Abstract
Phytoecdysteroids represent a class of naturally occurring substances known for their diverse biological functions, particularly their strong ability to stimulate protein anabolism. In this study, a computational machine learning-driven quantitative structure–activity relationship (QSAR) approach was applied to analyze the anabolic potential of 23 ecdysteroid compounds. The ML-based QSAR modeling was conducted using a combined approach that integrates Genetic Algorithm-based feature selection with Multiple Linear Regression Analysis (GA-MLRA). Additionally, structure optimization by semi-empirical quantum-chemical method was employed to determine the most stable molecular conformations and to calculate an additional set of structural and electronic descriptors. The most effective QSAR models for describing the anabolic activity of the investigated ecdysteroids were developed and validated. The proposed best model demonstrates both strong statistical relevance and high predictive performance. The predictive performance of the resulting models was confirmed by an external test set based on R2test values, which were within the range of 0.89 to 0.97.
1. Introduction
Phytoecdysteroids represent a class of natural bioactive compounds exhibiting multiple pharmacological activities [1,2,3]. These compounds are well-documented for their strong ability to stimulate protein synthesis and promote anabolic processes [2,3]. Anabolic processes contribute to the formation of organs and tissues by promoting cell growth, differentiation, and overall body enlargement through the synthesis of complex biomolecules. Ecdysteroids are hormones that regulate cell proliferation, growth, and developmental cycles in insects and other invertebrates. Recent studies suggest that the anabolic effect of ecdysterone—a naturally occurring steroid hormone believed to enhance physical performance—is mediated through its interaction with estrogen receptors (ERs) [2,3]. In comparison to banned anabolic agents like metandienone, plant-based ecdysterone demonstrated a notably strong anabolic effect in a recent study conducted on rats [2]. Nevertheless, extensive scientific investigations, particularly those involving human subjects, remain limited and scarcely available [1]. At the same time, ecdysteroids are commonly promoted as dietary supplements among athletes, with effects related to enhancing strength, supporting muscle growth during resistance training, reducing fatigue, and improving recovery. A literature review demonstrates that numerous studies have documented a broad spectrum of pharmacological effects of ecdysteroids in mammals, with the majority of these effects being beneficial to physiological function [3]. A considerable number of studies have investigated the growth-enhancing properties of ecdysterone across diverse animal species, such as rats, mice, Japanese quail, and cattle. As a result, growing concern has emerged regarding its potential misuse by athletes, prompting increased scrutiny from anti-doping authorities [2].
The literature indicates that ecdysteroids have been primarily isolated from plants such as Silene, Lyhnis, and Dianthus [4,5]. Given that some of these plants are endemic and do not grow in Uzbekistan, it was important to identify alternative local sources of phytoecdysteroids. An extensive search of the local flora revealed that plants of the Silene family are well-distributed in the region. The distribution of ecdysteroids among plant species remains poorly systematized, with no established rules governing their presence. These compounds are often detected in taxonomically diverse and unrelated plants, which complicates research efforts and suggests that their biological function is not yet clearly understood. To date, no definitive phytohormonal role has been confirmed. Nevertheless, a key observation is that certain plants can accumulate substantial amounts of ecdysteroids in specific parts of plants, such as leaves, roots, or seeds, during particular stages of their growth cycle. Plants containing ecdysteroids have been used in traditional medicine for centuries. One of the richest known sources is Leuzea carthamoides Iljin (family Asteraceae), a species native to Central Asia. This plant, which thrives under harsh environmental conditions, contains ecdysteroids in its dried roots and seeds at concentrations of 0.4% and 2%, respectively. Owing to anabolic, tonic, and other physiological effects, the pharmacological product derived from this plant, known as “Ecdisten”, has gained popularity among recreational athletes seeking rapid muscle growth and improved physical appearance [6]. In a 1996 study, Slama et al. isolated 96% pure 20-hydroxyecdysone (20E) from Leuzea seeds and tested its anabolic effects on Japanese quail. Administering 100 mg/kg of pure 20E led to a 115% increase in body mass, comparable to the 109.5% gain from seed-derived 20E equivalents. The results confirmed that Leuzea’s growth-promoting effects in vertebrates are primarily due to its ecdysteroid content [7].
