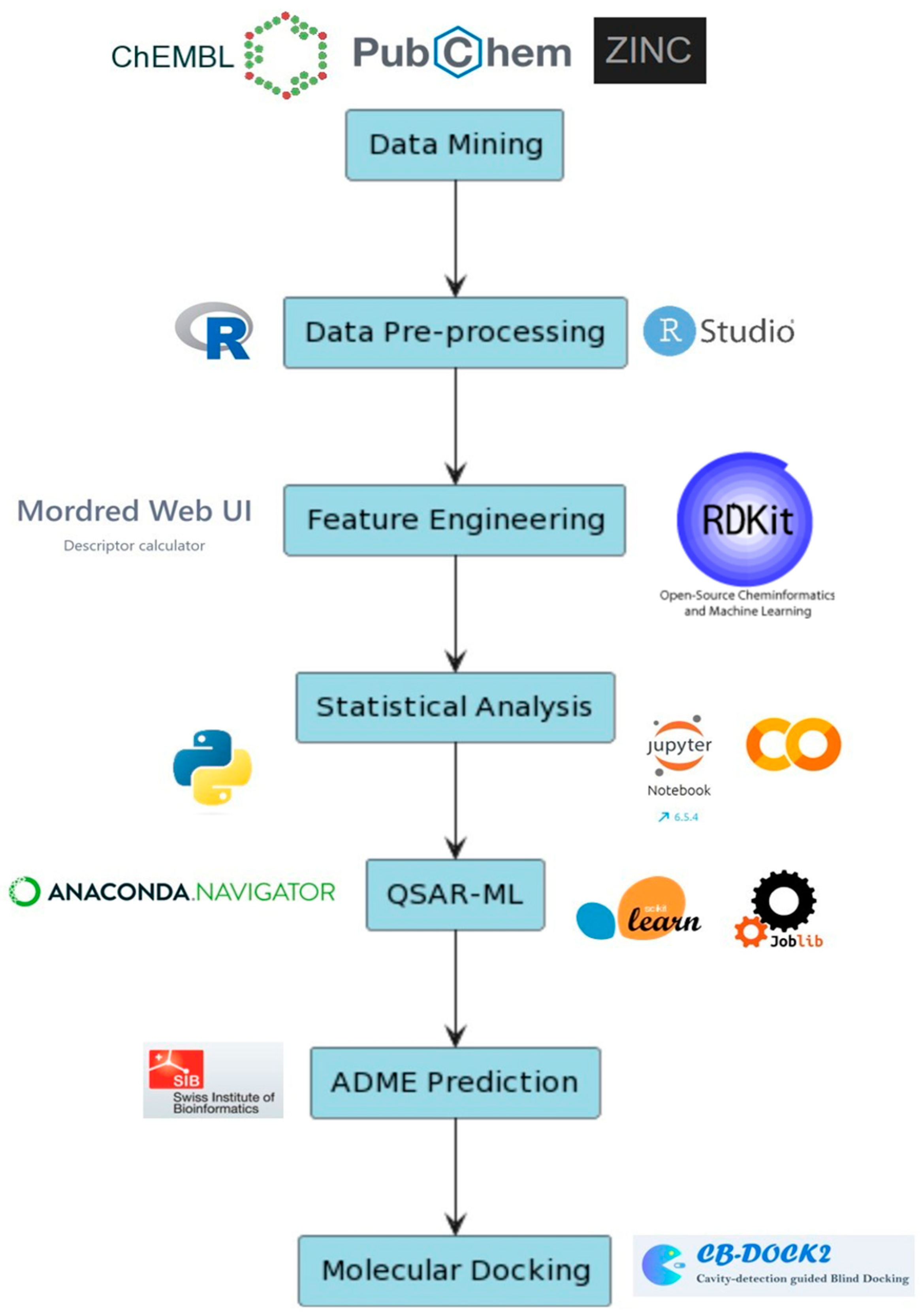
1. Introduction
Liver disease is regarded as a global health concern, contributing to a substantial amount of both mortality and morbidity worldwide. Within the Philippines, it has been implicated in 27.3 cases per 1000 deaths, underlining the gravity of the issue. Notably, hepatitis b, hepatitis c, and other viral infections targeting the liver inflict a substantial burden on low-income countries such as the Philippines. The nation faces challenges in this regard, including limited surveillance, epidemiological studies, and clinical practice guidelines, highlighting clinical and medical deficiencies in addressing liver diseases [
1]. Analysis of over 144,000 blood screening results for hepatitis c virus (HCV) revealed prevalence rates of 0.3% among hospital-based blood donors and 0.9% among overseas Filipino worker (OFW) candidates [
2]. These findings are in parallel with the global prevalence of viremic HCV infection ranging from 0.7% to 1.8% [
3,
4]. Additionally, risk factors such as unscreened blood transfusions, unsafe injections, and hemodialysis contribute to HCV infections [
5]. The use of serological tests, primarily utilizing anti-HCV immunoassays among blood donors, serves as the primary screening method [
1,
6]. It is advised to utilize direct-acting antivirals (DAAs) as the primary approach for managing HCV. Combinations of the antiviral medicine sofosbuvir/daclatasvir or sofosbuvir/velpatasvir were the recommended antiviral drugs in the country [
1,
7,
8]. Since HCV is a multistep process, it requires an entry factor. Various host-mediating factors play into the viral entry of HCV. The tight-junction protein (TJ) claudin-1 (CLDN1) plays a vital role in mediating HCV entrance. It is necessary for cell line infections in humans (hepatoma) and represents the first gateway protein that becomes vulnerable to HCV in cell line expressions (non-hepatocellular). It is also required in the late stage of the entry process [
9,
10]. Intestinal epithelial TJ proteins such as CLDN1 are responsible for these processes. Disruption of CLDN1 through pathogenic exposure relies on two main pathways; one of the pathways is rho-associated protein kinase 1 (ROCK1) as it regulates multiple cellular functions. Moderate activity of these kinases is linked closely to TJ protein formations and arrangement. Thus, excess activity of ROCK1/2 leads to defects in TJ protein pathogenesis such as inflammation and ethanol interruption [
11,
12,
13]. The FDA-approved drug for HCV treatment dasabuvir has anti-tumor effects and suppresses esophageal squamous cell carcinoma (ESCC) proliferation. It was also revealed that the drug is an inhibitor of the enzyme ROCK1. The drug dasabuvir inhibits the kinase activity of ROCK1 by reducing the activation of ERK1/2 through phosphate addition and decreasing the expression of CDK4 and CD1. This demonstrates dasabuvir’s effectiveness in blocking the ROCK1 signaling pathway [
14]. Understanding ROCK1’s structural information can lead to developing and adding potential inhibitors that can aid in therapeutic strategies, looking for specific molecule structures and relevant properties that may enhance and provide a more robust screening phase for selecting the compounds with accepted inhibitory properties against ROCK1. However, despite having DAAs, HCV infection still poses a major problem for liver diseases globally and domestically. Inhibition of these HCV entry factors can prevent chronic infection and prolonged liver damage. The continued search for inhibitor compounds and suppression of specific target pathways provides us with an approach to develop HCV antiviral strategies.
Older methods that manually isolate and characterize molecules of particular interest and evaluate their biological activity are tedious and consume considerable amounts of time, effort, and research expenses. Nevertheless, harnessing modern technology that is enabled by computers will help propel developments in drug discovery for HCV infections. Virtual screening is one of the most efficient and cost-effective techniques for finding such novel compounds. The identification of potential compounds from a chemical database is a characteristic of a structure-based drug development approach. These compounds are taken into consideration for their bioactivity and binding effects on the target protein structure. The number of compounds may be in the range of (
n > 1000); thus, computer-based screening techniques are employed to filter out undesirable compounds and would significantly reduce the cost and time it takes to pick out only the top compounds that may elicit inhibitory effects on the target of interest for drug discovery. Technological advancements have enabled computers to predict bioactivity through quantitative structure–activity relationship (QSAR)-based machine learning (ML) of novel compounds [
15,
16,
17]. In the context of the study, ROCK1’s half-maximal inhibitory concentration (IC
50) bioactivity is used for QSAR implementation. The relationship between the generated QSAR features can be established to effectively train a dataset using inhibitors for pattern recognition by use of machine learning (ML) techniques, and the bioactivity of the compounds can be predicted as a function of the features using ML models [
18]. These techniques demonstrated that developing a validated ML model has significance in drug discovery. Other studies have implemented QSAR for HCV, targeting inhibitors such as HCV NS5B polymerase, NS3, NS3/4A, and NS3 GT-3a protease inhibitors [
18,
19,
20,
21]. QSAR-ML is then combined with molecular docking as a way to further increase the study’s efficacy of effectively screening the potential compounds that were calculated from the predictive capabilities of the model from QSAR-ML.
