Abstract
Contemporary biological research (particularly in systems biology and the “omic” disciplines) is factually answering some of the poignant questions associated with the information concept and the limitations of information theory. Here, rather than emphasizing and persisting on a focalized discussion about the i-concept, an ampler conception of “informational entities” will be advocated. The way living cells self-produce, interact with their environment, and collectively organize multi-cell systems becomes a paradigmatic case of what such informational entities consist of. Starting with the fundamentals of molecular recognition, and continuing with the basic cellular processes and subsystems, a new interpretation of the global organization of the living cell must be assayed, so that the equivalents of meaning, value, and intelligence will be approached along an emerging “bioinformational” perspective. Further insights on the informational processes of brains, companies, institutions and human societies at large, and even the sciences themselves, could benefit from—and cross-fertilize with—the advancements derived from the informational approach to living systems. The great advantage fuelling the expansion of the bioinformation paradigm is that, today, cellular information processes may be defined almost to completion at the molecular scale (at least in the case of prokaryotic cells). This is not the case, evidently, with nervous systems and the variety of human organizational, cultural, and social developments. Concretely, the crucial evolutionary phenomenon of protein-domain recombination—knowledge recombination—will be analyzed here as a showcase of, and even as a model for, the interdisciplinary and multidisciplinary mixing of the sciences so prevalent in contemporary societies. Scientomics will be proposed as a new research endeavor to assist advancement. Informationally, the “society of enzymes” appears as a forerunner of the “society of neurons”, and even of the “society of individuals”.1. Introduction: Conceptions of Information Science
Some of the most difficult philosophical and scientific conundrums lurk under the discussion of what information is. Summarily approaching it as “distinction on the adjacent”, information becomes the first category, even prior to space and time notions, and at a pair with symmetry [1,2]. The apparent simplicity and depth of that statement should not beguile us into making implicit assumptions about the subject to which the distinction capability refers, the world on which such subject is situated, and the characteristics of the scientific observer as well as the disciplinary backgrounds to apply, conspire together against the framing of a coherent “catholic” discourse. A previous rhetorical consensus needs to be established on such matters. Otherwise definitions and classifications of information will continue to pile up, endlessly, without much commonality or congruence. As a matter of fact in the intervening years after the appearance of the information field, with Shannon, Wiener, Bateson, etc., a “cottage industry” on the definition of information has been established in the literature: in the hundreds and close to the thousands of definitions have been attempted [3].
Part of the problem lies in the restricted views about disciplinary development held by most researchers in the field. A new science is not established necessarily around a brand-new theory or after a successful theoretical unification. There are many ways to produce the birth of a discipline—actually any major discipline has established its own unrepeatable path along history, often around some “founding fathers” and a few “great books”. Think of the historical sequence that has decisively contributed to configuring the fundamental disciplines of Western science: natural (mathematical) science with Galileo's Dialogo, physics with Newton's Principia, chemistry with Lavoisier's Traité, biology with Darwin's Origins, and neuroscience with Cajal's Textura.
In our times, the view of isolated disciplines appearing and thriving around some central theory is untenable in most cases (at least, in the most interesting ones). Rather, disciplines coexist within “ecologies” of multiple fields and subfields of specialization—modules—that enter in communication with many other specialties or modules temporarily organizing domains of knowledge around important research topics. It is like an ecosystem of cognitive entities where “knowledge recombination” becomes the dominant mode of disciplinary communication, growth, and creation of new entities. Not many authors have recognized the multidisciplinary implications of knowledge recombination [4]. In that regard, as will be argued here, the cellular evolutionary paradigm on the evolution of protein domains—the “big bang” of protein evolution—appears as the genuine precedent of any knowledge recombination processes, which interestingly have also been proposed within neuronal realms [5].
The argument of this paper is framed under the above premises. It is an exercise of knowledge recombination—a very preliminary one indeed—basically performed between the classical notions of information science and the experimental facts of the cellular-biomolecular camp, in order to obtain overall directions for the bioinformation paradigm which could also be applied or discussed somewhere else, i.e., in the sciences themselves. As stated, we will march from genomics to scientomics.
Given that the knowledge recombination phenomenon also seems to occur in advanced neuronal systems (“neuronal workspace”) [5], the advancement of the bioinformation paradigm may apportion interesting insights and recombinatory impetus into a plurality of fields. Together with quantum information and cosmological information, as well as advanced artificial intelligence, these areas constitute the avant-garde producing most of the new answers and new results in the quest to establish a consolidated information science, beyond the classical—repetitive, self-centered—discussions.
In the extent to which the living cell is one of the central paradigms of the nascent information science, this discussion is also an inquiry about the essential cluster of concepts that should potentially apply to the analysis of other information-based entities [6]. As already stated, the way cells have evolutionarily handled their cognitive resources is more than a metaphor of the social handling of knowledge.