Although the precise mechanism of action remains uncertain, it is generally believed that the majority of ecdysteroid-induced responses are mediated through intracellular receptor complexes—namely, the ecdysone receptor (EcR) and ultraspiracle protein (USP)—both of which are members of the nuclear receptor superfamily [8], and that influence the transcriptional activity of targeted gene clusters. However, the last research demonstrates that this group of compounds may also present non-genomic activity [9]. Several studies have explored natural ecdysteroids, though typically using limited compound sets. Consequently, structure–activity relationship (SAR) analyses are susceptible to variables such as the selected invertebrate species, its developmental stage, and the configuration of the bioassay. Moreover, common challenges inherent to biological testing—such as the purity of the test compound, its absorption, distribution, and metabolic processing—further complicate interpretation. Despite these limitations, phytoecdysteroids (PEs) have consistently shown notable hormonal activity in a range of insect-based bioassay systems. Data obtained from both in vitro [10] experiments and various in vivo assays, particularly pupariation bioassays in Dipteran species such as Calliphora and Musca [11], have led to the identification of key structural features associated with high biological activity: (a) a cis fusion between the A and B rings; (b) the presence of a 7-en-6-one moiety; and (c) a fully intact sterol side chain bearing a 22R-oriented oxygen function, occasionally accompanied by additional alkyl substitutions at the 24α-position; (d) an oxygen-containing group, typically a 3β-hydroxyl (3β-OH); and (e) hydroxyl groups at C-14α and C-2β, with additional –OH groups often found at C-20 and C-25. A more recent comprehensive investigation of 20-hydroxyecdysone (20E) conjugates—including fatty acid and benzoic acid mono-, di-, and tri-esters, as well as glycosides and glycosidic esters—using standard bioassays in Sarcophaga (Diptera) and Galleria (Lepidoptera), demonstrated that both the 2-acetyl and 25-benzoate derivatives retained considerable biological activity. In contrast, the di- and tri-ester forms, as well as all glycosidic derivatives of 20E, exhibited significantly reduced or negligible activity compared to the parent compound [12]. PEs containing an 11α-hydroxyl group demonstrate a binding affinity to the ecdysteroid receptor of Chironomus tentans (Diptera) that is comparable to that of analogs lacking this group. The capacity of these compounds to elicit a hormonal response shows a strong correlation with their receptor-binding affinity, as indicated by the stimulation of acetylcholinesterase activity involved in cellular differentiation [13]. Nevertheless, studies in living organisms indicate that even slight differences in molecular structure can produce substantially different physiological effects [14].
As an alternative to traditional biological experiments, ML-based quantitative structure–activity relationship (QSAR) analysis has emerged as a powerful tool over the past three decades for exploring the biological activities and physicochemical properties of diverse organic and natural compounds, particularly those that are challenging to isolate in adequate amounts for experimental study [15,16,17,18,19,20,21,22]. Applying this approach, the set of ecdysteroids has been studied [23]. SAR was investigated using Comparative Molecular Field Analysis (CoMFA), resulting in the development of two predictive models with strong performance characteristics [23]. Based on these models, a pharmacophore hypothesis was proposed to describe ligand interaction with the ecdysteroid receptor. According to this hypothesis, receptor binding arises from the combined contributions of several molecular features, including heteroatoms at positions C-2, C-3, C-20, and C-22; a pronounced dipole at C-6; and a moderately bulky hydrophobic group positioned beyond C-22, all arranged at spatial angles and distances similar to those in 20-hydroxyecdysone (20E). While each feature enhances binding affinity, none is individually essential. Compared to earlier SAR models based on empirical observations, it is now evident that elements such as the cis A/B ring fusion, the 7-double bond, the 6-keto group, the complete eight-carbon side chain, and specific hydroxyl substitutions are not strictly required for biological activity. However, the absence of the 7-en-6-one moiety appears to significantly reduce activity. Additionally, the effect of adding or removing specific structural elements is not always additive; for instance, the presence or absence of hydroxyl groups at C-5 or C-25 can have varying impacts depending on the overall molecular context. Although it remains uncertain whether an entirely different molecular scaffold incorporating several or all of the proposed pharmacophore features would retain biological activity, the CoMFA-derived models are already being effectively used to predict the activity of novel ecdysteroid analogs before their synthesis. [24]. Additionally, computational studies of phytoecdysteroids based on ligand–receptor binding models have further advanced understanding of the ecdysteroid receptor [23]. In a separate survey, Ravi et al. employed 4D-QSAR methodologies using the same dataset previously used for CoMFA modeling [25,26].
The current study presents a combined computational and QSAR analysis of 23 ecdysteroids isolated from various plant sources, which were studied in vivo for the activity. The primary objective is to identify the structural descriptors underlying anabolic activity (AA) and to construct a predictive QSAR model that can help in guiding the design of potent and selective anabolic agents based on the new natural ecdysteroids and ecdysteroid scaffold.
2. Materials and Methods
2.1. Dataset and Biological Data
The dataset employed in this study comprises 23 ecdysteroid compounds (see Figure 1), with anabolic activity (AA) data sourced from our previous work [27]. The anabolic activity values were determined from in vivo experiments conducted on rat models, measuring radioactivity uptake (cpm/g) in target tissues following administration of each ecdysteroid compound at 5 mg/kg. The original activity values, determined at a dosage of 5 mg/kg, were converted into logarithmic molar units (log(AA)) for use as response variables in the QSAR analysis. The molecular structures of the compounds and their corresponding experimental log(AA) values are presented in Figure 1 and Table 1, respectively.
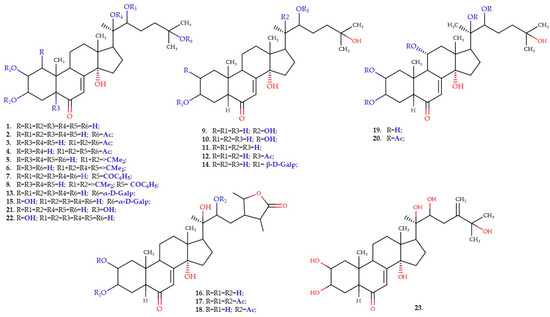
Figure 1.