In the case of ROCK1 inhibition, identification, and classification of these potential candidates for their desirable inhibitory properties to effectively screen the compounds in an abundant dataset, challenges such as emerging drug-resistant HCV variants are what fuel the search for continuous exploratory inhibitors of ROCK1. With these, we would enhance our current existing therapeutic capabilities, provide alternative methods of treatment, and lower the high costs of such treatments, especially in low-income countries such as the Philippines.
IC
50 is a term used to describe the drug dosage required to suppress a particular biological or biochemical activity by half. It varies based on the specific conditions of the assay used to measure it. This measurement is crucial in drug discovery as it offers an indication of the drug’s potency against its target. Methods in determining the IC
50 of a compound of pharmacological interest are based on the said assay conditions that use whole cell systems. IC
50 is a valuable point of information for drug potency; however, it is still limited to the experimental cell lines used and may not inhibit specific compound interactions [
22,
23]. A limited number of studies have been performed on actual drugs that specifically target ROCK1 for HCV treatment since most DAAs for HCV are used in conjunction with other drugs; also, a lot of the DAAs used for HCV treatment are against other targets.
The primary aim of this study is to efficiently narrow down and evaluate ROCK1 inhibitor compounds based on their bioactivity (pIC50) values within the context of HCV treatment. The objective is to gain insights into the type, structures, molecular properties, drug-likeness candidacy, and potential ROCK1 binding sites for drug affinity selection. The study employs QSAR-based ML modeling as a robust tool to uncover these findings. Utilizing feature engineering and ML techniques, the research screens and predicts the top compounds that exhibit optimal inhibitory effects, emphasizing reliable and critical features as key criteria for drug discovery. The QSAR-ML approach is complemented with ADME prediction and molecular docking of the compounds. This comprehensive methodology aims to solidify the assessment of whether these compounds possess drug-like properties, feature structures relevant for comparative analysis with established DAAs, and, crucially, if they prove to be effective ROCK1 inhibitors for targeting HCV. This varied approach enhances the understanding and selection of promising compounds with potential therapeutic efficacy in the treatment of HCV through ROCK1 inhibition.
4. Discussion
NS5B polymerase, a non-structural protein, functions as a viral polymerase for the replication process as a catalyst for synthesizing RNA based on an RNA template, generating new strands for viral replication. It converts the viral RNA into complementary RNA molecules within the replication process, thus allowing HCV to persist within the infected cells. Noviandy et al. used a QSAR-based stacked ensemble classifier to predict NS5B inhibitors using their IC
50 [
18]. NS3 protease, also a non-structural protein, alongside the viral cofactor NS4A peptide are also essential members for the HCV replication complexes. The NS3/4A protease has two domains: an N-terminal serine protease (AA 1-180) and an RNA helicase (AA 181-631) in the C-Terminal, which binds to the nucleic acid chains. The serine protease is considered one of the most promising HCV targets for drug development. Unfortunately, rapidly emerging resistant mutations within NS3/4A are reducing the sensitivity of drugs to protease inhibitors [
19]. Ghiasi et al. developed robust and reliable QSAR models using NS3/4A protease inhibitors for IC
50 prediction using the Monte Carlo technique [
20]. cAMP-dependent protein kinase (PKA) phosphorylation is vital for the regulation of SR-BI expression in the liver. The inhibition of PKA can lead to the redistribution of CLDN1 from the plasma membrane and a reduction in viral entry, thus confirming its importance [
30]. As described in the following study, inhibitors from NS5B polymerase (CHEMBL5375), NS3 (CHEMBL1293269), and NS3/4A (CHEMBL2095231) protease and the external validation dataset of cAMP-dependent protein kinase (PKA) enzyme are the basis for inclusion in building and externally validating the model.
In the context of HCV viral infection, a crucial aspect is the requirement for an entry factor. CLDN1 is highly present in the liver and is one of the key factors for entry, along with SRB1, CD81, and occludin. Evans et al. showed that expressing claudin-1 in non-hepatic cell lines makes them vulnerable to HCV infection. Claudin-1’s involvement is integral not only as an entry factor but also in the late-stage process following interaction with HCV and CD81 [
9,
10]. HCV particles specifically bind to the entry factors SR-BI and CD81 tetraspanin. The binding to CD81 triggers a diffusion of the virion-CD81 complex across the plasma membrane, heading towards sites for viral internalization. Consequently, claudin-1 plays a crucial role in the late-stage entry processes by interacting with CD81, especially in areas enriched with tetraspanin. CD81–claudin-1 complex actively participates in HCV endocytosis. These observations underscore the significant role of TJ proteins, particularly claudin-1, in the viral entry of HCV. However, much remains to be explored regarding the mechanisms of these proteins, and their regulation in the liver is yet to be fully understood. Limited information is available on claudin-1 trafficking and its interactions with other factors, such as CD81 and OCLN [
62]. One of the main pathways for TJ disruption is ROCK1; this is also called the Rho/ROCK pathway. ROCK1 and ROCK2 are both downstream effectors that regulate multiple cellular functions, e.g., cell migration, adhesion, polarity, and proliferation. When ROCK1 activity levels are moderate, the formation and arrangement of TJ proteins occur. However, elevated levels lead to TJ defects. This pathway is key in molecular signal transduction. They are involved in TJ formation, intestinal permeability, and inflammation. ROCK1 is thus considered a key protein in the said pathway [
29,
63,
64].
