Abstract
The current COVID-19 pandemic, caused by the rapid worldwide spread of the SARS-CoV-2 virus, is having severe consequences for human health and the world economy. The virus affects different individuals differently, with many infected patients showing only mild symptoms, and others showing critical illness. To lessen the impact of the epidemic, one problem is to determine which factors play an important role in a patient’s progression of the disease. Here, we construct an enhanced COVID-19 structured dataset from more than one source, using natural language processing to add local weather conditions and country-specific research sentiment. The enhanced structured dataset contains 301,363 samples and 43 features, and we applied both machine learning algorithms and deep learning algorithms on it so as to forecast patient’s survival probability. In addition, we import alignment sequence data to improve the performance of the model. Application of Extreme Gradient Boosting (XGBoost) on the enhanced structured dataset achieves 97% accuracy in predicting patient’s survival; with climatic factors, and then age, showing the most importance. Similarly, the application of a Multi-Layer Perceptron (MLP) achieves 98% accuracy. This work suggests that enhancing the available data, mostly basic information on patients, so as to include additional, potentially important features, such as weather conditions, is useful. The explored models suggest that textual weather descriptions can improve outcome forecast.
1. Introduction
The current COVID-19 pandemic, caused by the rapid worldwide spread of the SARS-CoV-2 virus, is affecting many aspects of society, in particular human health (at the time of writing, over 66 million diagnosed cases and 1.5 million deaths [1]), but also social issues [2,3], mental health, and the economy [4]. Researchers from different scientific fields, including immunology, genetics, and bioinformatics, are studying the pandemic to find ways to slow its progression.
Machine learning approaches are also part of this endeavor [5,6,7,8,9]. For example, Shahid et al. [10] use several models, including ARIMA, SVR, LSTM, and Bi-LSTM, for time series prediction of confirmed cases, deaths, and recoveries in ten major countries affected by COVID-19. Shreshth et al. [11] present a machine learning model to predict how the number of cases of COVID-19 will develop, and to forecast when a specific country can expect to see an end of the pandemic, using the FogBus framework. Other researchers have built machine learning models for the classification and diagnosis of COVID-19 that are based on medical images [12,13]. Further, Yan et al. [14] provide an interpretable mortality model that is based on a database of blood samples from 485 infected patients in the region of Wuhan, China. To date, most machine learning and deep learning research [15,16] on COVID-19 build a classification model on various types of data to investigate which might be the important features to predict a specific outcome. One potential difficulty when running such approaches on publicly available dataset is that the features are originally collected so as to fulfill the needs of the data provider, which then can be a source of bias, when the data is used to address other questions. In particular, features that have high predictive value for the outcome for an infected patient might be missing. Generally speaking, the presence or absence of features will impact the accuracy of a model.
The COVID-19 data provided by Xu et al. [17] contain a large number of samples, but limited features that mainly provide basic information on patients. Here, we seek to improve the usefulness of this data by adding a number of features that might help to increase the accuracy of a predictive model.
Research indicates that local climate plays a roles in pandemic outbreaks [18]. Lowen et al. [19] demonstrated that aerosol spread of the influenza virus is dependent upon both ambient relative humidity and temperature, using guinea pig as a model host. Tan et al. [20] investigated the effect of weather in four cities in China and concluded that SARS outbreaks were significantly associated with the temperature and its variations. For the SARS-CoV-2 virus, there are some contradicting findings. Initial studies suggested a negative correlation between temperature and COVID-19 infection [21], or temperature-independence [22], while other research detected a positive relation between temperature and COVID-19 cases at temperatures below 3 °C [23], and also relates temperature to decrease in spread parameters of the case dynamics [24]. Therefore, local weather factors should be taken into consideration.
Infection and mortality rates differ between countries, as does the response to the pandemic. A study on news platforms and social media indicates that more than half (52%) of all news headlines evoked negative sentiments [25], on the one hand, whereas public positive tweets outweighed negative tweets on the other hand [26]. Application of machine learning algorithms on such data indicates a growth in fear and negative sentiment [27]. To explore this further, in this study we assume that a researchers attitude toward COVID-19, optimistic or pessimistic, will reflect the situation in their country, to some extent, and might be detectable in their publications on the pandemic.
While most previous work focuses on a single data type, in this study, we combine multiple data types. While a number of papers focus on country-wise pandemic prediction [28,29,30], here we develop a classification model that is based on worldwide data.
We first built an initial structured dataset on patients that tested positive for the virus, based on the work in [17]. We then constructed an enhanced structured dataset by adding new features based on (1) the local weather conditions when the patient was probably infected, and (2) the average weighted average polarity score for research abstracts on the pandemic, per country.
Another reasonable hypothesis is that the specific genome sequence of the virus that affected a given patient may help predict the outcome for the patient. There is research that associates genomic variations with mortality rate of COVID-19 [31], and further research [32] shows that the SARS-CoV-2 virus carries 7.23 mutations per sample compared to the reference, on average. There is work that attempts to predict outcome using machine learning and deep learning methods [33,34]. Both NCBI [35] and GISAID [36,37] provide genomic data for the virus.
Ideally, we would have liked to further enhance the initial dataset by adding virus genome sequences to each sample. Unfortunately, these sequences are not available. So, to explore the use of genomic sequences, we created an additional sequence dataset that consists of unknown patients and their virus sequence, obtained from GISAID.
In this paper, we investigated the application of two algorithms—XGBoost and MLP—to build models both on the initial structured dataset and also on the enhanced structured dataset. In addition, we built a Bi-LSTM model on the sequence dataset. The applied analysis pipelines are summarized in Figure 1.