The structures of the compounds that were used in the study.

Table 1.
List of ecdysteroid compounds with experimentally determined anabolic activity values (log scale).
2.2. Computational Approach
In this work, HyperChem 8 software package is applied to draw chemical structures for further optimization [28]. The RM1 (Recife Model 1) semi-empirical quantum-chemical method is used for optimizing geometries of molecules to obtain minimal energy conformations [29]. To gain deeper insight into the experimental findings, a set of quantum-chemical descriptors was calculated and utilized in the cheminformatics analysis. It includes the energies of the highest occupied molecular orbital (HOMO, indication of nucleophilicity) and the lowest unoccupied molecular orbital (LUMO, indication of electrophilicity), dipole moment (including its X, Y, and Z components), total energy, LogP (lipophilicity index), refractivity, polarizability, and atomic charges. The numerical values of these descriptors are represented in Table 2.

Table 2.
Numerical values of quantum-chemical descriptors calculated in this work.
2.3. Cheminformatics Analysis
The initial selection of predictive models was conducted using the GA-MLRA technique, which combines a genetic algorithm (GA) with multiple linear regression analysis (MLRA) [30,31], as implemented in the QSARINS v2.2.3 software package [32,33]. To convert chemical structures into numerical representations, the optimized structures were used to calculate constitutional, topological, and molecular descriptors using DRAGON 6 software package [34]. The descriptor categories incorporated in the study included: (i) functional group-based descriptors, (ii) descriptors derived from atom-centered fragments, and (iii) topological indices such as molecular walk counts [35]. Following data curation and the removal of non-informative (zero-variance) descriptors, a final set of 384 distinct descriptors was retained to characterize the chemical diversity of the studied compounds and develop a final set of QSAR models. The most effective models were selected based on several performance criteria, including a high coefficient of determination (R2) for both the training and external validation sets, low standard deviation (s), and a minimal number of descriptors to ensure model simplicity. Additionally, a high Fisher statistic (F) and the absence of multicollinearity among descriptors were considered key factors in the selection process. The final QSAR models were further validated using the leave-one-out (LOO) cross-validation method, with predictive performance assessed by the cross-validated coefficient (Q2), calculated from the predictive residual sum of squares.
To detect redundancy and avoid overrepresentation of specific chemical features, pairwise correlation coefficients were calculated for all descriptors included in the model. Descriptors exhibiting high intercorrelation (r2 > 0.9) or constant values were excluded from further analysis to prevent bias in explaining the dependent variable. Additionally, descriptors with cross-correlation coefficients exceeding 0.6 were deliberately avoided during model construction to ensure descriptor independence and model robustness.
3. Results
For model training and validation, the complete set of 23 ecdysteroid compounds was divided in an 80:20 ratio, with 19 compounds allocated to the training set and 4 compounds reserved for external validation. This division was not performed randomly; instead, it was guided by structural diversity to ensure that at least one representative from each structural class present in the training set was also included in the test set. The application of the GA-MLRA method resulted in the identification of four statistically significant models, each incorporating different descriptor subsets, that demonstrated strong predictive capability for the anabolic activity (AA) of ecdysteroids. The statistical metrics for all developed models are summarized in Table 3.

Table 3.
Statistical characteristics of the one-, two-, three-, and four-variable models.
The equation below represents the two-descriptor QSAR model:
Log [AA] = 0.808 (±0.175) SIC0 − 0.582(±0.193) G3p + 6.491 (±0.142)
The model demonstrates strong performance, exhibiting high R2 and Q2 values for both the training and test sets. Graphical illustrations of the model’s predictive ability are presented in Figure 2. A comparison of experimental and predicted log(AA) values, based on Equation (1), is provided in Table 1.
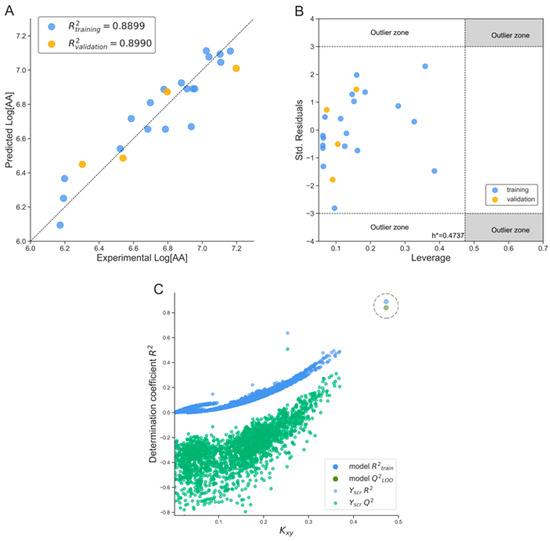
Figure 2.
Graphical evaluation of the statistical performance of the best two-descriptor QSAR model: (A) correlation plot of predicted versus experimental log(AA) values for training (blue) and validation (orange) sets; (B) Williams plot showing standardized residuals versus leverage values—all compounds lie within the model’s applicability domain (h * = 0.474); (C) Y-randomization (Y-scrambling) test confirming robustness of the model.