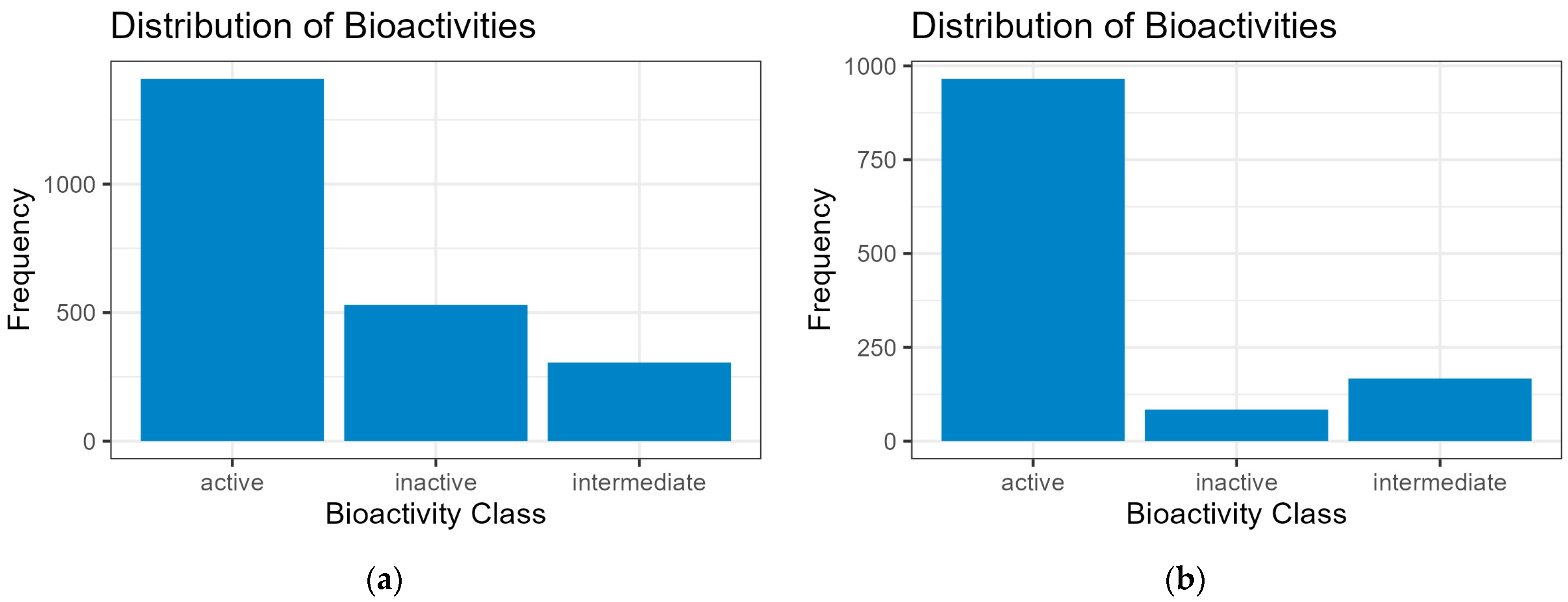
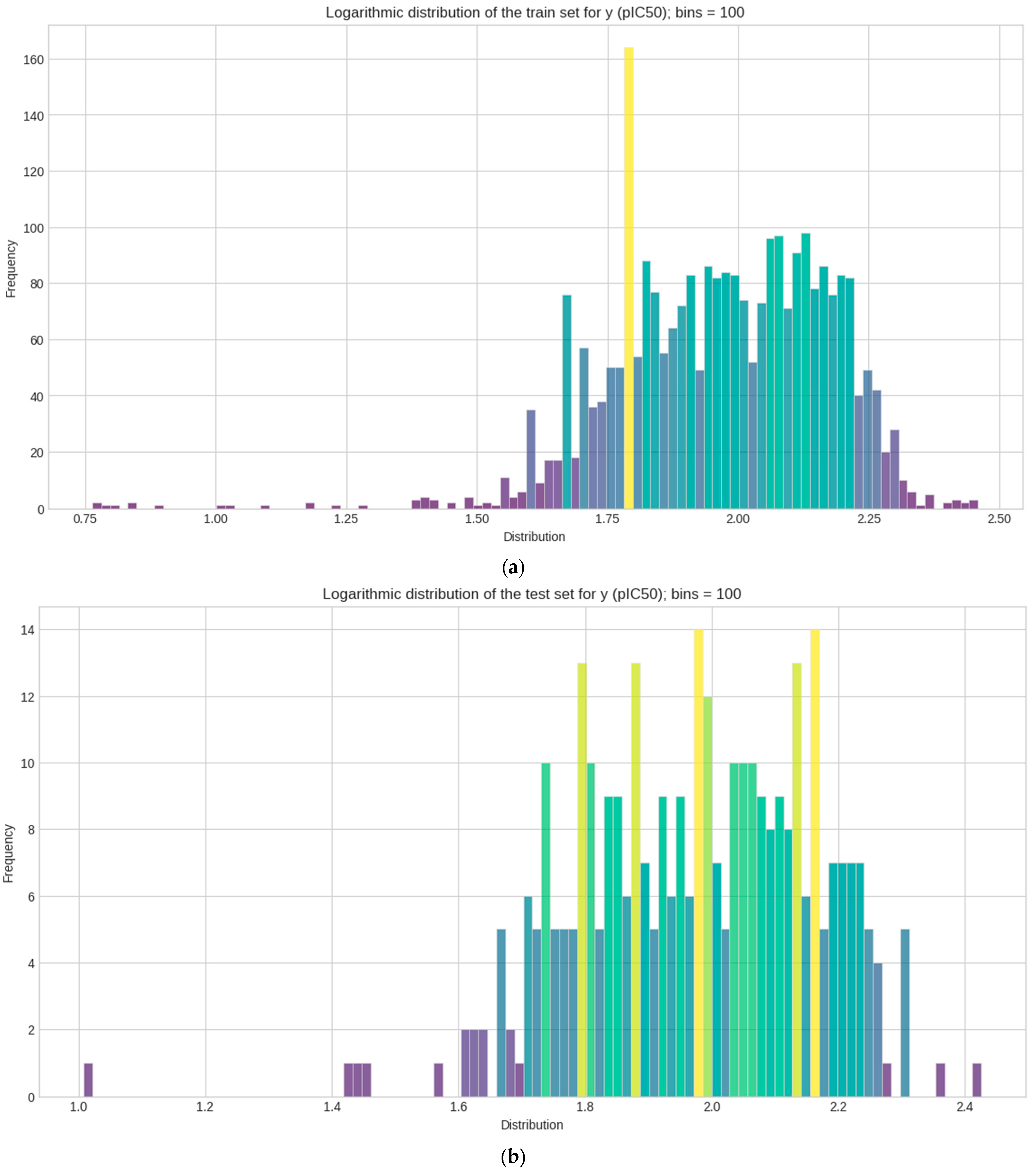
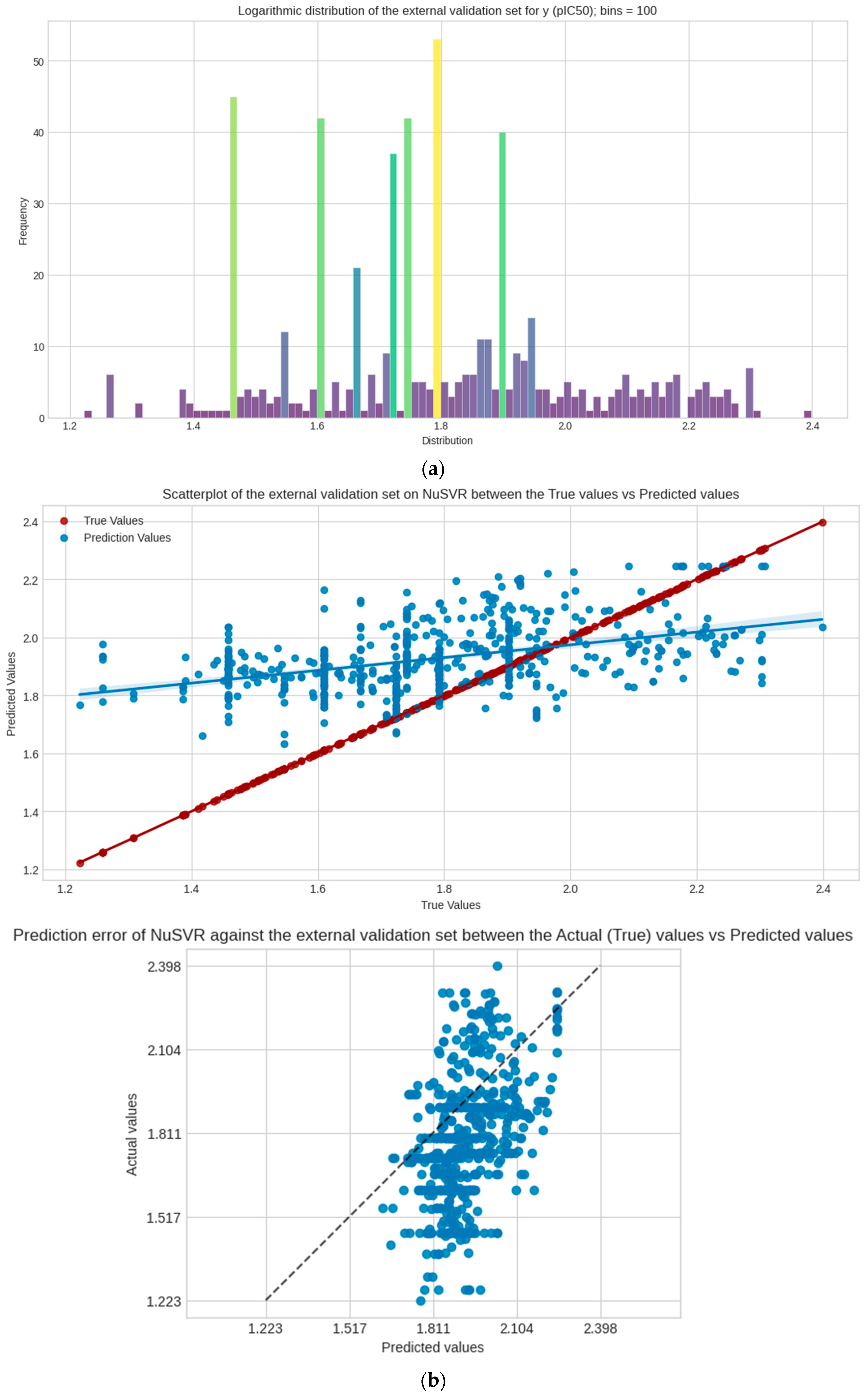
The negative skewed values of train, test, and validation sets in the statistical measurements indicate that the distribution of y in the sets have the characteristic of a longer tail on its left distribution; this is also representative in the histogram plot. Their distribution leans heavily on the right side of the curve. The distribution in the dataset can be considered leptokurtic, where a positive value (kurtosis > 0) shows that distribution has heavier tails and more peaked central regions. Outliers in the plot are also detected, as some values of y are clearly smaller or bigger than others. Initial analysis of y can help whether or not to check for these outliers and if the researcher chooses to remove them when inputting the data in ML models. In the following context, all values are kept for the prevention of the loss of data. These measurements of y are used for checking and comparative analysis with the external dataset (PKA).