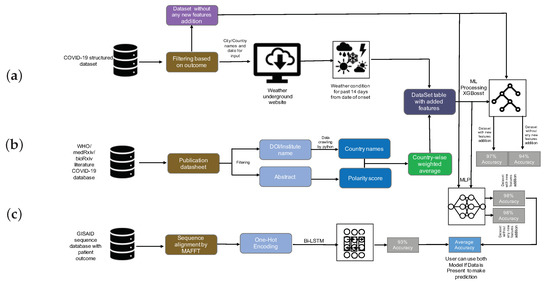
Figure 1.
Analysis summary. (a) The initial COVID-19 structured dataset was filtered for patients for which the outcome has been recorded, and then, for these items, the weather was determined using the Weather Underground website [38]. (b) The WHO, medRxiv, and bioRxiv COVID-19 literature database were filtered and preprocessed to extract author institute/address/country, and these were postprocessed so as to obtain a country-wise research sentiment polarity score. XGBoost and Multi-Layer Perceptron (MLP) were trained on both the initial and the enhanced structured data, and the accuracy of survival prediction was shown to be 94% and 97% (using XGBoost), and 98% and 98% (using MLP), respectively. (c) Bidirectional Long Short-Term Memory (Bi-LSTM) was used to train a classification model on the sequence dataset, the accuracy was 93%. Finally, the MLP model and Bi-LSTM models were stacked to jointly predict outcome.
Based on the initial dataset, we confirm that age is one of the most important factors for predicting survival. When considering the enhanced structured dataset, we find that the weather textual description, followed by local temperature, humidity, and age, arise as the most important features. On the enhanced data, we found that the Extreme Gradient Boosting (XGBoost) method achieved 97% accuracy in predicting a patient’s survival. We describe how to predict patient’s outcome using a combination of a Multi-Layer Perceptron (MLP) and Bidirectional Long Short-Term Memory (Bi-LSTM), using both the enhanced structured dataset, and the sequence dataset, respectively.
2. Materials
2.1. Data Collection
Data were collected from a number of sources.
2.1.1. COVID-19 Structured Dataset
We downloaded COVID-19 patient data provided by Xu et al. [17] from Github [39], on 21 August 2020 (file latestdata.csv). The dataset includes patient’s basic information features, including ID, age, sex, city, province, country, etc. All rows that do not contain a value in the outcome column were dropped, resulting in 307,382 patient data rows out of 2,676,311. The final dataset contained 301,363 patients from 46 countries. All further processing was performed on this dataset.
2.1.2. WHO, medRxiv, and bioRxiv COVID-19 Literature Database
We downloaded a database of literature on COVID-19 from the World Health Organization (WHO) website [40] on 13 April 2020. Of the 5354 downloaded entries, we kept only those whose Journal Name and DOI fields were not blank, which resulted in 4683 publications in 590 journals. This list was extended with COVID-19 SARS-CoV-2 preprints published on medRxiv [41] and bioRxiv [42]. For this we used the bioRxiv API [43] to download the paper information; a total of 8076 entries were downloaded on 27 August 2020. We then analyzed these publications to determine the authors’ institute and country; when no country was explicitly given, we used Google Maps [44] and Wikipedia [45] to determine the country in which the author’s institute is located. This gave rise to 9577 (1501 of 4683 WHO, 8076 of 8076 medRxiv and bioRxiv) entries. Finally, we merged the two datasets and removed all duplicates, obtaining 9542 (1484 of 1501 WHO, 8058 of 8076 medRxiv and bioRxiv, Additional File 1) entries in total.
2.1.3. GISAID CoV-19 Sequences Dataset
The GISAID sequence repository contains more than 244,000 genomic sequences for SARS-CoV-2. We downloaded all that were labeled as complete, with high coverage, and were found in a human host on 25 August 2020. This resulted in 4957 genome sequences (with metadata). Further, we included the reference SARS-CoV-2 Wuhan genome (NCBI Accession MN908947.3 [46]) to the dataset and collected the patient information from the publication [47]. Finally, we removed all those sequences that did not have a patient status in the metadata file. Our final dataset contained 4720 sequences (Additional File 2).
2.2. COVID-19-Enhanced Structured Dataset
In this paper, we present an enhanced COVID-19 structured dataset, which is based on the above described initial COVID-19 structured dataset. These data were enhanced by adding features that reflect the weather situation in the location of the infected person, and the research sentiment in units of country, as described in the following.
2.3. Addition Feature Construction
It has been demonstrated that there is a link between environmental factors and the development of COVID-19 [48]. It is reasonable to assume that weather plays a role in disease progression. Therefore, we collected temperature, humidity, and textual description of the weather for the city where the patient lives from the Weather Underground website [38]. Assuming that the incubation period of the virus is approximately 14 days, we collected weather data from 14 days before the patient exhibited relevant symptoms (as recorded in the initial structured dataset).
We also wanted to explore the assumption that researchers’ attitudes toward COVID-19, either optimistic or pessimistic, reflect the situation in each country, to some extent, and might be detectable in their publications on the pandemic. Therefore, we collected journal publications from the WHO and from the medRxiv and bioRxiv COVID-19 literature database. For each abstract, we determined the author’s institution with the help of the paper’s DOI and address by institute name. We applied sentiment analysis to obtain a polarity score on each abstract, and then calculated an weighted average polarity score for each country. Figure 2 displays the weighted average polarity score inferred for different countries.
