
. If we retain the idea of weighted contributions for each input segmentation, we may write
, where
measures how well the segment
A “fits” into the segmentation
. As a minimal requirement, for any given interval
A, the score
must attain its minimum value if the interval
A is a segment in
. Since two segmentations in general do not have segments or breakpoints in common, measures are required that are more fine-grained than the distinction between identical and distinct segments or breakpoints.
, thus, are similar to a measure of overlap, between
A and the segments of
that are covered by
A. Clearly, the potential-based measures can be understood in this manner.
An interesting class of dissimilarities utilizes the distance between break points instead of the lengths of intersections between segments. For a segmentation with segments , , we define and set , i.e., the segments are . By slight abuse of notation, we write , i.e., we now specify a segmentation in terms of its breakpoints. Moreover, we write to mean that s represents a breakpoint in the segmentation .
The
“boundary movers distance” was introduced in References [
24,
33] as
where
is some distance function between the positions
s and
c on
. The dissimilarity measure
is not symmetric and satisfies
whenever
is a refinement of
. The segmentation aggregation problem that minimizes
, therefore, is solved by the union segmentation
, while
is minimized by the indiscrete segmentation
. As noted in References [
24,
33], these measures, thus, are only useful with additional constraints on the number or size of allowed segments.
The symmetrized version of
, however, has attractive properties for our purposes, as we shall see: Clearly,
, where
and
delimit the segment of
within which
s resides. If
or
, the contribution vanishes; hence, we can write
This term individually penalizes a segment
of
for containing boundary points of
in its interior. On the other hand, we can rewrite
in terms of segments of
by simply splitting the contribution of each boundary between the two adjacent segments:
Here, we have used that the lower bounds of the first segments and the upper bounds of the last segments necessarily coincide (they are the boundary of the interval on which our segmentations live); therefore, they do not contribute to the distance. Again, the minima are only taken over two alternative breakpoints of
for each given value of
or
, namely those delimiting the segments of
harboring the breakpoints
and
of the consensus segment. It is not difficult to see that
vanishes only if
. Furthermore,
can be written as a sum of contributions
for each of the segments
and each segment of the input segmentations
. Clearly,
depends only on the input segmentations
and the boundary breakpoints
and
, i.e., an individual segment in the consensus
. The segmentation aggregation problem with the symmetrized boundary movers distance, therefore, can again be solved by dynamic programming recursion Equation (
8). A more in-depth analysis of this distance function is the subject of ongoing research.
4. Concluding Remarks
In this work, we have extended and generalized previous work [
24,
33] on the segmentation aggregation problem. We showed that for the class of boundedly convex potential functions, including negentropies and powers
with
, all consensus breakpoints are breakpoints in at least one of the contributing segmentations. Furthermore, we showed that for all superadditive potentials, consensus segments cannot be longer than twice the longest input segment. This bound allows a further reduction of the computational effort.
Consensus segmentations as described here pertain to two major application scenarios: (i) Reconciliation of segmentations of multi-dimensional data, comprising, e.g., independent measurements, such as biological or technical replicates, or measurements of different quantities, e.g., different histone modifications. (ii) Reconciliation of segmentations of the same data set produced with different similarity measures. In principle, it is also possible to compute the consensus segmentation of different segmentations produced, e.g., with randomized algorithms or different heuristics using the same similarity measures. A major advantage of consensus segmentations is that they can be computed without specific information about the data underlying the input segmentations. Such knowledge is not needed because the segmentation aggregation problem depends only on the distance function D as a “parameter”. Empirically, we found that variations of the distance functions have only very moderate consequence on the consensus segmentation.
In simulations, we found strong support for the Refinement Conjecture. This provides support for approaches that utilize a dynamic programming segmentation method to select be best segmentation from the union of segmentations that are computed with different heuristics. Such a scheme has been proposed in Reference [
42]. In this manner, one can potentially achieve a substantial gain in computational efficiency compared to the full dynamic programming segmentation. The
C-KS approach [
12] also restricts itself to the union segmentation.
We considered two very different application scenarios. In applications to transcriptome data consensus segmentations have the potential to substantially improve annotations. A particular strength of the consensus approach is, by highlighting differences that are consistent between data tracks, the ability to identify processing-related boundaries. This is of particular interest in organisms with operons, poly-cistronic primary transcripts, or no expression-free gaps between genes. In all these cases, it becomes difficult and often impossible to distinguish transcriptional units from patterns of mapped RNA-seq reads alone. Here, we have used data from yeast strain IFO 0233, which has been used previously to illustrate transcriptome segmentation in Reference [
11]. We have seen that the consensus segmentation provides a robust prediction of transcriptomic units from a moderate number of individual segmentations with very different parameters. We obtained thousands of segments that may correspond to the non-coding transcripts in
S. cerevisae IFO 0233. Since the present contribution is intended to describe the method of consensus segmentation and its mathematical justification, we will report elsewhere on a comprehensive analysis of the IFO 0233 transcriptome.