2. The Bioinformation Paradigm
From the information science point of view, the living cell incorporates the highest trove of informational phenomena that one can think of at the molecular scale. It is a micro-world teeming up with millions of specific molecular recognition events, genetic codes, transcription and translation processes, molecular machines and self-assembling complexes, signaling systems, messengers, transducers, second messengers, regulators, effectors, connectivity networks, interferences, etc. Conspicuously, the information metaphor has become the natural way of talking about biomolecular phenomena, almost from the very beginning, and even more along the current omic and bioinformatic revolution [6].
Biologically, the phenomenon of information involves far more than standard metrics on well-defined signaling spaces or heterogeneous structures. It has to deal with the whole happenstances occurring in the communication processes between autonomous life cycles—in the connection between networked entities themselves in the making, “in formation”. This implies self-production organization, inner generation of functional “voids”, signal creation, encoding, channel transmission, reception, decoding, meaning elaboration, self-modification, and production of an adaptive response. Indeed what information supports is a new mode of adaptive existence in contrast with inanimate nature. Rather than resorting to those especial terms coined during the 1970s and 1980s (self-transcendence, autopoiesis, autogenesis, autocatakinesis, self-production, etc.) we may just say that the living existence is informational.
What are the fundamental informational characteristics of the living cell? The need to consistently sketch them has been argued in [7]. Three basic themes were introduced there. Fundamentally, it was argued that molecular recognition varieties should be taken as the starting point of the bio-information analysis. Further it was argued that the multidomain embodiment of function in enzymes and proteins allows for configuration of a new type of collective computation within an evolvable quasi-universal problem-solving “engine”, or a quasi-universal “constructor” more properly. Finally, the advancement of the cellular cycle in its adaptive coupling with the environment—signaling system mediated—had to be taken as the semiotic reference frame (with the appearance of meaning, value, fitness, and intelligence), thereby bringing closure to the informational “cellular theory”. These ideas will be amplified in what follows.
2.1. The starting Point: Molecular Recognition
Molecular recognition is the key element from which the whole biochemical and evolutionary universe is constructed. Like any other chemical reaction, recognition between molecules is based on the “making and breaking of bonds”. This—and only this—is what makes the mutual recognition and the formation of complexes between biomolecular partners possible.
By far, it is in the highly heterogeneous molecules that constitute the molecular “soup” of any living cell where molecular recognition reaches its maximal universality, ubiquity, and combinatory capabilities [8]. Myriads of recognition encounters at the cytoplasm of a bacterium are taking place in a highly organized and systematic way: no “insulating wires” are needed to organize complex networked functions involving specific contacts among multiple molecular partners; and this “wirelessness” is one of the most remarkable information processing resources of the living cell—a wired cell would be unthinkable at the molecular scale!
The problem with biomolecular recognition instances is that they involve an amazing variety and combinatorics of almost any type of chemical interaction: hydrogen bonds, hydrophobic/hydrophilic forces, dipole forces, van der Waals forces, ionic Coulombian forces, etc. Dozens or even hundreds of weak bonds participate, for instance, in the formation of a protein-protein specific complex. Quite probably, measuring molecular recognition and establishing its crucial parameters and variables can only be realized biologically on a case-by-case basis. At least this is the current trend in most molecular biological and molecular dynamic approaches. A few references, however, could provide some interesting insights about molecular-recognition generalities. First, W. Meggs [9] about “biological homing”, mainly from a Coulombian “lock and key” combinatory point of view; then S.K. Lin [10] about the changes in thermodynamic entropy of mixing derived from molecular similarity changes; and finally M. Carlton [11], with original proposals for measuring the information content of any complex molecular system.
The usefulness and depth of symmetry considerations in molecular recognition phenomena, as emphasized by S.K. Lin [10], are self-evident. Symmetry allows a direct classification of biomolecular recognition occurrences by means of three ordering categories: identity, complementarity and supplementarity. They respectively mean: recognition by sharing identical molecular properties (e.g., self-organization of phospholipids in membranes), recognition by means of complementary properties of the molecular partners (e.g., moieties, or the nucleic acids' double helix), and recognition through a quasi-universal capability to wrap or envelop any molecular shape by building a complex molecular scaffold of weak bonds around the target (e.g., enzymatic active sites, protein complexes). In the latter supplementarity case (not contemplated by Lin's approach), the partial surfaces involved are inherently sloppy in their specificity and display a variable affinity with respect to the very clean and holistic matching between molecular fractions (complementarity), but they are highly tunable by evolutionary procedures. The three mentioned categories are easily distinguishable in the architectural arrangements of the cell: structural support, memory banks, and functional agents.
Another way to understand molecular recognition is the mutual information approach, as shown by J.D. Wichard [12]. The mutual information measures the dependence of two random variables, being zero when they are statistically independent. The central concept of this approach is that molecules sharing a high similarity to existing ligands have an enhanced probability to share the biological profile (e.g., DNA encoding). In the coupling of receptors and messengers, antibodies and antigens, or in complementary surfaces of proteins, the mutual information between the respective sequences precisely peaks when the highest degree of specific recognition is achieved. That way, knowledge of one of the variables reduces the uncertainty of the other, and vice versa. The point in molecular recognition is how much interdependence is necessary to exclude other bonding possibilities. Information theory and mutual information provide a general rule about how many positions in a sequence must be selected to evaluate this.