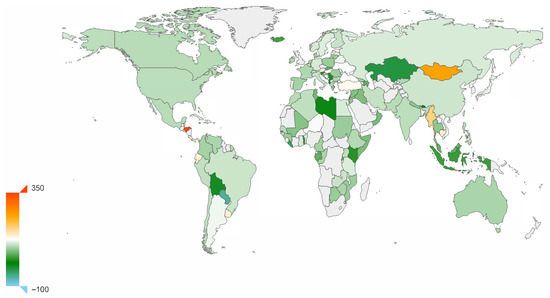
Figure 2.
Sentiment polarity score. Average research sentiment polarity score of research, for different countries. Based on a sentiment analysis of abstracts of papers published on COVID-19. One-thousand times the real value.
The weather and sentiment features were added to the initial structured dataset so as to produce the enhanced structured dataset, as outlined in Figure 1.
2.4. Data Processing
2.4.1. Structured Data
The features present in the initial COVID-19 structured dataset include both categorical variables and discrete variables. Each sample in the dataset contains the variables sex, age, the time interval between the patient’s onset date, confirmed infected date and admission date, symptoms description, presence of chronic disease, and outcome.
To this initial data, we then added local weather variables (temperature, humidity, and climate description) and the weighted polarity score of the country’s scientific research sentiment. The result of this is called the enhanced structured dataset.
To prepare the datasets for building classification models using both XGBoost and MLP (as discussed below), we performed the following steps. We encoded all multi-value text features, such as symptom description (values such as fever, cough, and sputum) or climate description (values such as fair, light rain shower, and cloudy) into three-dimensional embedding vectors, using label encoding on categorical variables such as sex and history of chronic disease (Additional File 3).
We assigned the constant −999 to all missing values. After filtering for samples that have a valid outcome value and city record, we obtained 301,363 samples. Additionally, when we ran MLP, we treated sex and binary chronic disease as categorical features and all others as numerical features, and we normalized all numerical features.
2.4.2. Sequence Data
We performed multiple sequence alignment of the sequence dataset using MAFFT [49], run as follows.
mafft --retree 2 --maxiterate 1000 --thread 48 DeathAndAliveForMafft.fasta
>DeathAndAliveForMafftAlignment1000Iterate.fasta
The program required 589 walk-clock minutes to align the 4720 virus genome sequences. The resulting alignment length was 32,015 (Additional File 4).
Furthermore, we applied character-level one-hot encoding on each sequence, mapping each position to a six-dimensional vector (one dimension for each of the four nucleotides, one for the gap character, and one for all ambiguity codes). Each sequence was padded to a fixed length of 33,100 (a multiple of 100), so as to allow us to use 100 time steps in the model described below.
2.5. Data Statistics
We built both a XGBoost model and an MLP model on both the initial structured dataset and on the enhanced structured dataset, respectively.
To evaluate the methods, we split each dataset into a training set and test set in proportion 8:2. Further, to prevent overfitting, we used cross-validation on our training datasets, instead of splitting additional validation sets from the original dataset. As shown in Table 1, the original dataset is typically imbalanced. To address this, we applied the Synthetic Minority Oversampling Technique (SMOTE) [50] to the minority group of each training set, attaining a ratio of positive to negative samples of 10:1. Note that here positive samples refer to patients that survive.

Table 1.
Sampling statistics. For the enhanced structured dataset, we report the number of positive and negative samples both in the training set and test set, both before and after oversampling, respectively.
3. Methods and Experiment
3.1. Sentiment Analysis
A number of papers have studied the forecasting of pandemics using natural language processing on data obtained from various social media [51,52,53]. Along these lines, we performed sentiment analysis on the abstracts of research papers (associated with COVID-19) using the Python package Textblob [54], which operates by analyzing text content and assigning emotional values to words based on matches to a built-in dictionary.
3.2. Machine Learning Algorithm
Our focus was on the performance of prediction of survival of the infection, based on either the initial or the enhanced structured dataset.
Here, we use the Extreme Gradient Boosting (XGBoost) [55] method to build a prediction model. XGBoost is a powerful member of the gradient boosting family, which is designed to perform well on sparse features, and is known to perform well on Kaggle tasks. This approach avoids overfitting using its built-in L and L regularization on the target function:
As an additive model, XGBoost consists of k base models, and in most cases we choose the tree model as its base model. Suppose that, for the k-th of t iterations, we train the tree model , then
is the estimated result for the ith sample after t iterations. During construction of each tree, XGBoost minimizes the objective function, with the regularization term show in Equation (1) in the split phase of each node. In each tree, we calculate the Gain of the feature and choose the tree that has the biggest value as the leaf node to be split:
3.3. Deep Learning Algorithms
To broaden our research and to allow a comparison of methods, we also built deep learning models on both the initial and enhanced structured datasets, together with the sequence dataset, respectively (Figure 3).
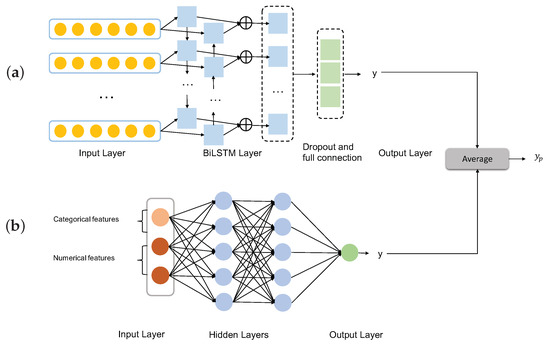
Figure 3.
Ensemble deep learning model. (a) The MLP is trained on the structured dataset. (b) The Bi-LSTM model is trained on the sequence dataset. The two models are stacked in the prediction step.
3.3.1. Multi-Layer Perceptron
As indicated in Figure 3b, we use a simple Multi-Layer Perceptron (MLP) as neural network structure, which has an input layer, hidden layer, and output layer, to build a classification model on the structured dataset.
