Theoretical Investigation on the Selective Hydroxyl Radical–Induced Decolorization of Methylene-Blue-Dyed Polymer Films
Abstract
1. Introduction
2. Methods
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Temeepresertkij, P.; Yenchit, S.; Iwaoka, M.; Iwamori, S. Interactions between Methylene Blue and Pullulan According to Molecular Orbital Calculations. IEEJ Trans. Fundam. Mater. 2020, 140, 529–533. [Google Scholar] [CrossRef]
- Temeepresertkij, P.; Iwaoka, M.; Iwamori, S. Molecular Interactions between Methylene Blue and Sodium Alginate Studied by Molecular Orbital Calculations. Molecules 2021, 26, 7029. [Google Scholar] [CrossRef] [PubMed]
- Yenchit, S.; Yamanaka, H.; Temeeprasertkij, P.; Oda, Y.; Kanie, O.; Okamura, Y.; Inazu, T.; Iwamori, S. Chemical Stability of a Colorimetric Indicator Based on Sodium Alginate Thin Film and Methylene Blue Dye upon Active Oxygen Species Exposure. Jpn. J. Appl. Phys. 2020, 59, SDDF09. [Google Scholar] [CrossRef]
- Yenchit, S.; Tadokoro, Y.; Iwamori, S. Measuring Active Oxygen Species across a Nonwoven Fabric Using a Pullulan-Mixed Methylene Blue Thin Film and Electron Spin Resonance. IEEJ Trans. Sens. Micromach. 2019, 139, 54–60. [Google Scholar] [CrossRef]
- Kumar, A.; Pottiboyina, V.; Sevilla, M.D. Hydroxyl Radical (OH•) Reaction with Guanine in an Aqueous Environment: A DFT Study. J. Phys. Chem. B 2011, 115, 15129–15137. [Google Scholar] [CrossRef]
- Çakmak, E.; Isin, D.O. A Theoretical Evaluation on Free Radical Scavenging Activity of 3-Styrylchromone Derivatives: The DFT Study. J. Mol. Model. 2020, 26, 98. [Google Scholar] [CrossRef]
- Hassaan, M.A.; Nemr, A.E.; Madkour, F.F. Testing the advanced oxidation processes on the degradation of Direct Blue 86 dye in wastewater. Egypt. J. Aquat. Res. 2017, 43, 11–19. [Google Scholar] [CrossRef]
- Miklos, D.B.; Remy, C.; Jekel, M.; Linden, K.G.; Drewes, J.E.; Hübner, U. Evaluation of advanced oxidation processes for water and wastewater treatment. Water Res. 2018, 139, 118–131. [Google Scholar] [CrossRef]
- Kuang, Y.; Zhang, X.; Zhou, S. Adsorption of Methylene Blue in Water onto Activated Carbon by Surfactant Modification. Water 2020, 12, 587. [Google Scholar] [CrossRef]
- Yoshino, K.; Matsumoto, H.; Iwasaki, T.; Kinoshita, S.; Noda, K.; Oya, K.; Iwamori, S. Investigation of a Sterilization System Using Active Oxygen Species Generated by Ultraviolet Irradiation. Biocontrol Sci. 2015, 20, 11–18. [Google Scholar] [CrossRef]