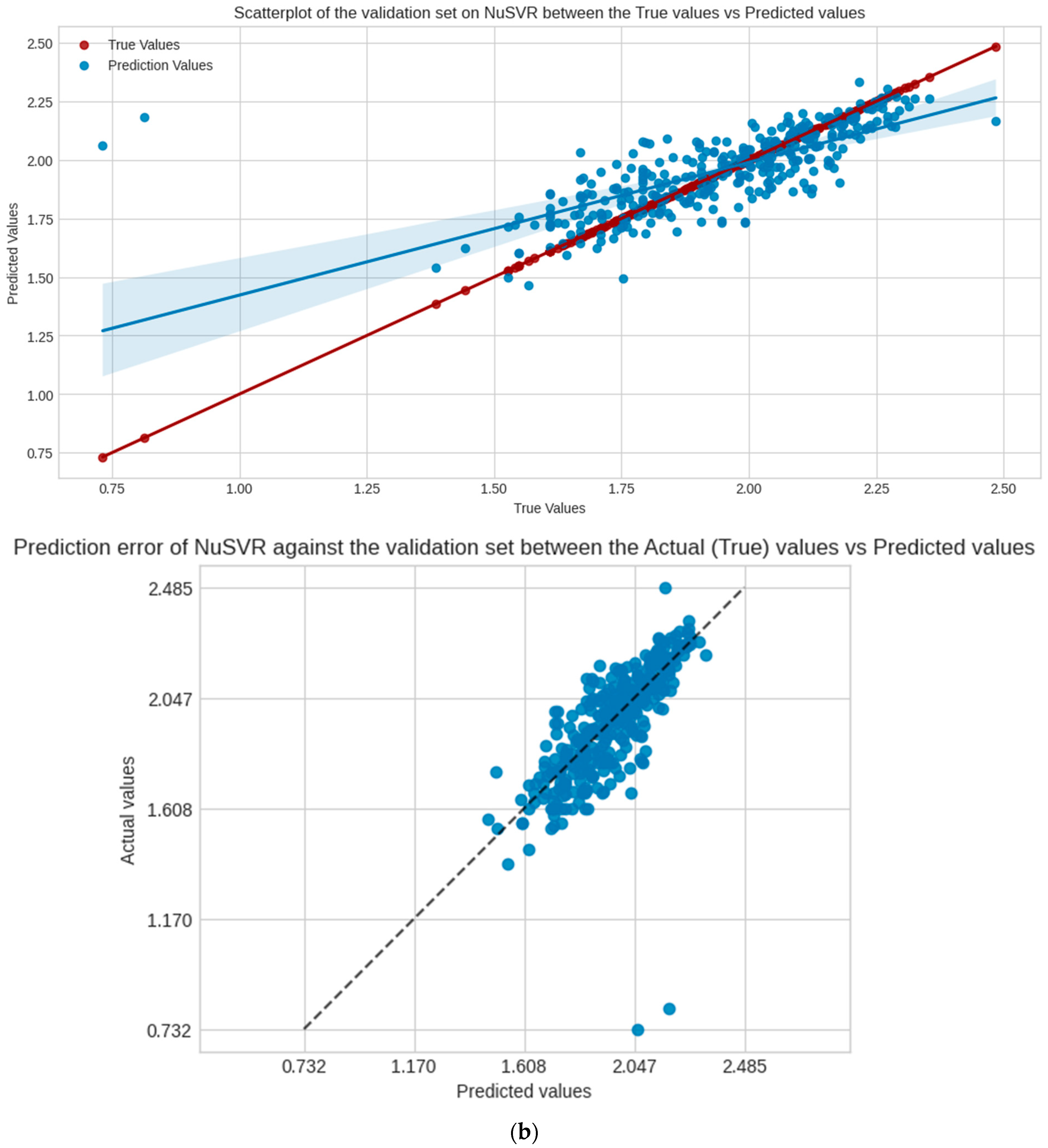
NuSVR, as the chosen model, is a type of support vector machine for regression. This ML algorithm focuses on controlling the margin of errors by using support vectors. These support vectors are data points that have the most impact in determining the most optimal regression line. NuSVR uses the parameter nu to control the number of these support vectors, where nu is an upper bound on the fraction of training errors and a lower bound on the fraction of support vectors [
26,
43,
46,
48,
49]. When the external dataset is compared with the HCV-ROCK1 set, the distribution of y in the following dataset is more skewed to the right, as indicated in its positive skewness value, and a negative kurtosis value indicates that this external dataset expresses a lighter tail and a flatter distribution, which is considered a platykurtic distribution; see
Table 1. The R
2 performance for this dataset displayed a negative score of −0.21870. This is slightly expected as support vector machines like NuSVR are non-linear regression models; this outcome arises when the predictions made on this set, compared to the actual outcome, have not been derived from model fitting using these data. This negative case may still arise since NuSVR uses a non-linear function in its algorithm. The chosen model still poses a great challenge for generalizability in using external datasets, including the following: ensuring all parameters are accounted for, such as tuning to the most optimized hyperparameters for support vector machines; the size of the training data; the compatibility of the chosen model and its decision space algorithm with the external dataset; and possibility that the response variable (pIC
50) is only loosely associated with the features (e.g., molecular descriptors and fingerprints). Since the study has limited computational resources in its capabilities, these challenges are met whenever any ML model requires good transferability to other datasets.