Descriptors definitions: SIC0—Structural Information Content index of order 0, representing the neighborhood symmetry of atoms; part of the class of information indices and G3p—Third principal component directional WHIM descriptor, weighted by atomic polarizability; belongs to the WHIM (Weighted Holistic Invariant Molecular) descriptor family. This model highlights the positive contribution of the first descriptor and the negative contribution of the second descriptor to the predicted anabolic activity (log[AA]), with a high level of statistical confidence in the coefficients.
In addition to the two-variable model, two other models with three and four descriptors were also developed.
The model with three descriptors is represented by Equation (2) as follows:
Log [AA] = 0.412 (±0.217) GATS5e − 0.454 (±0.219) G3p − 0.830 (±0.210) ALOGP2 + 6.957(±0.182)
This model exhibits excellent fit for both training and test sets. A graphical representation is provided in Figure S1 of the Supplementary Information.
Equation (3) represents a model with four descriptors as follows:
Log [AA] = 0.643 (±0.367) Mp − 0.595 (±0.923) TIE − 0.480 (±0.169) G3p − 1.188 (±0.629) ALOGP + 7.337 (±0.116)
Among all developed models, the four-variable model yields the highest R2 value for the training set and performs well in predicting the external test set. The corresponding plot is shown in Figure S2 of the Supplementary Information.
Overall, all models demonstrate similarly strong statistical performance and predictive accuracy. However, the three-variable model (Equation (2)) provides the highest external predictive ability with Rext2 = 0.97. Nevertheless, it is important to highlight that the two-variable model (Model 1) achieved the best y-randomization (y-scrambling) validation results, indicating superior robustness. Therefore, Model 1 may be considered the most reliable and generalizable model for predicting the anabolic activity of ecdysteroids.
4. Discussion
In the context of the developed QSAR models, a detailed examination of the selected molecular descriptors provides insight into the key structural and physicochemical features that govern anabolic activity in ecdysteroids. As presented in the Results section, the most statistically significant models incorporated the following descriptors: SIC0, a measure of structural information content that reflects zero-order neighborhood symmetry; G3p, a third-order WHIM (Weighted Holistic Invariant Molecular) descriptor associated with molecular symmetry and weighted by atomic polarizability; and Mp, representing the mean atomic polarizability normalized to a carbon atom, falling under the category of constitutional descriptors. Additionally, the TIE descriptor captures E-state topological information, accounting for the electronic environment of atoms in the molecular topology. GATS5e is a 2D autocorrelation descriptor indicating the spatial distribution of Sanderson electronegativity at a lag of 5 bonds. Finally, the lipophilicity of molecules is characterized by ALOGP (Ghose-Crippen logP) and ALOGP2 (its squared form), both of which relate to the molecule’s hydrophobic character and are essential for membrane permeability and receptor binding. These descriptors collectively highlight the relevance of electronic distribution, molecular shape, symmetry, and hydrophobicity in modulating the anabolic activity of ecdysteroids.
The first selected descriptor, SIC0, belongs to the class of information content indices. These descriptors are derived from the molecular graph and quantify structural complexity by calculating the distribution of equivalence classes within the molecule [35]. This information-based descriptor incorporates neighborhood symmetry as well as data related to neighbor degree and edge multiplicity within the molecular graph. According to the regression coefficient, an increase in the SIC0 value correlates positively with enhanced anabolic activity (AA) in ecdysteroids. In the selected QSAR model, SIC0 emerges as a key positive contributor to biological activity. Notably, the lead compounds (19, 21, and 22) exhibit the highest SIC0 values, aligning with their superior anabolic potential. In contrast, compound 6, which demonstrates the lowest biological activity, is characterized by a significantly smaller SIC0 value.
As observed, the descriptor G3p is included in all three of the discussed models, and its contribution is substantial based on the magnitude of its regression coefficient. G3p refers to the third component, directional WHIM index, weighted by atomic polarizability, and is categorized under the WHIM (Weighted Holistic Invariant Molecular) descriptors. These descriptors capture the three-dimensional molecular geometry and encode information about molecular size, shape, symmetry, and atom-related properties—such as polarizability—which are crucial for understanding interactions with biological targets [36]. WHIM descriptors are molecular descriptors based on statistical indices calculated on the projections of the atoms along principal axes [37]. WHIM descriptors are designed to capture essential three-dimensional (3D) molecular information such as size, shape, symmetry, and atomic distribution relative to fixed reference axes. Specifically, directional WHIM symmetry descriptors γl, γ2 and γ3, represent the degree of symmetry along each principal axis of the molecule. These are calculated as the mean information content associated with atomic distribution relative to the center of the score values [38]. The symmetry index for the m-th component is calculated using the following formula:
where ns is the number of atoms symmetrically distributed along the m-th component, na is the number of asymmetrically distributed atoms, and n is the total number of atoms in the molecule. A higher γm value implies greater symmetry, and these values are used as part of the WHIM descriptor set to encode structural features relevant to molecular recognition and activity. Thus, according to the model and regression coefficient, the increase in G3p value in ecdysteroids decreases the AA potency.