- Oya, K.; Watanabe, R.; Soga, Y.; Ikeda, Y.; Nakamura, T.; Iwamori, S. Effect of humidity conditions on active oxygen species generated under ultraviolet light irradiation and etching characteristics of fluorocarbon polymer. J. Photochem. Photobiol. A Chem. 2015, 298, 33–39. [Google Scholar] [CrossRef][Green Version]
- Murakami, Y.; Oguchi, T.; Hashimoto, K.; Nakamura, A.; Sakai, Y.; Ando, H. Density functional study of the phenylethyl + O2 reaction: Kinetic analysis for the low-temperature autoignition of ethylbenzenes. Int. J. Quantum Chem. 2012, 112, 1968–1983. [Google Scholar] [CrossRef]
- Tokmakov, I.V.; Kim, G.-S.; Kislov, V.V.; Mebel, A.M.; Lin, M.C. The reaction of phenyl radical with molecular oxygen: A G2M study of the potential energy surface. J. Phys. Chem. A 2005, 109, 6114–6127. [Google Scholar] [CrossRef]
- Yan, J.; Li, Z.; Zhang, J.; Qiao, C. Preparation and Properties of Pullulan Composite Films. Adv. Mater. Res. 2012, 476–478, 2100–2104. [Google Scholar] [CrossRef]
- Li, Y.; Yokoyama, W.; Wu, J.; Ma, J.; Zhong, F. Properties of Edible Films based on Pullulan–Chitosan Blended Film-Forming Solutions at Different pH. R. Soc. Chem. 2015, 5, 105844–105850. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, H.; Zhu, Z. Study on the Blends of Silk Fibroin and Sodium Alginate: Hydrogen Bond Formation, Structure and Properties. Polymer 2019, 163, 144–153. [Google Scholar] [CrossRef]
- Lan, W.; He, L.; Liu, Y. Preparation and Properties of Sodium Carboxymethyl Cellulose/Sodium Alginate/Chitosan Composite Film. Coatings 2018, 8, 291. [Google Scholar] [CrossRef]
- Harnsilawat, T.; Pongsawatmanit, R.; McClements, D.J. Characterization of β-Lactoglobulin–Sodium Alginate Interactions in Aqueous Solutions: A Calorimetry, Light Scattering, Electrophoretic Mobility and Solubility Study. Food Hydrocoll. 2006, 20, 577–585. [Google Scholar] [CrossRef]
- Pascoe, D.J.; Ling, K.B.; Cockroft, S.L. The Origin of Chalcogen-Bonding Interactions. Am. Chem. Soc. 2017, 139, 15160–15167. [Google Scholar] [CrossRef]
- Balachandar, S.; Dhandapani, M.; Enoch, I.V.M.V.; Suganthi, S. Structural Analysis, Molecular Docking and DFT Calculations of Bis (Pyrazolium Picrate) Monohydrate Interaction with Calf Thymus DNA and Microbes. ChemistrySelect 2017, 2, 9298–9311. [Google Scholar] [CrossRef]