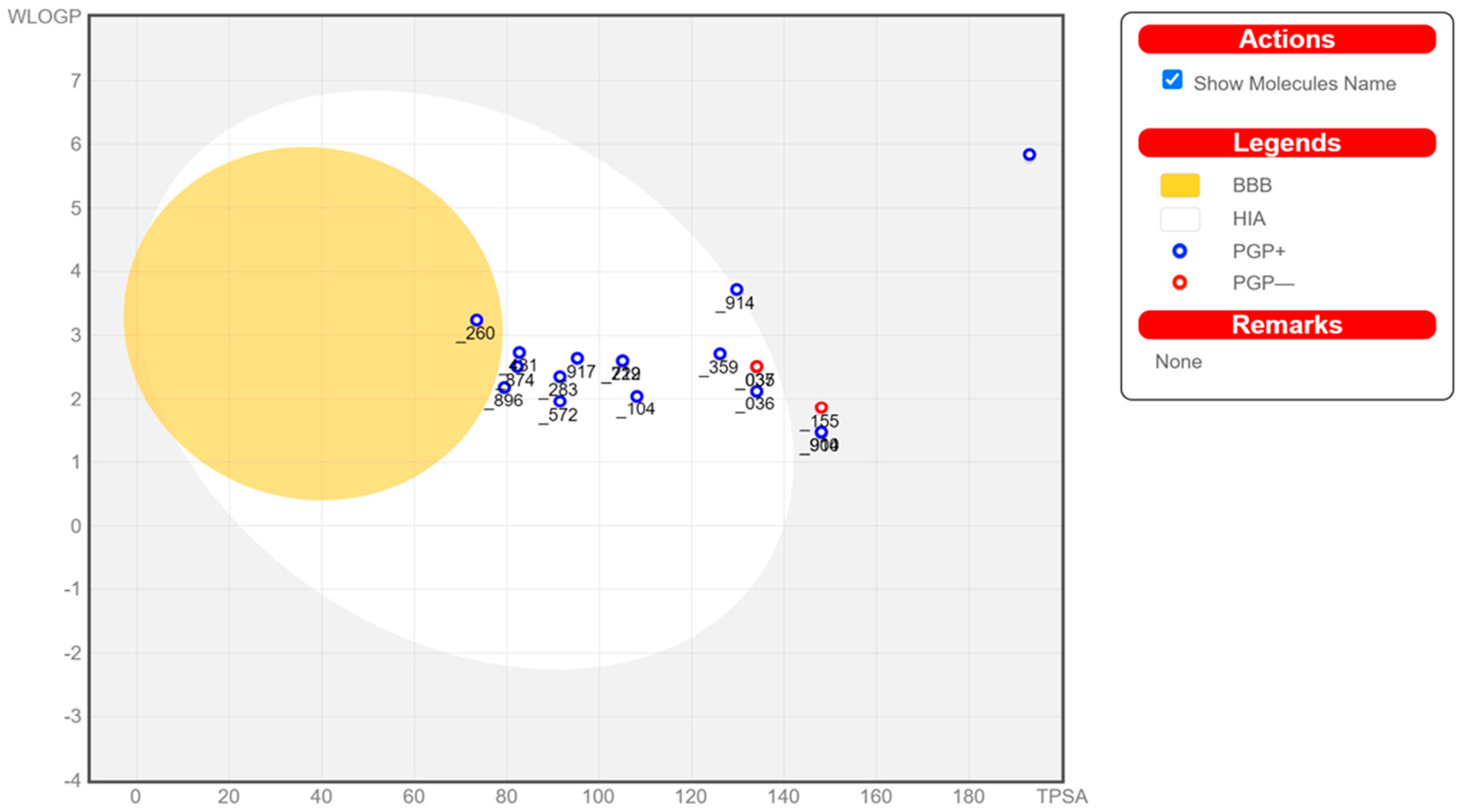
For the compound to be an effective drug, it must reach its target in sufficient amounts of concentration and stay in its bioactive form long enough to elicit the expected biological events to happen. The molecules must exhibit high biological activity and low toxicity in the body. SwissADME reports the bioavailability radar to provide an overview of the drug likeness of the compounds of interest. Each property represents the following: LIPO (lipophilicity between −0.7 and +5.0 for XLOGP3), SIZE (MW between 150 and 500 g/mol), POLAR (polarity between 20 and 130 Å
2 for TPSA), INSOLU (solubility logS not higher than 6), INSATU (fraction of carbons sp3 hybridized not less than 0.25), and FLEX (rotatable bonds must not exceed 9) [
52]. The compounds that did not pass in this section are excluded in molecular docking analysis in
Table 5; complete details of ADME properties are provided in the
Supplementary Material (Figure S1: Bioavailability Radar). Many drug discovery and development failures are due to pharmacokinetics and bioavailability; gastrointestinal (GI) and brain access are behaviors that must be taken into account for estimation in the stages of such discovery processes.
The general scoring function of AutoDock Vina is represented in Equation (6).
where the sum of all pairs of atoms (
i) that can move relative to each other, normally excluding atoms separated by three covalent bonds. This is assigned as type (
ti), with a symmetric set of interaction functions (