While descriptors such as SIC0 and G3p are mathematical constructs derived from molecular graphs and 3D geometry, they can be interpreted in a chemically meaningful way. SIC0 is a measure of structural information content, which increases with greater atomic diversity and topological complexity—often associated with branching or asymmetry in the molecule. Thus, analogs with greater substitution patterns or higher degrees of structural variation near active sites may exhibit elevated SIC0 values. Conversely, a lower G3p value implies reduced 3D symmetry or non-uniform polarizability distribution. Therefore, in practical terms, designing analogs with asymmetrical polarizable groups, such as selective acylation, esterification, or hydroxyl substitutions, reduces G3p while increasing SIC0. These structural modifications are feasible via semisynthetic derivatization of natural ecdysteroids. Hence, although abstract in formulation, these descriptors point toward tangible molecular changes that can be implemented and tested in future analog design efforts.
Mp is a mean atomic polarizability descriptor (scaled on carbon atom) is among constitutional indices [39]. The descriptor is synergistic with the G3p descriptor, as G3p is also weighted by polarizability.
The TIE descriptor represents an electrotopological state (E-state), i.e., a topological parameter and belongs to the class of topological indices. It encodes information about the molecule’s topology and atomic electronic environment, contributing to the model by capturing features related to molecular connectivity and electronic distribution [40].
Next descriptor, GATS5e, is Geary autocorrelation of lag 5 weighted by the Sanderson electronegativity descriptor, belongs to 2D autocorrelation descriptors [41]. The presence of this descriptor confirms that electronegativity plays an important role in anabolic activity exhibited by ecdysteroid compounds. These results are in accordance with those displayed in the dendrogram in Figure 3. As can be seen in the picture there are three main clusters of compounds in the dendrogram. In the first cluster (left side) most compounds are with the lowest activity and sharing a maximum common substructure (MSC) displayed as MCS1. In the case of the third cluster (right side) the compounds with the highest activity are included, and the MSC3 has the same features as the MSC_ecdysteroids for the whole dataset. In the bottom part are included the two lowest active compounds (6 and 11) and the two highest active natural products (18 and 20) where the last ones have the main differences with the MSC related to the presence of (O-C=O) groups showing a positive relationship to the activity corroborating that the increase in electronegativity given by the amount of carbonyl groups plays a fundamental role in the activity.
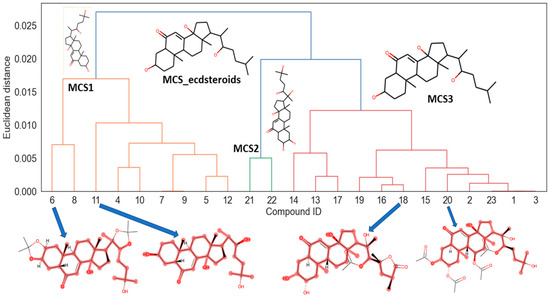
Figure 3.
Hierarchical cluster analysis of the dataset and maximum common substructure search. MCS1: Maximum common substructure in cluster 1. MCS2: Maximum common substructure in cluster 2. MCS3: Maximum common substructure in cluster 3.
Two other descriptors—ALOGP and ALOGP2 (squared ALOGP)—are octanol-water partition descriptors that encode the lipophilicity property of molecules, which are responsible for the solubility of compounds in water [42,43]. The presence of these types of descriptors confirms the importance of lipophilicity factors or water solubility in the exhibition of anabolic activity by ecdysteroid compounds.
Interestingly, none of the quantum-mechanical (QM) descriptors, such as HOMO, LUMO, dipole moments, or total energy, were retained in any of the final regression models. This outcome suggests that the variability in anabolic activity across the studied ecdysteroids is more effectively captured by topological, structural, and physicochemical descriptors rather than electronic properties or processes. One main explanation is the significant influence of structural factors. Furthermore, many QM descriptors tend to be highly correlated with other types of structural descriptors, such as WHIM or polarizability-based indices, causing them to be excluded by the genetic algorithm during variable selection. Thus, while QM descriptors provide valuable molecular insights, their exclusion from the final models highlights that simpler structural/topological descriptors offer sufficient predictive power for this particular chemical series.
Overall, it can be concluded that the polarizability, electronegativity, and lipophilicity properties of the molecules, together with structural factors, play a significant role in the AA potency exhibition of investigated ecdysteroid compounds. Future efforts will focus on sourcing or synthesizing novel ecdysteroid analogs that optimize key molecular features, such as high polarizability, favorable lipophilicity, and balanced topological symmetry, based on the present QSAR findings. Further in vivo and in vitro validation studies are planned to assess their anabolic potential and explore additional pharmacological properties.
5. Conclusions
A comprehensive QSAR study was conducted on a set of 23 ecdysteroid compounds to investigate and predict their anabolic activity (AA). The modeling framework employed genetic algorithms for descriptor selection and multiple linear regression analysis for model construction. Molecular mechanics and quantum-chemical computations were initially employed to optimize molecular structures and generate physicochemical descriptors for further modeling.