3.3.2. Bidirectional Long Short-Term Memory
Each sample in our sequence dataset has length 33,100 after alignment and data processing. We can interpret each sequence as a time-series, where is the data associated with the tth time point. Recurrent neural networks (RNN) proposed by Elman [56] are commonly used for time series; however, they are not suitable for our task due to the length of the alignments. Long short-term memory (LSTM) [57] is a special variant of RNN. It uses a gate structure in the hidden layer of each time step to protect and control the cell state.
An LSTM cell employs three gates, namely, a forget gate, an input gate, and an output gate, operating as shown in Figure 4. An LSTM learns to memorize and forget specific information during the training step. It provides the ability to capture long-term dependency relationships.
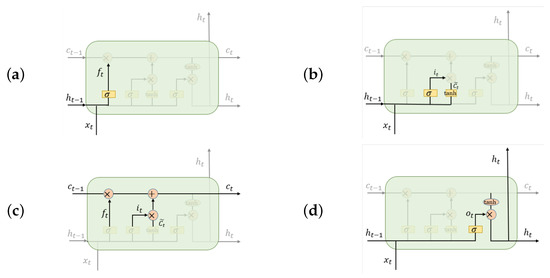
Figure 4.
Operation of gates in an LSTM cell. The LSTM determines the hidden state and cell state at the present sequence location as follows. (a) A forget gate controls the input of the ()th hidden state, (b) an input gate controls the input of , (c) a transitional phase calculates the tth positions cell state, and then, finally, (d) an output gate returns the tth position’s hidden state .
Each gate employs a sigmoid function that aims at producing output values of 0 or 1, defined as
An LSTM does not encode the information in inverse order, so it does not capture the impact of later words on previous words. A bidirectional long short-term memory (Bi-LSTM) overcomes this problem by combining a forward LSTM with a backward LSTM in each time step. This design addresses the issue of bidirectional semantic dependency during model building.
Therefore, we use a Bi-LSTM on our sequence data. Assume we are given a sequence , where reflects the one-hot encoding. The hidden state of each time point is
In summary, this allows us to consider the impact of the virus sequence information on the patient’s condition.
Finally, we stacked the MLP and Bi-LSTM deep learning classification models to jointly predict whether the infected patient will survive.
3.4. Implementation
3.4.1. Machine Learning Algorithms
In this study, we ran the XGBoost algorithm both on the initial structured dataset and also on the enhanced structured dataset, the latter additionally containing local weather and research sentiment. To determine the model parameters with the best capacity for prediction, we used GridSearchCV (a function of sklean) to systematically traverse multiple parameter combinations and determine the best parameters through cross-validation. Each subtree in our model is a complicated tree whose maximum depth is 10. Based on the result of model tuning, we set the learning rate to 0.05 and eta to 0.2. Further, we used 1500 estimators, and gamma, alpha, and lambda equal to 0.01, 0.5, and 0.8, respectively.
Each tree was trained on half of the features and half of the samples, chosen at random.
3.4.2. Deep Learning Algorithms
In Figure 3a we show the architecture of the model that accepts aligned sequences. It is a single Bi-LSTM with 128 hidden units and 100 time steps. After randomly dropping 1% of neurons, we use a fully connected layer and ReLU (rectified linear unit) activation function. Output is passed through a sigmoid function.
To model datasets that include both categorical features and normalized numerical features (Figure 3b), we used a 2-layer full connected neural network with 256 hidden units for each layer. To prevent model overfitting, we dropped a neuron with 5% probability during the forward propagation. A sigmoid function was used to determine output.
During training of both models, we split validation set from training set as proportion 1:3, and to moderate bias created by imbalanced data distribution, we set the class weight ratio between positive samples and negative samples to 1:10. After training as described above, we stacked the two models together so as to obtained average probability, passed through a sigmoid function (Figure 3).
4. Results
We evaluated the algorithms’ performance using multiple metrics (Table 2).

Table 2.
Performance measures. We report accuracy (Acc.), area under the curve (AUC), F1 score, recall, and precision (Prec.) for the named models and datasets. To compare the performance of the models using the initial or enhanced structured datasets, superior values are shown in bold. (for confusion matrices see Additional file 5).
4.1. Machine Learning Model
The accuracy of the model created by using the initial structured dataset (no added features) is 94%, whereas using the enhanced structured dataset (with added features), the model’s accuracy is 97%. As accuracy on an imbalanced dataset is limited, we display the receiver operating characteristic (ROC) curve of both datasets in Figure 5 to provide a further comparison. The enhanced structured dataset has significantly higher area under the ROC curve (AUC) scores than the model built on the initial structured dataset. There also exist tiny differences between the F1 score, recall, and precision of the two models. The method we chose to evaluate the importance score of feature is based on counting the number of times that a feature occurred in a tree. The feature importance for both datasets is shown in Figure 6. For the initial structured dataset, age plays a more important role than other features. For the model based on the enhanced structured dataset, the weather description, temperature, and humidity are more important than age; moreover, the level of importance of weather is higher than that of age. We visualized the frequency of the textual weather description on survivors and non-survivors, respectively (Figure 7). The weighted average research sentiment polarity score does not have an exceptional f score.