- Okutsu, N.; Shimamura, K.; Shimizu, E.; Shulga, S.; Danilov, V.I.; Kurita, N. DFT Study on the Attacking Mechanisms of H and OH Radicals to G-C and A-T Base Pairs in Water. In Proceedings of the AIP Conference Proceedings 1709, Aichi, Japan, 22–23 October 2015; AIP Publishing LLC: Huntington, NY, USA, 2016; pp. 020006-1–020006-14. [Google Scholar]
- Boys, S.F.; Bernardi, F. The calculation of small molecular interactions by the differences of separate total energies. Some procedures with reduced errors. Mol. Phys. 1970, 19, 553–566. [Google Scholar] [CrossRef]
- Simon, S.; Duran, M.; Dannenberg, J.J. How does basis set superposition error change the potential surfaces for hydrogenbonded dimers? J. Chem. Phys. 1996, 105, 11024. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H. Gaussian 16; Revision C.01; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Dennington, R.; Keith, A.T.; Millam, M.J. GaussView; Version 6.1; Semichem Inc.: Shawnee, KS, USA, 2016. [Google Scholar]
- Batsanov, S.S. Calculation of van der Waals radii of atoms from bond distances. J. Mol. Struct. THEOCHEM 1999, 468, 151–159. [Google Scholar] [CrossRef]
- Parr, R.G.; Pearson, R.G. Absolute Hardness: Companion Parameter to Absolute Electronegativity. Am. Chem. Soc. 1983, 105, 7512–7516. [Google Scholar] [CrossRef]
- Pearson, R.G. Absolute Electronegativity and Hardness: Application to Inorganic Chemistry. Inorg. Chem. 1988, 27, 734–740. [Google Scholar] [CrossRef]
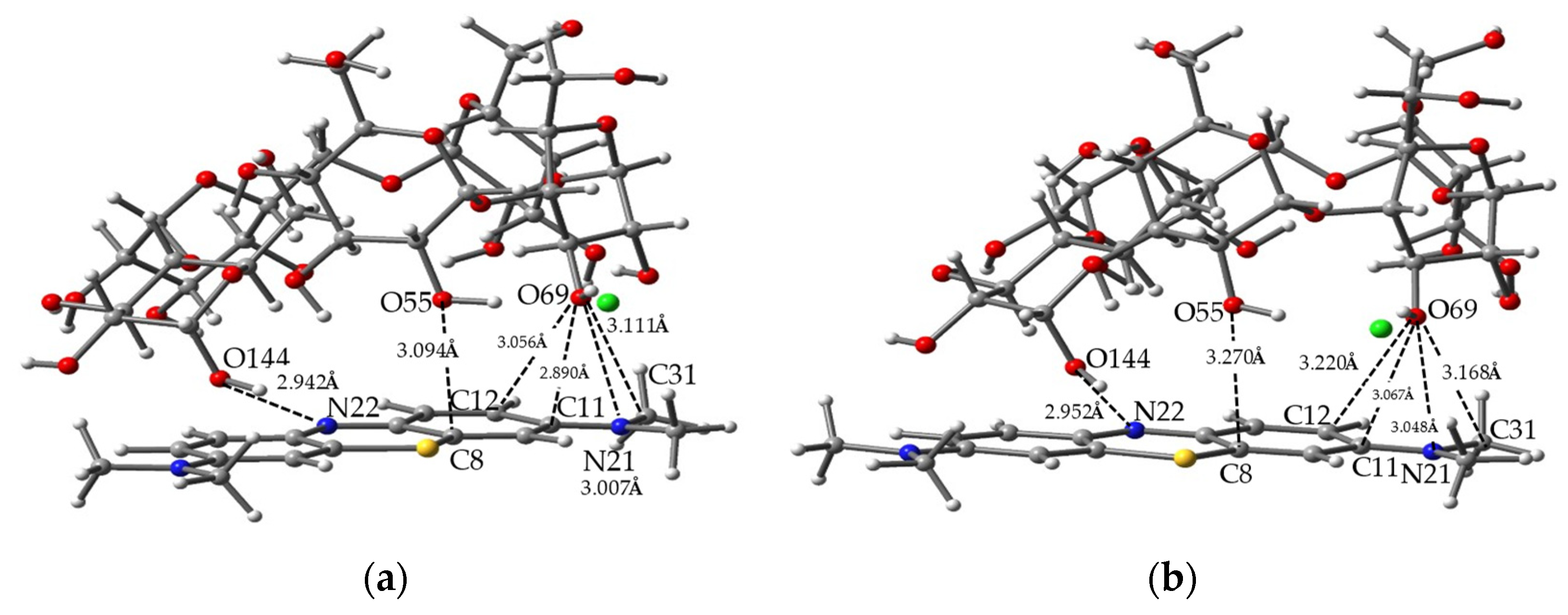

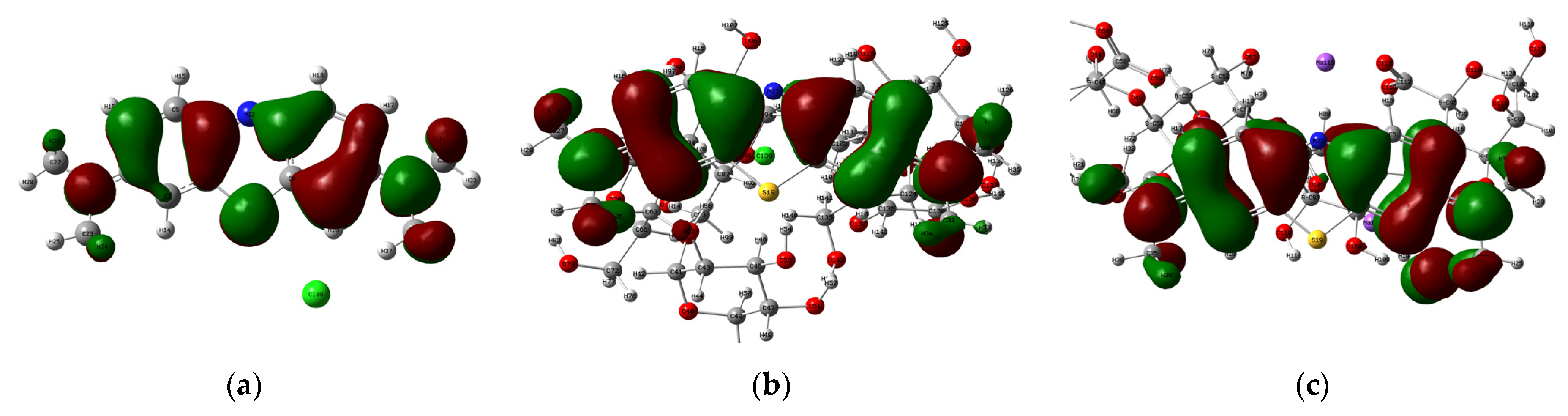
| MB/P | Reactive Energy (kcal/mole) | Complexation Energy (kcal/mole) | |||
|---|---|---|---|---|---|
| B3LYP/6-31G(d)//B3LYP/6-31G(d) | B3LYP/6-31+G(d,p)//B3LYP/6-31G(d) | B3PW91/6-31+G(d,p)//B3LYP/6-31+G(d,p) | |||
| GD3 | in Water (SCRF) | GD3+BSSE | |||
| 1 | 0.00 | 0.00 | 0.00 | 0.00 | −66.67 |
| 2 | 12.80 | 11.86 | 7.32 | 2.33 | −59.18 |
| 3 | 14.39 | 17.96 | 9.83 | 1.66 | −39.49 |
| 4 | 13.82 | 14.17 | 10.03 | 2.17 | −57.37 |
| 5 | 21.42 | 22.29 | 24.29 | 9.48 | −22.22 |
| 6 | 21.16 | 19.45 | 25.04 | 18.79 | −41.39 |
| 7 | 30.89 | 27.08 | 25.17 | 14.30 | −48.87 |
| 8 | 23.34 | 23.51 | 28.59 | 10.25 | −19.11 |
| 9 | 26.62 | 23.82 | 29.41 | 8.38 | −17.15 |
| MB/P | N···O | C···O | ||||||
|---|---|---|---|---|---|---|---|---|