Three best predictive models were developed using two, three, and four descriptors, respectively. The two-variable model demonstrated the best balance of transparence, simplicity and predictive power, yielding squared correlation coefficients of R2 = 0.89 for the training set and R2 = 0.84 for the test set. William’s plot confirmed that all compounds fall within the model’s applicability domain; furthermore, y-scrambling validation verified the robustness and reliability of the model. The two key descriptors identified in the optimal model were SIC0, a structural information content index reflecting molecular symmetry, and G3p, a directional WHIM descriptor weighted by polarizability. These descriptors were found to be significant contributors to anabolic potency, offering structural-level insights into the biological activity of the compounds.
Overall, the results of this study indicate that polarizability, electronegativity, lipophilicity, and topological symmetry are critical molecular features influencing anabolic activity. The two-variable model (Model 1) thus represents a robust and interpretable QSAR tool for estimating the anabolic potential of newly synthesized or virtual ecdysteroid derivatives belonging to this functional class.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/computation13080195/s1, Figure S1. Graphical representation of statistical performance of 3-variable model; Figure S2. Graphical representation of statistical performance of the 4-variable model.
Author Contributions
Conceptualization, D.U., G.M.C.-M., V.S. and B.R.; methodology, D.U., U.Y., G.M.C.-M. and B.R.; software, D.U., U.Y. and G.M.C.-M.; validation, D.U., U.Y. and G.M.C.-M.; formal analysis, D.U., U.Y., V.S., G.M.C.-M. and B.R.; investigation, D.U., U.Y., G.M.C.-M. and B.R.; resources, D.U., V.S. and B.R.; data curation, D.U., U.Y. and G.M.C.-M.; writing—original draft preparation, D.U. and U.Y.; writing—review and editing, D.U., U.Y., V.S., G.M.C.-M. and B.R.; visualization, D.U., U.Y. and G.M.C.-M.; supervision, V.S. and B.R.; project administration, V.S. and B.R.; funding acquisition, V.S. and B.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported in part by National Science Foundation (NSF) [MRI Award number 2019077].
Data Availability Statement
The original contributions presented in this study are included in the article. Further inquiries can be directed to the corresponding author(s).
Acknowledgments
D.U. thank the Ministry of Innovation Development of the Republic of Uzbekistan and the Fund to Support Innovative Development and Innovative ideas for the travel grant provided. Authors thank Paola Gramatica for generously providing a free license for the QSARINS software. This work used resources of the Center for Computationally Assisted Science and Technology (CCAST) at North Dakota State University, which was made possible in part by the National Science Foundation (NSF) [MRI Award number 2019077]. Supercomputing support provided by the CCAST HPC System at NDSU is gratefully acknowledged. D.U. thanks the Department of Coatings and Polymeric Materials (NDSU) for providing training and computer facilities to perform this project.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Isenmann, E.; Ambrosio, G.; Joseph, J.F.; Mazzarino, M.; de la Torre, X.; Zimmer, P.; Kazlauskas, R.; Goebel, C.; Botrè, F.; Diel, P.; et al. Ecdysteroids as non-conventional anabolic agent: Performance enhancement by ecdysterone supplementation in humans. Arch. Toxicol. 2019, 93, 1807–1816. [Google Scholar] [CrossRef]
- Parr, M.K.; Botrè, F.; Naß, A.; Hengevoss, J.; Diel, P.; Wolber, G. Ecdysteroids: A novel class of anabolic agents? Biol. Sport 2015, 32, 169–173. [Google Scholar] [CrossRef]
- Syrov, V.N. Comparative experimental investigation of the anabolic activity of phytoecdysteroids and steranabols. Pharm. Chem. J. 2000, 34, 193–197. [Google Scholar] [CrossRef]
- Yusupova, U.Y.; Usmanov, D.A.; Ramazonov, N.S. Phytoecdysteroids from the Plant Dianthus helenae. Chem. Nat. Compd. 2019, 55, 393–394. [Google Scholar] [CrossRef]
- Yusupova, U.Y.; Usmanov, D.A.; Ramazonov, N.S. Phytoecdysteroids from the Aerial Part of Silene popovii. Chem. Nat. Compd. 2020, 56, 562–563. [Google Scholar] [CrossRef]