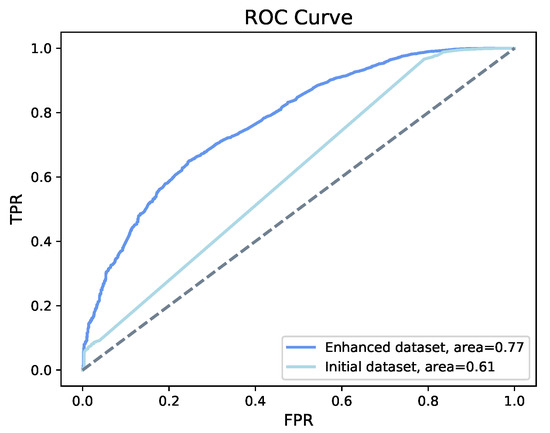
Figure 5.
ROC of XGBoost. XGBoost shows an the accuracy of 94% on the initial structured dataset and an accuracy 97% on the enhanced structured dataset, with an increase of the area under the curve from 61% to 77%.
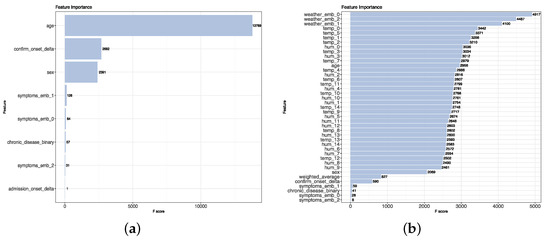
Figure 6.
Feature scores on the enhanced structured dataset. (a) XGBoost processing of the initial structured dataset identified age as an important feature. (b) XGBoost processing of the enhanced structured dataset identified in the weather as an important feature.

Figure 7.
Textual weather description. (a) Word cloud visualization of the frequency of textual weather description for survivors. (b) Word cloud visualization of the frequency of textual weather description for non-survivors.
4.2. Deep Learning Model
As shown in Table 2, on both the initial and enhanced structured datasets, the MLP method demonstrated higher accuracy than the XGBoost method. For both datasets, the accuracy using MLP is 98%. However, the ROC curve (Figure 8) indicates that the model shows a better classification ability on the enhanced structured dataset.
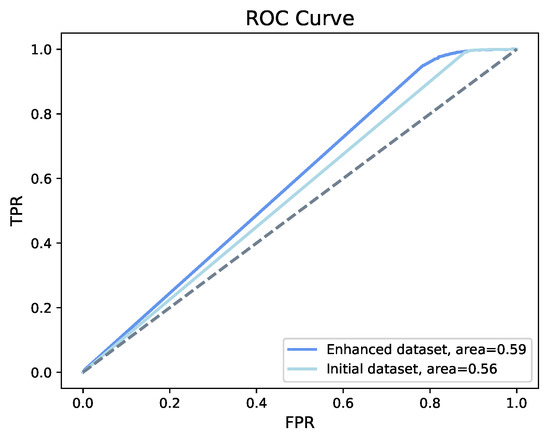
Figure 8.
ROC of MLP. MLP shows an accuracy of 98% on both the initial and the enhanced structured dataset, with an increase in area under the curve from 56% to 59%.
Taking sequence data into account, we obtained 93% accuracy and the area under the ROC curve is 0.73, as shown in Figure 9. Among all the models we built, the AUC score was highest when using a Bi-LSTM on the sequence data.
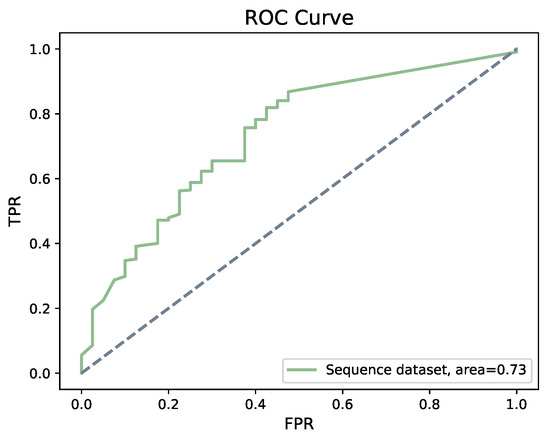
Figure 9.
ROC of Bi-LSTM. Bi-LSTM shows an accuracy of 93% on sequence dataset, with an area under the curve of 0.73.
5. Discussion and Conclusions
The performance of machine learning and deep learning methods depends on the amount and quality of available features. Our analysis illustrates that current publicly available data can be enhanced, so as to increase the accuracy of survival prediction by 3% along with positive changes in other model validating metrics, such as AUC (16%), F1 score (2%), and Recall (3%) in case of XGBoost. For MLP the accuracy, F1 score, Recall, and Precision remained the same both for the initial and enhanced structured dataset, but the AUC increased by 3%.
To further evaluate the capability of the proposed models, we repeated the construction of all models on the same datasets, however, with the roles of positive and negative samples reversed, that is, this time considering patients who did not survive as positive samples. We observed that for XGBoost and MLP, the models based on the enhanced structured dataset perform better than those based on initial structured dataset in all aspects except recall (see Table 3). Further, it can be observed that even the best model has really poor performances in detecting patients who did not survive, as witnessed by the F1 score of 0.20.

Table 3.
Performance measures (predicting death). Considering patients that die as positive samples, we report performance as in the previous Table (for confusion matrices see Additional file 5).
Our study shows how one might enhance a dataset by adding informative features that are not available in the original dataset. Here we demonstrated this for local weather and country-wise research sentiment. Local weather conditions has been implicated as an important feature previous studies.
Our analysis also shows that age is an important factor for survival of COVID-19 as well. However, in the data considered here, the total number of deaths above age 60 were 793 and 2887 survived or were still alive, while in the age group between 40 and 60 there were 421 deaths and 10,346 alive or survived. Therefore, linking mortality to a particular age group is not appropriate based on the current data.
While this analysis suggests that elderly have a higher risk of death, which has already been observed [58,59], saying that mortality is associated with old age is probably generally true for any infectious disease. Age is one of the confounding factors that could be responsible for an increased COVID-19 mortality rate [60,61].