| Distance (Å) | Distance (Å) | |||||||
| Position | B3LYP/ 6-31G(d) | B3LYP/ 6-31+G(d,p) | Difference | Position | B3LYP/ 6-31G(d) | B3LYP/ 6-31+G(d,p) | Difference | |
| 1 | none | – | – | – | C2,O55 | 3.141 | 3.503 | +0.36 |
| C35,O138 | 3.191 | 5.165 | +1.97 | |||||
| 2 | none | – | – | – | C3,O144 | 3.253 | 3.311 | +0.06 |
| C23,O69 | 3.313 | 3.333 | +0.11 | |||||
| 3 | none | – | – | – | C13,O55 | 3.183 | 3.350 | +0.17 |
| C2,O120 | 3.191 | 3.349 | +0.16 | |||||
| C1,O120 | 3.224 | 3.758 | +0.53 | |||||
| C11,O144 | 3.229 | 3.393 | +0.16 | |||||
| 4 | N22,O144 | 2.942 | 2.952 | +0.01 | C11,O69 | 2.890 | 3.067 | +0.18 |
| N21,O69 | 3.007 | 3.048 | +0.04 | C12,O69 | 3.056 | 3.220 | +0.16 | |
| C8,O55 | 3.094 | 3.270 | +0.18 | |||||
| C31,O69 | 3.111 | 3.168 | +0.06 | |||||
| 5 | N22,O69 | 2.990 | 2.974 | −0.02 | C5, O69 | 3.159 | 3.226 | +0.07 |
| N22,O55 | 3.050 | 3.081 | +0.03 | C31,O138 | 3.191 | 3.522 | +0.33 | |
| 6 | none | – | – | – | C27,O123 | 3.255 | 3.722 | +0.47 |
| C23,O123 | 3.284 | 3.372 | +0.09 | |||||
| 7 | N20,O132 | 3.205 | 3.341 | −0.14 | none | – | – | – |
| 8 | N22,O144 | 2.817 | 2.838 | +0.02 | none | – | – | – |
| 9 | N22,O53 | 2.954 | 2.844 | −0.11 | none | – | – | – |
| MB/SA | Reactive Energy (kcal/mole) | Complexation Energy (kcal/mole) | |||
|---|---|---|---|---|---|
| B3LYP/6-31G(d)//B3LYP/6-31G(d) | B3LYP/6-31+G(d,p)//B3LYP/6-31G(d) | B3PW91/6-31+G(d,p)//B3LYP/6-31G(d) | |||
| GD3 | in Water (SCRF) | GD3+BSSE | |||
| 1 | 0.00 | 0.00 | 0.00 | 0.00 | −93.19 |
| 2 | 5.53 | 4.35 | 9.25 | 4.17 | −69.54 |
| 3 | 6.75 | 5.66 | 8.68 | 1.55 | −68.69 |
| 4 | 8.56 | 1.19 | 16.71 | 0.67 | −78.52 |
| 5 | 12.16 | 8.18 | 13.00 | 2.75 | −87.91 |
| 6 | 15.90 | 9.59 | 28.00 | 14.48 | −57.42 |
| 7 | 17.12 | 10.05 | 31.90 | 17.45 | −52.18 |
| 8 | 21.29 | 13.97 | 27.32 | 8.86 | −53.13 |
| 9 | 24.24 | 17.80 | 29.93 | 9.52 | −60.17 |
| MB/SA | N···O | C···O | S···O | |||
|---|---|---|---|---|---|---|
| B3LYP/6-31G(d) | ||||||
| Position | Distance (Å) | Position | Distance (Å) | Position | Distance (Å) | |
| 1 | none | none | C3,O106 | 3.032 | S19,O106 | 3.215 |
| C27,O121 | 3.223 | |||||
| 2 | N20,O117 | 2.961 | C2,O113 | 3.127 | none | none |
| C35,O59 | 3.236 | |||||
| C1,O117 | 3.254 | |||||
| 3 | none | none | C2,O113 | 3.200 | none | none |
| C35,O59 | 3.201 | |||||