- Wilborn, C.D.; Taylor, L.W.; Campbell, B.I.; Kerksick, C.; Rasmussen, C.J.; Greenwood, M.; Kreider, R.B. Effects of methoxyisoflavone, ecdysterone, and sulfo-polysaccharide supplementation on training adaptations in resistance-trained males. J. Int. Soc. Sports Nutr. 2006, 3, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Sláma, K.; Koudela, K.; Tenora, J.; Maťhová, A. Insect hormones in vertebrates: Anabolic effects of 20-hydroxyecdysone in Japanese quail. Experientia 1996, 52, 702–706. [Google Scholar] [CrossRef] [PubMed]
- Yao, T.-P.; Forman, B.M.; Jiang, Z.; Cherbas, L.; Chen, J.D.; McKeown, M.; Cherbas, P.; Evans, R.M. Functional ecdysone receptor is the product of EcR and Ultraspiracle genes. Nature 1993, 366, 476–479. [Google Scholar] [CrossRef]
- Tomaschko, K.-H. Nongenomic effects of ecdysteroids. Arch. Insect Biochem. Physiol. 1999, 41, 89–98. [Google Scholar] [CrossRef]
- Cherbas, L.; Yonge, C.D.; Cherbas, P.; Williams, C.M. The morphological response of Kc-H cells to ecdysteroids: Hormonal specificity. Wilhelm Roux’s Arch. Dev. Biol. 1980, 189, 1–15. [Google Scholar] [CrossRef]
- Lafont, R.; Dauphin–Villemant, C.; Warren, J.T.; Rees, H. Ecdysteroid Chemistry and Biochemistry. Compr. Mol. Insect Sci. 2005, 3, 125–195. [Google Scholar] [CrossRef]
- Sláma, K.; Abubakirov, N.K.; Gorovits, M.B.; Baltaev, U.A.; Saatov, Z. Hormonal activity of ecdysteroids from certain asiatic plants. Insect Biochem. Mol. Biol. 1993, 23, 181–185. [Google Scholar] [CrossRef]
- Spindler-Barth, M.; Quack, S.; Rauch, P.; Spindler, K.-D. Biological effects of muristerone A and turkesterone on the epithelial cell line from Chironomus tentans (Diptera: Chironomidae) and correlation with binding affinity to the ecdysteroid receptor. Eur. J. Entomol. 1997, 94, 161–166. [Google Scholar]
- Clément, C.Y.; Bradbrook, D.A.; Lafont, R.; Dinan, L. Assessment of a microplate-based bioassay for the detection of ecdysteroid-like or antiecdysteroid activities. Insect Biochem. Mol. Biol. 1993, 23, 187–193. [Google Scholar] [CrossRef]
- Jagiello, K.; Grzonkowska, M.; Swirog, M.; Ahmed, L.; Rasulev, B.; Avramopoulos, A.; Papadopoulos, M.G.; Leszczynski, J.; Puzyn, T. Advantages and limitations of classic and 3D QSAR approaches in nano-QSAR studies based on biological activity of fullerene derivatives. J. Nanoparticle Res. Interdiscip. Forum Nanoscale Sci. Technol. 2016, 18, 256. [Google Scholar] [CrossRef]
- Juretic, D.; Kusic, H.; Dionysiou, D.D.; Rasulev, B.; Loncaric Bozic, A. Modeling of photooxidative degradation of aromatics in water matrix; combination of mechanistic and structural-relationship approach. Chem. Eng. J. 2014, 257, 229–241. [Google Scholar] [CrossRef]
- Rasulev, B. Recent Developments in 3D QSAR and Molecular Docking Studies of Organic and Nanostructures. In Handbook of Computational Chemistry; Springer: Dordrecht, The Netherlands, 2016; pp. 1–29. [Google Scholar] [CrossRef]
- Turabekova, M.A.; Rasulev, B.F.; Dzhakhangirov, F.N.; Salikhov, S.I. Aconitum and Delphinium alkaloids: “Drug-likeness” descriptors related to toxic mode of action. Environ. Toxicol. Pharmacol. 2008, 25, 310–320. [Google Scholar] [CrossRef]
- Turabekova, M.A.; Rasulev, B.F.; Dzhakhangirov, F.N.; Leszczynska, D.; Leszczynski, J. Aconitum and Delphinium alkaloids of curare-like activity. QSAR Anal. Mol. Docking Alkaloids Into AChBP. Eur. J. Med. Chem. 2010, 45, 3885–3894. [Google Scholar] [CrossRef]
- Toropova, A.P.; Toropov, A.A.; Rasulev, B.F.; Benfenati, E.; Gini, G.; Leszczynska, D.; Leszczynski, J. QSAR models for ACE-inhibitor activity of tri-peptides based on representation of the molecular structure by graph of atomic orbitals and SMILES. Struct. Chem. 2012, 23, 1873–1878. [Google Scholar] [CrossRef]
- Toropov, A.A.; Toropova, A.P.; Rasulev, B.F.; Benfenati, E.; Gini, G.; Leszczynska, D.; Leszczynski, J. Coral: QSPR modeling of rate constants of reactions between organic aromatic pollutants and hydroxyl radical. J. Comput. Chem. 2012, 33, 1902–1906. [Google Scholar] [CrossRef] [PubMed]
- Usmanov, D.; Rasulev, B.; Syrov, V.; Yusupova, U.; Ramazonov, N. Structure-Hepatoprotective Activity Relationship Study of Iridoids. Int. J. Quant. Struct.-Prop. Relatsh. 2020, 5, 108–118. [Google Scholar] [CrossRef]
- Wurtz, J.M.; Guillot, B.; Fagart, J.; Moras, D.; Tietjen, K.; Schindler, M. A new model for 20-hydroxyecdysone and dibenzoylhydrazine binding: A homology modeling and docking approach. Protein Sci. A Publ. Protein Soc. 2000, 9, 1073–1084. [Google Scholar] [CrossRef] [PubMed]