For the model based on the enhanced structured dataset, the weather textual description, followed by local temperature, humidity, and age, appear as the most important features and account for the increase in the accuracy of the model. The most apparent difference in the weather attributes for survivors and non-survivors (Figure 7) is “smoke”. This suggests that environmental conditions, in particular air pollution, may play a role in determining the outcome of the disease.
In contrast, in our investigation, the research sentiment score did not show the importance that we had suspected. The values of this feature are never particular high or low, and the highest value of this feature is only 0.35, and thus the difference between the highest score and lowest score is also small. We assume that one of the reasons for this is that academic writing aims for a neutral tone.
The model that we developed on the virus genome dataset failed to provide added predictive power. We suspect that virus genome data would be much more useful, if it were available for the large, structured dataset. However, our study may provide a starting point for further work.
Further, this analysis confirms that enhancing a dataset, rather than just analyzing the originally given features, might lead to a better prediction of a particular outcome. Along with some of the features which should be paid more attention while collecting the data.
There are a number of possible directions for future work. As more viral genomes become available, more powerful Deep Learning methods can be applied to them to help predict patient survival. Additional features such as patient health status, weight, height, medical history should also be integrated. The effect of climate on patient survival warrants more investigation. Finally, methods such as a Recurrent Neural Network-based LSTM might help to study how mutations influence the transmissibility of the virus [62].
Supplementary Materials
Additional files, datasets and models analyzed during our study along with the supplementary materials (like scripts) can be accessed at https://github.com/husonlab/covid19paper. Additional file 1: Merge and processed publication data downloaded from WHO, medRxiv and bioRxiv COVID-19 literature database; Additional file 2: Sequences used for MAFFT alignment downloaded from GISAID and NCBI; Additional file 3: COVID-19 Enhanced structured dataset; Additional file 4: Aligned sequence used for Bi-LSTM; Additional file 5: Confusion matrices for all the build models.
Author Contributions
D.H.H. proposed and guided the project. D.H.H., W.Z., and A.G. wrote the manuscript. W.Z. and A.G. designed the work, wrote the code for data generation, conducted processing, and carried out the analysis. W.Z. carried out the machine learning and deep learning model development and analysis. All authors read and approved the final manuscript.
Funding
This work was supported by the BMBF-funded de.NBI Cloud within the German Network for Bioinformatics Infrastructure (de.NBI) (031A537B, 031A533A, 031A538A,031A533B, 031A535A, 031A537C, 031A534A, 031A532B). Furthermore, we acknowledge support by the Open Access Publishing Fund of University of Tübingen.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Publicly available datasets were analyzed in this study. These datasets can be found here: Xu et al. dataset. [https://doi.org/10.1038/s41597-020-0448-0], preprints from medRxiv and bioRxiv were accessed by using API: [ https://api.biorxiv.org/covid19/help], sequence dataset was download from GISAID: [https://www.gisaid.org/] and NCBI: [https://www.ncbi.nlm.nih.gov/search/all/?term=MN908947] and remaining processed and generated dataset can be downloaded from [https://github.com/husonlab/covid19paper].
Acknowledgments
We would like to thank Caner Bagči for helpful discussions on sequence analysis.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| COVID-19 | coronavirus disease |
| SARS-CoV-2 | severe acute respiratory syndrome coronavirus 2 |
| ARIMA | Autoregressive Integrated Moving Average model |
| Bi-LSTM | Bidirectional Long Short-Term Memory |
| LSTM | Long Short-Term Memory |
| MLP | Multi-Layer Perceptron |
| NLP | Natural Language Processing |
| WHO | World Health Organization |
| NCBI | National Center for Biotechnology Information |
| GISAID | Global initiative on sharing all influenza data |
| RNN | Recurrent Neural Network |
| SVR | Support Vector Regression |
| XGBoost | Extreme Gradient Boosting |
References
- WHO Coronavirus Disease (COVID-19) Dashboard. Available online: https://covid19.who.int/ (accessed on 5 January 2021).