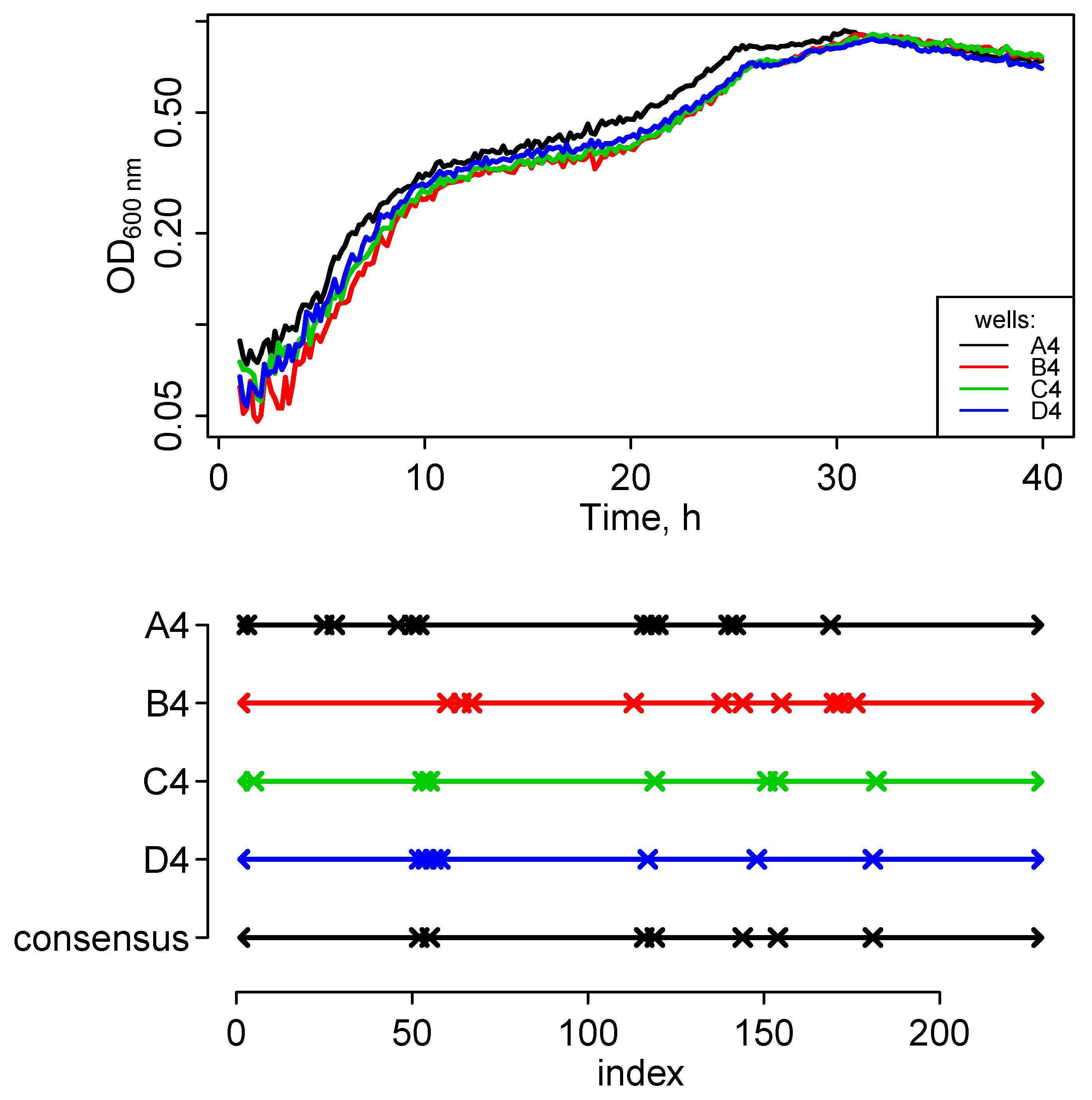
Consensus formation is also of use to aggregate data from biological replicates. As an example, we showed that consensus segmentations of growth curves can be used to robustly determine distinct growth phases.
The consensus segmentation methods incorporate weights that refer to input segmentations. This feature can be used, for instance, to weight individual transcriptome data by coverage. In the case of growth functions, weights may be chosen to decrease with average measurement error, quantified, e.g., as average deviation from the linear fit. It would also be of interest to associate weights with individual segments. This can certainly be done in the context of the Boundary Mover’s distance. Whether this can be also be achieved in the potential-based approach, and to what extent the mathematics results of this contribution will remain intact, however, is a question for future research.
We observed that consensus segmentations are quite robust w.r.t. to the choice of the potential on real data, while we observed a trend towards shorter consensus segments with increasing for the power potentials on i.i.d. random data. Conceptually, consensus segmentations based on the comparison of segments are designed to handle essentially arbitrary heterogeneity along the time or genome coordinate, while breakpoint-centered approaches, such as C-KS, need to rely on statistical regularities of true breakpoints. In order to assess the utility of different potentials and dissimilarity measures , and to compare the segment-centered dynamic programming approach with breakpoint-centered alternatives, a principled way of benchmarking consensus segmentation methods will be necessary. This will require, in particular, the development of a simulator for correlated segmentations that mimic characteristics of different types of underlying data. At present, such tools are not available.
Our analysis of consensus segmentations suggests several avenues for future research. From a theoretical point of view, the most immediate open problem is the Refinement Conjecture, and the characterization of the potential function and—more generally—the dissimilarity measures for which the union segmentation refines the consensus segmentation. This is also of practical relevance since the recursions can be restricted to breakpoints of the union segmentation in such cases; see Equation (
15). In addition, a more detailed understanding of the consensus segmentation would be useful. For instance, are there potentials or dissimilarities that guarantee a breakpoint in the consensus within every interval that contains a breakpoint in every input segmentation, or in the (weighted) majority of the input segmentations? Such results could immediately be used to limit the scope of
j in Equation (
8). More generally, one may ask whether the idea of consensus segmentation can give rise to useful ways of measuring the local accuracy or reliability of consensus and/or input break points.