ftitj) of the distance (interatomic) (
rij) defined. The optimization algorithm used finds the global minimum of c and other low-scoring confirmations and is ranked; then, the predicted free energy binding is calculated from the intermolecular part of the lowest-scoring confirmation in Equation (7).
where
g can be any arbitrary strictly increasing smooth (possibly linear) function. CB-Dock2 uses the updated version of Vina scoring from version 1.2.0 of AutoDock Vina. The search algorithm continues to be used, but not with new features from AutoDock4 (AD4), such as batch processing and simultaneous docking of ligands. The search algorithm of Vina uses a Monte Carlo (MC) iterative search combined with the Broyden–Fletcher–Goldfarb–Shanno (BFGS) method for local optimization as a gradient-based local optimization. This allows MC to explore a broader conformational space, which is refined by BFGS to converge to a more energetically favorable structure.
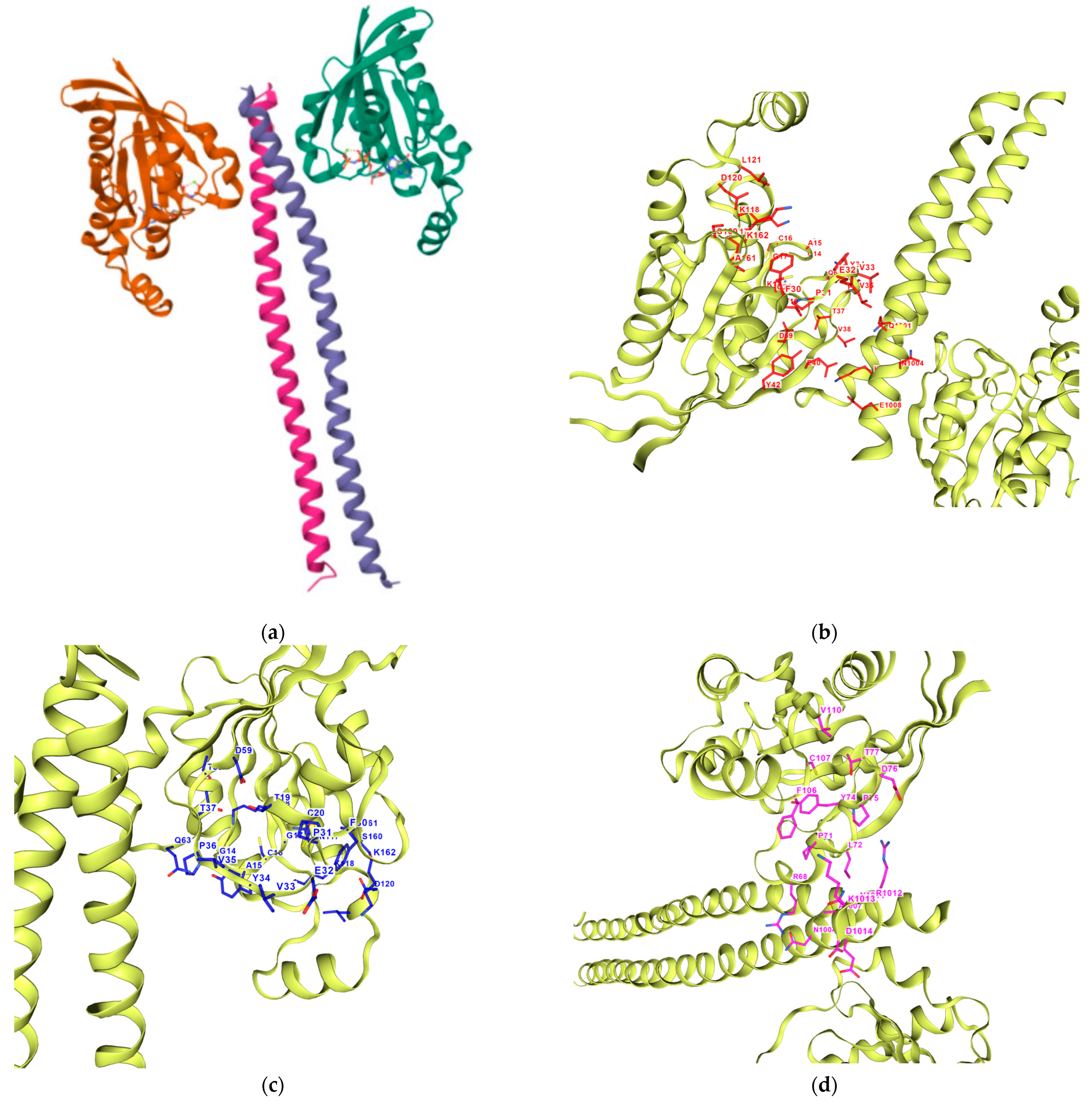
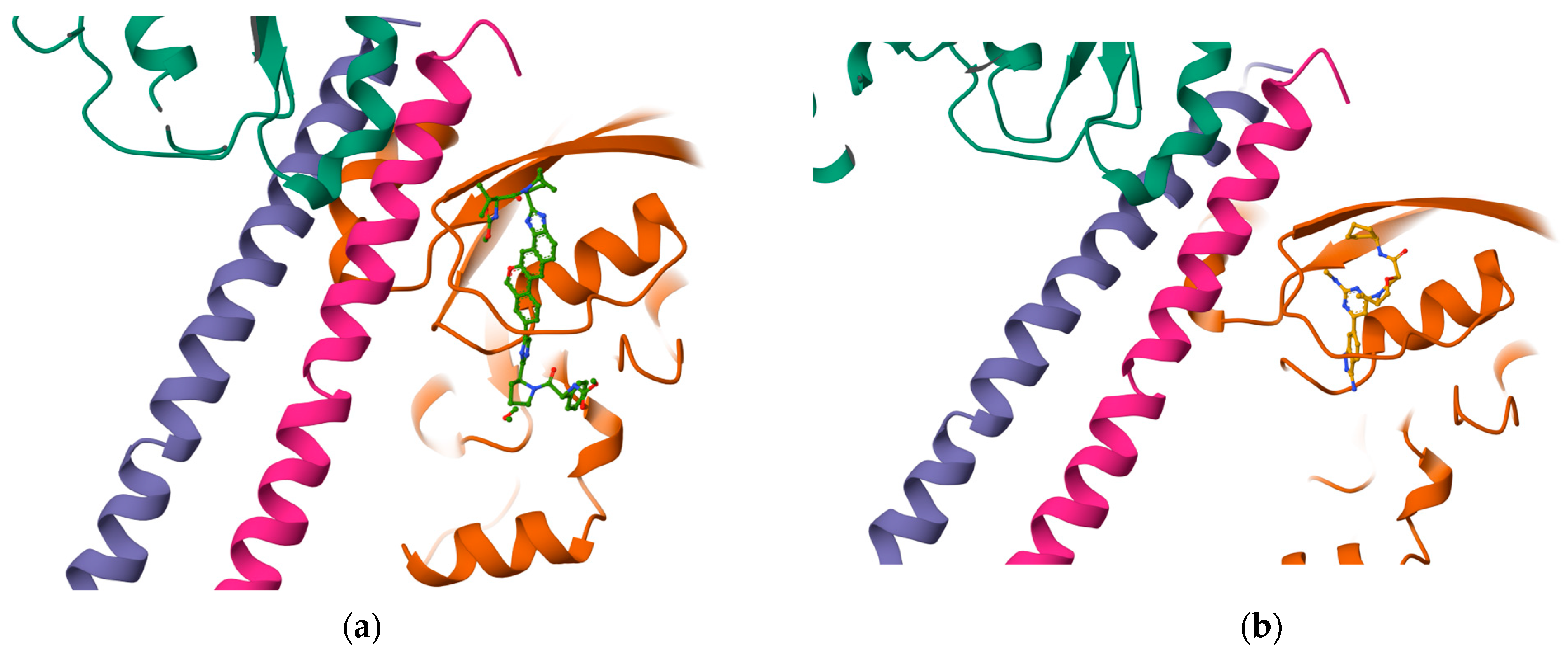
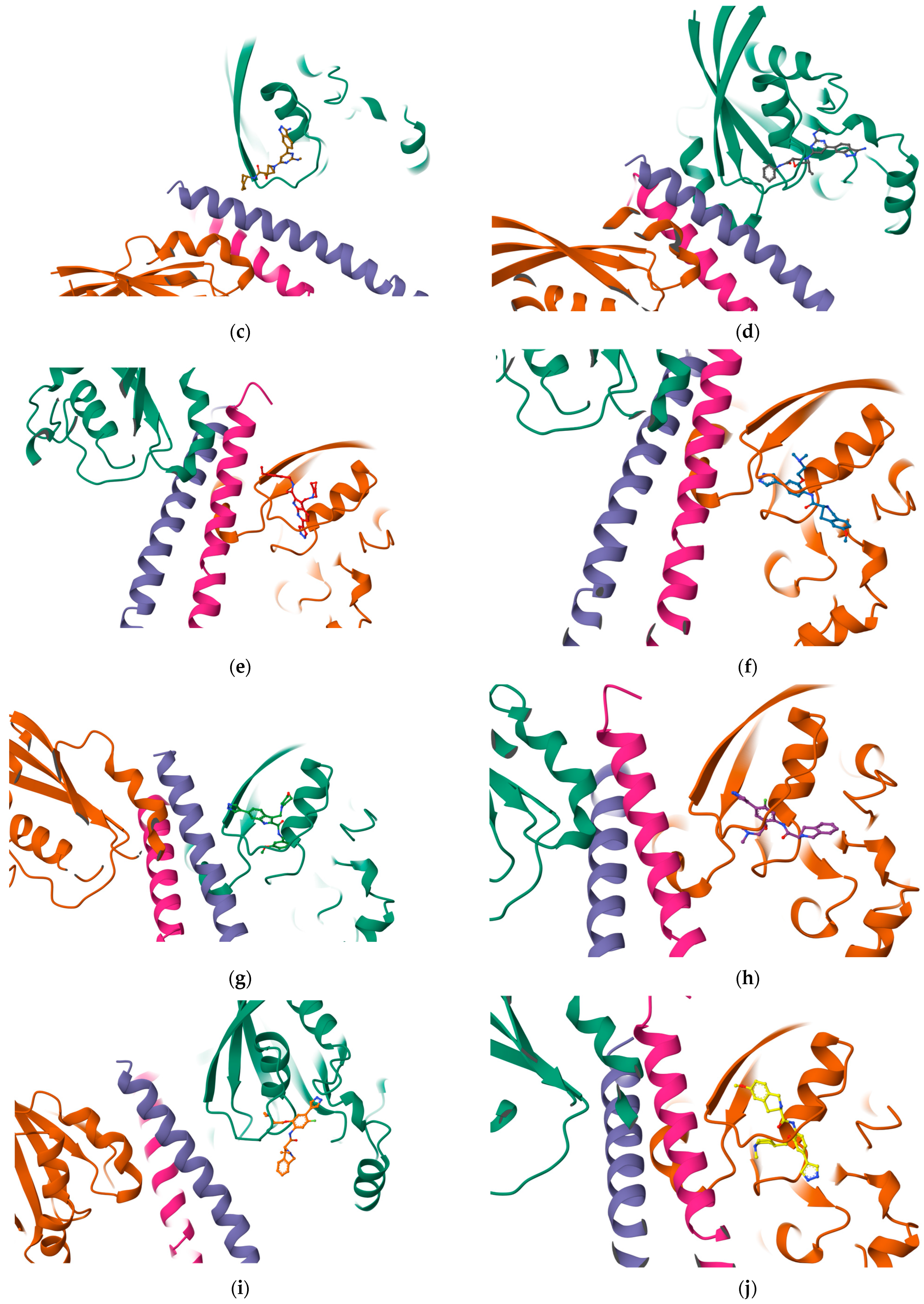
The results reported in
Table 7 suggest that C2 is the best-scoring pocket for velpatasvir. For the protein–ligand complexes, the following pocket IDs were observed: 1S1C-ZINC000071318464 at C2, 1S1C-ZINC000071296700 at C1, 1S1C-ZINC000071315829 at C1, 1S1C-ZINC000073170040 at C2, 1S1C-ZINC000058568630 at C2, 1S1C-ZINC000073196364 at C1, 1S1C-ZINC000058591055 at C2, 1S1C-ZINC000058568675 at C1, 1S1C-ZINC000058574949 at C2.