- Torales, J.; O’Higgins, M.; Castaldelli-Maia, J.M.; Ventriglio, A. The outbreak of COVID-19 coronavirus and its impact on global mental health. Int. J. Soc. Psychiatry 2020, 31, 0020764020915212. [Google Scholar] [CrossRef]
- Singh, J.; Singh, J. COVID-19 and its impact on society. Electron. Res. J. Soc. Sci. Humanit. 2020, 2, 102–105. [Google Scholar]
- Holmes, E.A.; O’Connor, R.C.; Perry, V.H.; Tracey, I.; Wessely, S.; Arseneault, L.; Ballard, C.; Christensen, H.; Silver, R.C.; Everall, I.; et al. Multidisciplinary research priorities for the COVID-19 pandemic: A call for action for mental health science. Lancet Psychiatry 2020, 7, 547–560. [Google Scholar] [CrossRef]
- Lalmuanawma, S.; Hussain, J.; Chhakchhuak, L. Applications of machine learning and artificial intelligence for Covid-19 (SARS-CoV-2) pandemic: A review. Chaos Solitons Fractals 2020, 139, 110059. [Google Scholar] [CrossRef] [PubMed]
- Ramchandani, A.; Fan, C.; Mostafavi, A. Deepcovidnet: An interpretable deep learning model for predictive surveillance of covid-19 using heterogeneous features and their interactions. IEEE Access 2020, 8, 159915–159930. [Google Scholar] [CrossRef]
- Wang, P.; Zheng, X.; Li, J.; Zhu, B. Prediction of epidemic trends in COVID-19 with logistic model and machine learning technics. Chaos Solitons Fractals 2020, 139, 110058. [Google Scholar] [CrossRef] [PubMed]
- Mirri, S.; Delnevo, G.; Roccetti, M. Is a COVID-19 Second Wave Possible in Emilia-Romagna (Italy)? Forecasting a Future Outbreak with Particulate Pollution and Machine Learning. Computation 2020, 8, 74. [Google Scholar] [CrossRef]
- Alakus, T.B.; Turkoglu, I. Comparison of deep learning approaches to predict COVID-19 infection. Chaos Solitons Fractals 2020, 140, 110120. [Google Scholar] [CrossRef]
- Shahid, F.; Zameer, A.; Muneeb, M. Predictions for COVID-19 with deep learning models of LSTM, GRU and Bi-LSTM. Chaos Solitons Fractals 2020, 140, 110212. [Google Scholar] [CrossRef]
- Tuli, S.; Tuli, S.; Tuli, R.; Gill, S.S. Predicting the Growth and Trend of COVID-19 Pandemic using Machine Learning and Cloud Computing. Internet Things 2020, 11, 100222. [Google Scholar] [CrossRef]
- Elaziz, M.A.; Hosny, K.M.; Salah, A.; Darwish, M.M.; Lu, S.; Sahlol, A.T. New machine learning method for image-based diagnosis of COVID-19. PLoS ONE 2020, 15, e0235187. [Google Scholar] [CrossRef] [PubMed]
- Barstugan, M.; Ozkaya, U.; Ozturk, S. Coronavirus (Covid-19) classification using ct images by machine learning methods. arXiv 2020, arXiv:2003.09424. [Google Scholar]
- Yan, L.; Zhang, H.-T.; Goncalves, J.; Xiao, Y.; Wang, M.; Guo, Y.; Sun, C.; Tang, X.; Jing, L.; Zhang, M. An interpretable mortality prediction model for COVID-19 patients. Nat. Mach. Intell. 2020, 2, 283–288. [Google Scholar] [CrossRef]
- Narin, A.; Kaya, C.; Pamuk, Z. Automatic detection of coronavirus disease (COVID-19) using X-ray images and deep convolutional neural networks. arXiv 2020, arXiv:2003.10849. [Google Scholar]
- Magar, R.; Yadav, P.; Farimani, A.B. Potential neutralizing antibodies discovered for novel corona virus using machine learning. arXiv 2020, arXiv:22003.08447. [Google Scholar]
- Xu, B.; Gutierrez, B.; Mekaru, S.; Sewalk, K.; Goodwin, L.; Loskill, A.; Cohn, E.L.; Hswen, Y.; Hill, S.C.; Cobo, M.M.; et al. Epidemiological data from the COVID-19 outbreak, real-time case information. Sci. Data 2020, 7. [Google Scholar] [CrossRef]
- Lin, K.; Fong, D.Y.T.; Zhu, B.; Karlberg, J. Environmental factors on the SARS epidemic: Air temperature, passage of time and multiplicative effect of hospital infection. Epidemiol. Infect. 2006, 134, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Lowen, A.C.; Mubareka, S.; Steel, J.; Palese, P. Influenza virus transmission is dependent on relative humidity and temperature. PLoS Pathog. 2007, 3, 151. [Google Scholar] [CrossRef]
- Tan, J.; Mu, L.; Huang, J.; Yu, S.; Chen, B.; Yin, J. An initial investigation of the association between the SARS outbreak and weather: With the view of the environmental temperature and its variation. J. Epidemiol. Community Health 2005, 59, 186–192. [Google Scholar] [CrossRef]
- Prata, D.N.; Rodrigues, W.; Bermejo, P.H. Temperature significantly changes COVID-19 transmission in (sub)tropical cities of brazil. Sci. Total. Environ. 2020, 729, 138862. [Google Scholar] [CrossRef]
- Jamil, T.; Alam, I.; Gojobori, T.; Duarte, C.M. No evidence for temperature-dependence of the COVID-19 epidemic. Front. Public Health 2020, 8, 436. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Zhu, Y. Association between ambient temperature and COVID-19 infection in 122 cities from China. Sci. Total. Environ. 2020, 724, 138201. [Google Scholar] [CrossRef] [PubMed]
- Demongeot, J.; Flet-Berliac, Y.; Seligmann, H. Temperature decreases spread parameters of the new COVID-19 case dynamics. association between ambient temperature and COVID-19 infection in 122 cities from China. Biology 2020, 9, 94. [Google Scholar] [CrossRef]
- Aslam, F.; Awan, T.M.; Syed, J.H.; Kashif, A.; Parveen, M. Sentiments and emotions evoked bynews headlines of coronavirus disease (covid-19) outbreak. Humanit. Soc. Sci. Commun. 2020, 7, 1–9. [Google Scholar] [CrossRef]
- Hung, M.; Lauren, E.; Hon, E.S.; Birmingham, W.C.; Xu, J.; Su, S.; Hon, S.D.; Park, J.; Dang, P.; Lipsky, M.S. Social network analysis of covid-19 sentiments: Application of artificial intelligence. J. Med. Internet Res. 2020, 22, e22590. [Google Scholar] [CrossRef]
- Samuel, J.; Ali, G.G.; Rahman, M.; Esawi, E.; Samuel, Y. Covid-19 public sentiment insights and machine learning for tweets classification. Information 2020, 11, 314. [Google Scholar] [CrossRef]
- Souza, F.S.H.; Hojo-Souza, N.S.; Santos, E.B.; Silva, C.M.; Guidoni, D.L. Predicting the disease outcome in COVID-19 positive patients through Machine Learning: A retrospective cohort study with Brazilian data. medRxiv 2020. [Google Scholar] [CrossRef]
- Pinter, G.; Felde, I.; Mosavi, A.; Ghamisi, P.; Gloaguen, R. COVID-19 Pandemic Prediction for Hungary: A Hybrid Machine Learning Approach. Mathematics 2020, 8, 890. [Google Scholar] [CrossRef]
- Arora, P.; Kumar, H.; Panigrahi, B.K. Prediction and analysis of COVID-19 positive cases using deep learning models: A descriptive case study of India. Chaos Solitons Fractals 2020, 139, 110017. [Google Scholar] [CrossRef]
- Toyoshima, Y.; Nemoto, K.; Matsumoto, S.; Nakamura, Y.; Kiyotani, K. SARS-CoV-2 genomic variations associated with mortality rate of COVID-19. J. Hum. Genet. 2020, 65, 1075–1082. [Google Scholar] [CrossRef]
- Mercatelli, D.; Giorgi, F.M. Geographic and genomic distribution of SARS-CoV-2 mutations. Front. Microbiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Bhonde, S.; Bhati, M.; Prasad, J. Predictive Analytics to Combat with COVID-19 Using Genome Sequencing. 2020. Available online: https://ssrn.com/abstract=3580692 (accessed on 5 January 2021).