- Dinan, L. Ecdysteroid Structure-Activity Relationships. Bioact. Nat. Prod. 2003, 29 Pt. J, 3–71. [Google Scholar] [CrossRef]
- Dinan, L.; Sarker, S.D.; Bourne, P.; Whiting, P.; Ik, V.; Rees, H.H. Phytoecdysteroids in seeds and plants of Rhagodia baccata (Labill.) Moq. (Chenopodiaceae). Arch. Insect Biochem. Physiol. 1999, 41, 18–23. [Google Scholar] [CrossRef]
- Dinan, L.; Hormann, R.E.; Fujimoto, T. An extensive ecdysteroid CoMFA. J Comput Aided Mol Des 1999, 13, 185–207. [Google Scholar] [CrossRef]
- Syrov, V.N.; Saatov, Z.; Sagdullaev, S.S.; Mamatkhanov, A.U. Study of the Structure—Anabolic Activity Relationship for Phytoecdysteroids Extracted from Some Plants of Central Asia. Pharm. Chem. J. 2001, 35, 667–671. [Google Scholar] [CrossRef]
- Coleman, W.F.; Arumainayagam, C.R. HyperChem 5 (by Hypercube, Inc.). J. Chem. Educ. 1998, 75, 416. [Google Scholar] [CrossRef][Green Version]
- Rocha, G.B.; Freire, R.O.; Simas, A.M.; Stewart, J.J.P. RM1: A reparameterization of AM1 for H, C, N, O, P, S, F, Cl, Br, and I. J. Comput. Chem. 2006, 27, 1101–1111. [Google Scholar] [CrossRef]
- Davis, L. Handbook of Genetic Algorithms; Van Nostrand Reinhold: New York, NY, USA, 1991. [Google Scholar]
- Devillers, J. Genetic Algorithms in Computer-Aided Molecular Design. In Genetic Algorithms in Molecular Modeling; Academic Press: Cambridge, MA, USA, 1996; pp. 1–34. [Google Scholar] [CrossRef]
- Gramatica, P.; Cassani, S.; Chirico, N. QSARINS-chem: Insubria datasets and new QSAR/QSPR models for environmental pollutants in QSARINS. J. Comput. Chem. 2014, 35, 1036–1044. [Google Scholar] [CrossRef]
- Gramatica, P.; Chirico, N.; Papa, E.; Cassani, S.; Kovarich, S. QSARINS: A new software for the development, analysis, and validation of QSAR MLR models. J. Comput. Chem. 2013, 34, 2121–2132. [Google Scholar] [CrossRef]
- Toddeschini, R.; Consonni, V.; Mauri, A.; Pavan, M. Dragon Software for the Calculation of Molecular Descriptors, Version 6 for Windows; Talete SRL: Milan, Italy, 2014. [Google Scholar]
- Roy, A.B.; Basak, S.C.; Harriss, D.K.; Magnuson, V.R. NEIGHBORHOOD COMPLEXITIES AND SYMMETRY OF CHEMICAL GRAPHS AND THEIR BIOLOGICAL APPLICATIONS. In Mathematical Modelling in Science and Technology; Pergamon Press: Oxford, UK, 1984; pp. 745–750. [Google Scholar] [CrossRef]
- Najafi, A.; Sobhanardakani, S.; Marjani, M. Exploring QSAR for Antimalarial Activities and Drug Distribution within Blood of a Series of 4-Aminoquinoline Drugs Using Genetic-MLR. J. Chem. 2013, 2013, 560415. [Google Scholar] [CrossRef]
- Todeschini, R.; Lasagni, M.; Marengo, E. New molecular descriptors for 2D and 3D structures. Theory. J. Chemom. 1994, 8, 263–272. [Google Scholar] [CrossRef]
- Todeschini, R.; Gramatica, P. 3D-modelling and Prediction by WHIM Descriptors. Part 5. Theory Development and Chemical Meaning of WHIM Descriptors. Quant. Struct.-Act. Relatsh. 1997, 16, 113–119. [Google Scholar] [CrossRef]
- Akbar, J.; Iqbal, S.; Batool, F.; Karim, A.; Chan, K.W. Predicting retention times of naturally occurring phenolic compounds in reversed-phase liquid chromatography: A Quantitative Structure-Retention Relationship (QSRR) approach. Int. J. Mol. Sci. 2012, 13, 15387–15400. [Google Scholar] [CrossRef]
- Voelkel, A. Structural descriptors in organic chemistry—New topological parameter based on electrotopological state of graph vertices. Comput. Chem. 1994, 18, 1–4. [Google Scholar] [CrossRef]
- Ambre, P.; Wavhale, R.; Coutinho, E. New Horizons in Antimalarial Drug Discovery in the Last Decade by Chemoinformatic Approaches. Comb. Chem. High Throughput Screen. 2015, 18, 129–150. [Google Scholar] [CrossRef]
- Ghose, A.K.; Crippen, G.M. Atomic physicochemical parameters for three-dimensional-structure-directed quantitative structure-activity relationships. 2. Modeling dispersive and hydrophobic interactions. J. Chem. Inf. Comput. Sci. 1987, 27, 21–35. [Google Scholar] [CrossRef]
- Ghose, A.K.; Crippen, G.M. Atomic Physicochemical Parameters for Three-Dimensional Structure-Directed Quantitative Structure-Activity Relationships I. Partition Coefficients as a Measure of Hydrophobicity. J. Comput. Chem. 1986, 7, 565–577. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).