The results suggest that C2 is the best-ranked pocket ID for the complexes. The ligands for C2 are as follows: ZINC000071318464, ZINC000073170040, ZINC000058568630, ZINC000058591055, and ZINC000058574949. The ligand ZINC000071318464 is the only one that is considered as an intermediate in its bioactivity class; the rest are active (see
Table 5). The five ligands that have the best scoring Vina for C2 all have the following structural properties embedded in their MACCS keys gathered from feature importance on the selected model.
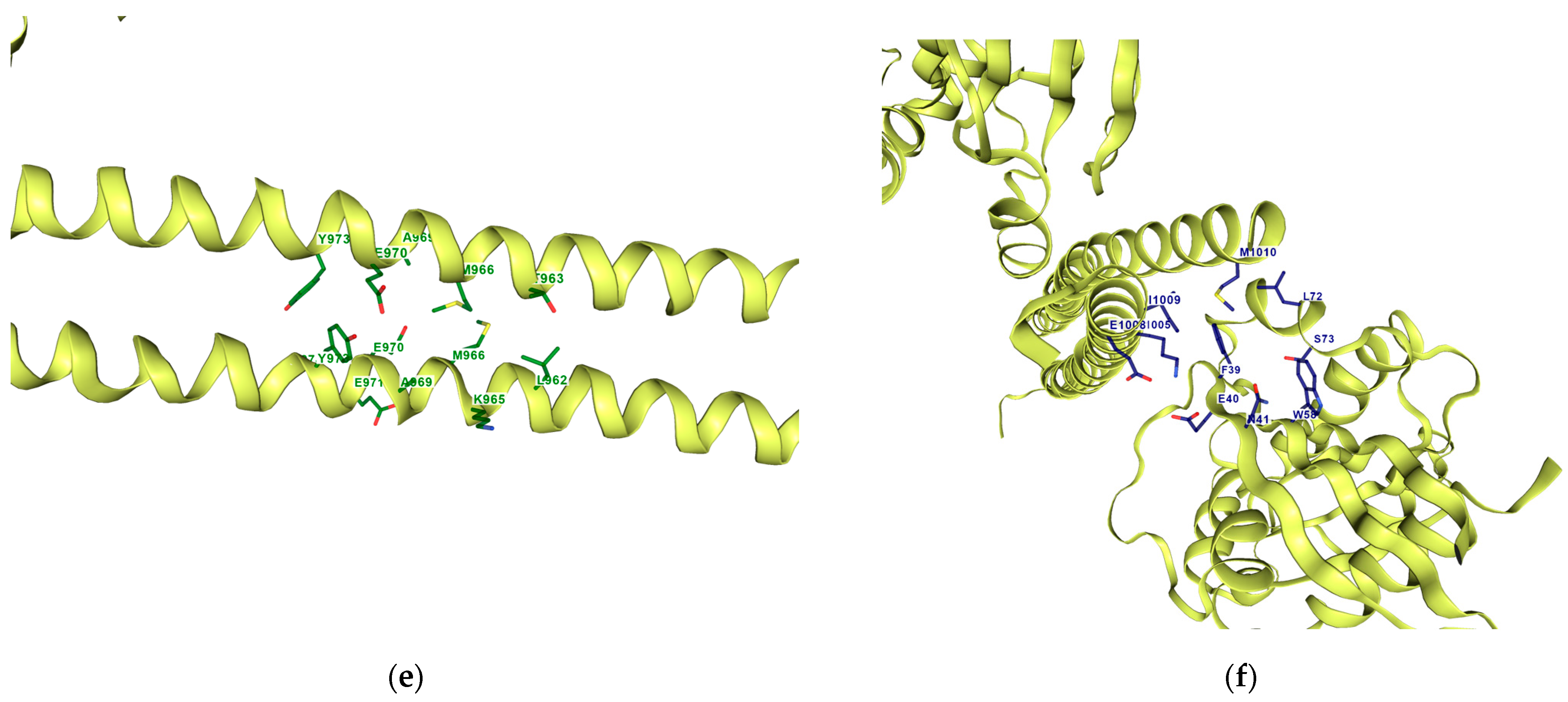
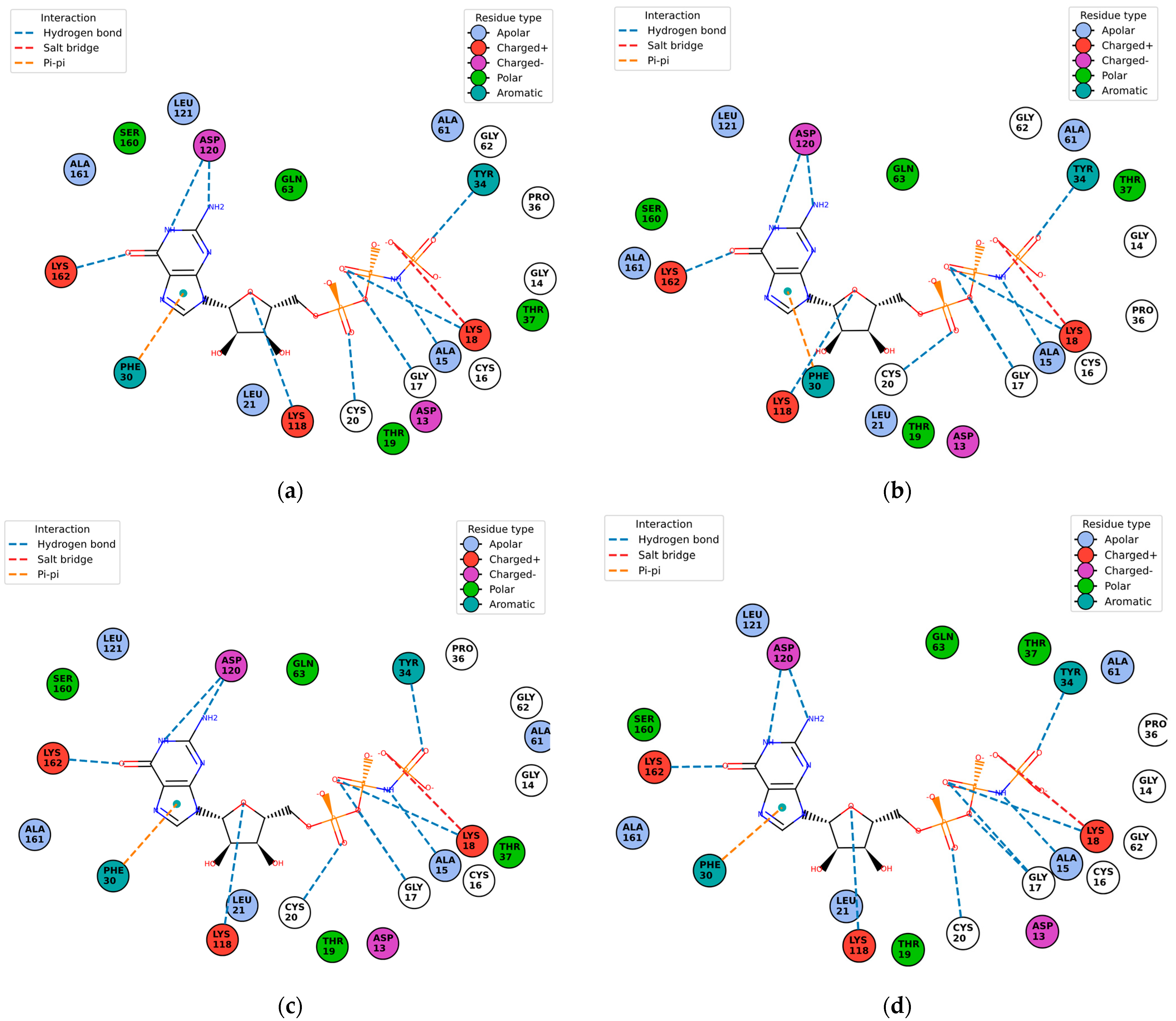
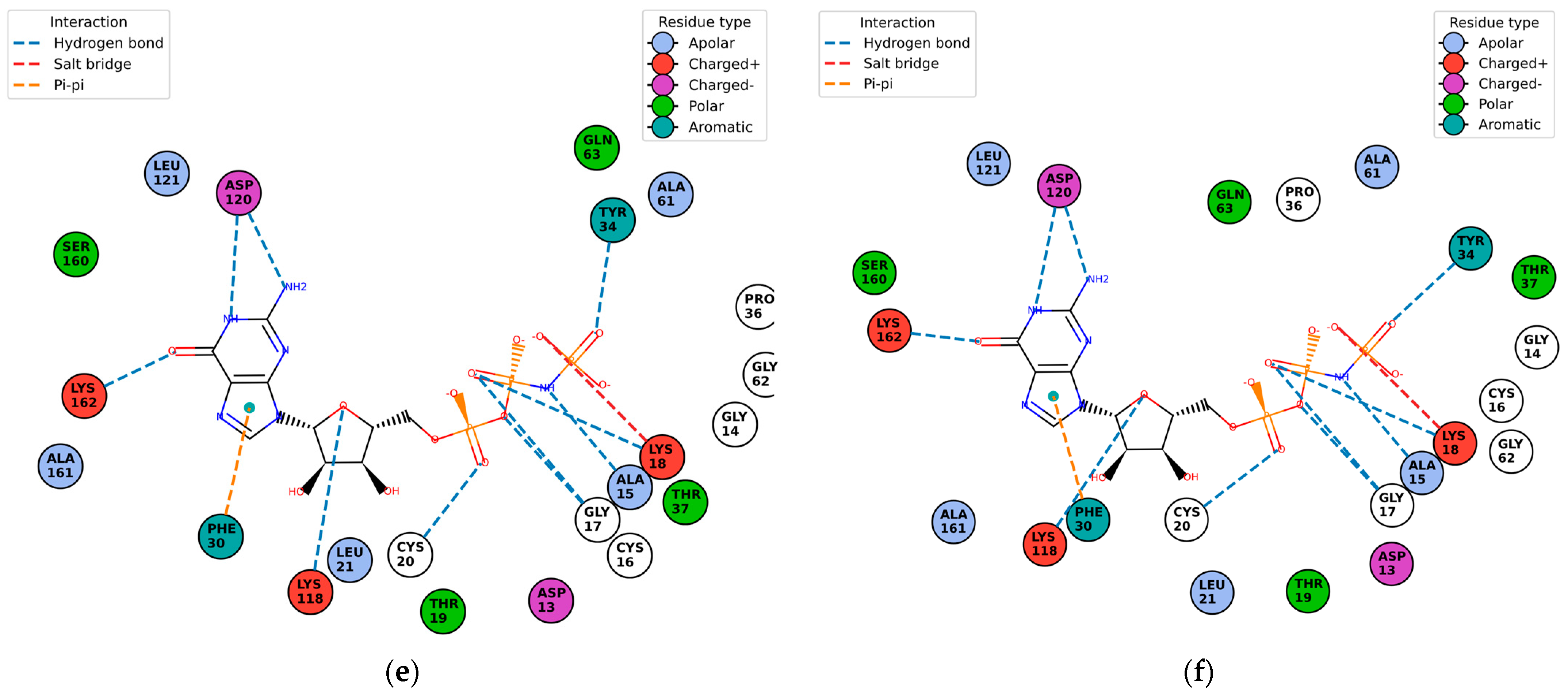
Figure 10 shows the contact residues of the complexes at the docked C2 site, which shows the H-bond, π–π, and salt bridge interactions between the residues and ligands [
65]. The following residues interacting for velpatasvir at chain sequence B revealed the H-bond interactions at −ASP120, +LYS162, +LYS118, CYS20, GLY17, non-polar ALA15, +LYS18, and aromatic TYR34. π–π interaction was observed on the aromatic ring for PHE30. Hydrophobic interactions for the residues ALA161, LEU121, ALA61, LEU21 GLY62, and PRO36 were also observed. H-bond interactions are key in binding specificity while hydrophobic interactions contribute to binding affinity. The mobility and functional orientation of the hydrogen bonding hydrogens are of great pharmacological interest for influencing ligand bindings [
66]. For the rest of the five screened ligands that show favorable energy at C2, such as ZINC000071318464, consistent residues paralleling that of velpatasvir on chain sequences B and Y were observed. Immune cells that are activated are dependent on enough SER and differentiation of T cells. GLN is essential for cell proliferation and immune system cells as GLN is a precursor in amino sugars and nucleotides. CYS availability has a key role in T cell functions because T cells do not have enzymes converting MET to CYS [
67].
It is worth noting that these structural properties (e.g., an aromatic nitrogen atom that is part of an aromatic ring system, the presence of a quaternary ammonium cation, the presence of a quaternary nitrogen atom (-N) that is bonded to three other atoms, etc.) can be used as a screening criterion for finding other compounds of interest that may have the same embedded molecular fingerprints from QSAR-ML and molecular docking findings in the study. Selecting a value of pIC
50 > 5 can also be taken into consideration for this inhibitor property (see
Table 5). Drug–drug interactions can be predicted using similarity tests such as the cosine method, used to calculate the feature similarity of drugs and compounds where a high dimensionality vector is constructed; the five compounds were subjected to the following method to further probe their structural similarities alongside the already approved DAA for HCV treatments;
Supplementary Material (Supplement S1) [
68].
Currently, DAA combinations of sofosbuvir/daclatasvir and sofosbuvir/velpatasvir are the recommended antiviral treatments in the Philippines [
1,
8]. Dasabuvir is also another drug used to treat HCV in combination with other antivirals such as ombitasvir, paritprevir, and ritonavir. However, unlike sofosbuvir, dasabuvir is not used in the country for treatment [