- Machine Learning for Biology: How Will COVID-19 Mutate Next? Available online: https://towardsdatascience.com/machine-learning-for-biology-how-will-covid-19-mutate-next-4df93cfaf544 (accessed on 5 January 2021).
- National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 5 January 2021).
- Elbe, S.; Buckland-Merrett, G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob. Challenges 2017, 1, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Shu, Y.; McCauley, J. GISAID: Global initiative on sharing all influenza data–from vision to reality. Eurosurveillance 2017, 22, 13. [Google Scholar] [CrossRef] [PubMed]
- Weather Underground. Available online: https://www.wunderground.com/ (accessed on 5 January 2021).
- nCoV2019. Available online: https://github.com/beoutbreakprepared/nCoV2019/tree/master/latest_data (accessed on 5 January 2021).
- Global Research on Coronavirus Disease (COVID-19). Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/global-research-on-novel-coronavirus-2019-ncov (accessed on 5 January 2021).
- medRxiv. Available online: https://www.medrxiv.org/ (accessed on 5 January 2021).
- bioRxiv. Available online: https://www.biorxiv.org/ (accessed on 5 January 2021).
- API Summary for the Collection of COVID-19 SARS-CoV-2 Preprints from medRxiv and bioRxiv. Available online: https://api.biorxiv.org/covid19/help (accessed on 5 January 2021).
- Google Map. Available online: https://www.google.com/maps/ (accessed on 5 January 2021).
- WIKIPEDIA. Available online: https://www.wikipedia.org/ (accessed on 5 January 2021).
- NCBI Accession MN908947.3. Available online: https://www.ncbi.nlm.nih.gov/search/all/?term=MN908947 (accessed on 5 January 2021).
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265269. [Google Scholar] [CrossRef]
- Triplett, M. Evidence that higher temperatures are associated with lower incidence of COVID-19 in pandemic state, cumulative cases reported up to March 27, 2020. medRxiv 2020. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2020, 16, 321–357. [Google Scholar] [CrossRef]
- Alessa, A.; Faezipour, M. A review of influenza detection and prediction through social networking sites. Theor. Biol. Med Model. 2018, 15, 2. [Google Scholar] [CrossRef]
- Lee, K.; Agrawal, A.; Choudhary, A. Forecasting influenza levels using real-time social media streams. In Proceedings of the 2017 IEEE International Conference on Healthcare Informatics (ICHI), Park City, UT, USA, 23–26 August 2017; pp. 409–414. [Google Scholar]
- Wang, Y.; Xu, K.; Kang, Y.; Wang, H.; Wang, F.; Avram, A. Regional influenza prediction with sampling Twitter data and PDE model. Int. J. Environ. Res. Public Health 2020, 17, 678. [Google Scholar] [CrossRef]
- TextBlob. Available online: https://github.com/sloria/TextBlob (accessed on 5 January 2021).
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd ACM Sigkdd International Conference on Knowledge Discovery and Data Mining; ACM: New York, NY, USA, 2016; pp. 785–794. [Google Scholar]
- Elman, J.L. Finding structure in time. Cogn. Sci. 1990, 14, 179–211. [Google Scholar] [CrossRef]
- Hochreiter, S.; Schmidhuber, J. Long short-term memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef] [PubMed]
- Verity, R.; Okell, L.C.; Dorigatti, I.; Winskill, P.; Whittaker, C.; Imai, N.; Cuomo-Dannenburg, G.; Thompson, H.; Walker, P.G.; Fu, H.; et al. Estimates of the severity of coronavirus disease 2019: A model-based analysis. Lancet Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Glynn, J.R. Protecting workers aged 60–69 years from COVID-19. Lancet Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Wang, H.; Li, T.; Barbarino, P.; Gauthier, S.; Brodaty, H.; Molinuevo, J.L.; Xie, H.; Sun, Y.; Yu, E. Dementia care during COVID-19. Lancet 2020, 395, 1190–1191. [Google Scholar] [CrossRef]
- Armitage, R.; Nellums, L.B. COVID-19 and the consequences of isolating the elderly. Lancet Public Health 2020, 5, e256. [Google Scholar] [CrossRef]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking changes in SARS-CoV-2 Spike: Evidence that D614G increases infectivity of the COVID-19 virus. Cell 2020, 182, 812–827. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).