| 4 | none | none | C2,O63 | 3.243 | S19,O59 | 3.256 |
| S19,O63 | 3.118 | |||||
| 5 | none | none | C8,O108 | 3.034 | S19,O108 | 3.477 |
| C31,O62 | 3.244 | |||||
| C12,O62 | 3.299 | |||||
| 6 | none | none | None | none | none | none |
| 7 | N20,O57 | 3.176 | C27,O57 | 3.193 | none | none |
| 8 | none | none | C8,O60 | 2.956 | S19,O60 | 3.269 |
| C7,O60 | 3.165 | |||||
| 9 | none | none | C35,O60 | 3.194 | none | none |
| Property | Formula | AOS | ||
|---|---|---|---|---|
| OH Radical | H2O2 | O3 | ||
| LUMO energy (eV) | ELUMO a | −6.59 a | 0.57 | −4.90 |
| HOMO energy (eV) | EHOMO | −7.26 | −6.71 | −8.98 |
| Energy gap (eV) | Eg = ELUMO − EHOMO | 0.67 | 7.27 | 4.08 |
| Hardness | η = Eg/2 | 0.34 | 3.64 | 2.04 |
| Softness | σ =1/η | 2.98 | 0.27 | 0.49 |
| Electronegativity | χ = −(ELUMO + EHOMO)/2 | 6.93 | 3.07 | 6.94 |
| Property | Structure | ||||
|---|---|---|---|---|---|
| MB | Pullulan | Sodium Alginate | MB/Pullulan | MB/Sodium Alginate | |
| LUMO energy (eV) | −3.23 | 0.81 | −2.05 | −3.49 | −3.21 |
| HOMO energy (eV) | −5.72 a | −6.73 | −5.40 | −5.97 | −5.63 |
| Energy gap (eV) | 2.49 | 7.54 | 3.35 | 2.49 | 2.41 |
| Hardness | 1.25 | 3.77 | 1.68 | 1.24 | 1.21 |
| Softness | 0.80 | 0.27 | 0.60 | 0.80 | 0.83 |
| Electronegativity | 4.48 | 2.96 | 3.73 | 4.73 | 4.42 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Temeeprasertkij, P.; Iwaoka, M.; Iwamori, S. Theoretical Investigation on the Selective Hydroxyl Radical–Induced Decolorization of Methylene-Blue-Dyed Polymer Films. Computation 2022, 10, 169. https://doi.org/10.3390/computation10100169
Temeeprasertkij P, Iwaoka M, Iwamori S. Theoretical Investigation on the Selective Hydroxyl Radical–Induced Decolorization of Methylene-Blue-Dyed Polymer Films. Computation. 2022; 10(10):169. https://doi.org/10.3390/computation10100169
Chicago/Turabian StyleTemeeprasertkij, Pasika, Michio Iwaoka, and Satoru Iwamori. 2022. "Theoretical Investigation on the Selective Hydroxyl Radical–Induced Decolorization of Methylene-Blue-Dyed Polymer Films" Computation 10, no. 10: 169. https://doi.org/10.3390/computation10100169
APA StyleTemeeprasertkij, P., Iwaoka, M., & Iwamori, S. (2022). Theoretical Investigation on the Selective Hydroxyl Radical–Induced Decolorization of Methylene-Blue-Dyed Polymer Films. Computation, 10(10), 169. https://doi.org/10.3390/computation10100169