7]. In the context of ROCK1 inhibition, using these drugs as our metric can be beneficial for finding other drug compounds for HCV treatment. Identifying potential ROCK1 inhibitors could expand HCV therapeutic strategies. The structural information of ROCK1 can be exploited to develop these inhibitors. Screening compounds from chemical libraries for their impact on the protein structure and their bioactivity may lead to promising candidates [
15].
5. Conclusions
The main motivation behind this study lies in the ever-increasing prevalence of drug-resistant HCV variants, propelling explorations of novel molecules specifically targeting enzymes like ROCK1, for which minimal studies have been conducted. A consideration to take when performing QSAR-ML is the limited research on ROCK1 and its inhibitors. The usage of datasets that did not use ROCK1 as its bioactivity assay for determining pIC50 represents a challenge, as the correlation between the following parameter and features of the inhibitors may vary. Moreover, the sample size is a challenge, resulting in a less generalized ML model. The presented results show the criteria a compound must meet in drug screening for ROCK1. Docking results highlight C2 as the best-ranked pocket for ligand binding. The ligands that exhibited the best scores on C2, paralleling those in the control (velpatasvir), are as follows: ZINC000071318464, ZINC000073170040, ZINC000058568630, ZINC000058591055, and ZINC000058574949. They have satisfied all of the requirements for their ADME properties, and the qualities they have exhibited can be considered drug-like. The structural properties of the compounds presented by the QSAR-ML with molecular docking provide valuable findings for identifying other potential candidates that inhibit ROCK1. The results in the study offer substantial utility in discovering compounds for ROCK1 inhibition. This information can guide the identification of significant binding properties and can contribute to the still ongoing drug discovery efforts involving ROCK1 inhibitors.
To enhance the generalizability of the ML model, researchers in future studies may opt for improved hyperparametization in the model, a technique that allows for the exploration of improved search spaces and optimization of model parameters. Additionally, employing multiple iterative processes to determine feature importance can contribute to a more refined model. While the present study faces constraints in terms of computational resources, future research endeavors in the domain of QSAR-ML, molecular docking and molecular dynamics may leverage the techniques and protocol outlined herein and further refine them according to their unique requirements. A substantial amount of attention should be dedicated to meticulously preparing the dataset, as the challenges encountered in this study primarily stem from the pre-processing of raw data sourced from chemical databases. The actions taken by researchers during the pre-processing stages wield significant influence over the subsequent phases of their investigation. As such, meticulous dataset preparation is imperative for laying a robust foundation for the entirety of the study. Another validation method involves performing molecular dynamics simulation on the protein–ligand complexes. This approach can validate the energy stability using the QM/MM inputs during simulation runs, providing a more accurate evaluation.