2.2. Self-production: Transcription and Signaling
Essentially the cellular game is about a collective problem-solving dynamics applied to self-production of their own structures—implying both synthesis and degradation—which is performed by a “network society” of specialized enzyme and protein agents, continuously exchanging information about their specific activities thanks to the especial solvent properties of the water matrix. In response to communicational signals of the environment, thousands of constitutive enzymes and proteins, “nanomolecular processors” endowed with a peculiar modular structure, are synthesized (and also degraded) from the sequential generative information of the DNA and RNA “data bases”, which are themselves incessantly subject to evolutionary combinatory games [13].
The elements of the constitutive architecture (“diluted” enzymes and proteins) are all of them coded into the generative architecture (“sequential” DNA and RNA), and the functional control of the latter by the former provides the core self-production and self-modification capabilities of the system—how gene expression is controlled by transcription factors. Traditionally most studies have focused on the expression of individual genes and not in the overall network and systemic instances of control. Currently, however, massive transcriptional regulatory networks are built for different prokaryotic microorganisms and for eukaryotic cellular functions and specialized cell-types.
As an instance of such networks, the authors have compiled a large-scale M. tuberculosis transcriptional regulatory (TR) network, which has been built upon a previously published TR network [14] the largest to date, with further addition of different kinds of resources pertaining to publicly available sources: DNA microarrays, operons, orthology approaches, and synthetic biology experiments [15]. See Figure 1.
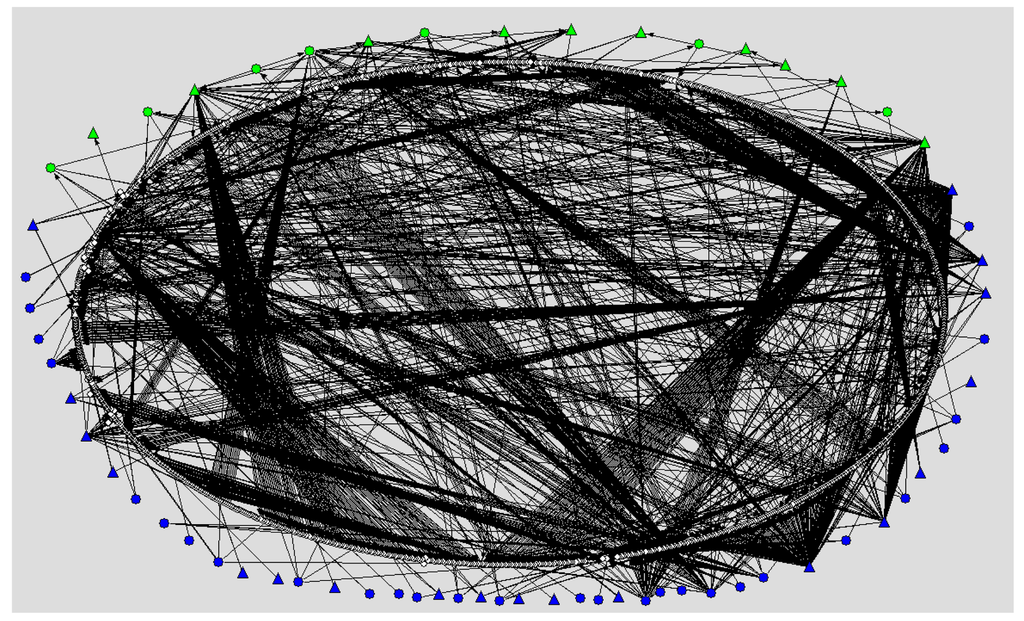
Of the 1400 network nodes represented, all correspond to specific genes of M. tuberculosis and their protein products, while the 2304 links correspond to gene expression regulatory interactions by 94 transcription factors. The network shows a clear organization in structural levels that correspond with the complex functions and life-cycle stages of this highly sophisticated pathogen. Overall, the genome of the bacillus contains more than 4000 genes and close to 190 transcription factors. In general, an increased number of transcription factors per genome translate into greater genetic network connectivity, which is correlated with increased complexity of the microorganism structures and life cycle [17].
By itself the transcription network pictured is “blind”. In other words, the coupling between the sequential and the diluted architectures needs the injection of further adaptive capability to respond to environmental demands. This is done by means of signaling guidance, which allows deploying the genetic circuits in response to relevant happenstances of the environment or from within the cell. The topological governance of the transcription regulatory network, the decision of what parts should be activated or what particular circuits should be inhibited, is achieved thus by the cellular signaling system or signalome.
In prokaryotes, a variety of molecular systems are involved in the signalome, ranging from simple transcription-sensory regulators (a single protein comprising two domains), such as the well-known embR, alkA or furB, to those systems of two, three or multiple components and interconnected pathways that regulate key stages of the cell cycle, such as latency, pathogenesis, replication, and dispersion. A basic taxonomy of bacterial signaling systems was proposed elsewhere [18]; it was centered on “the 1-2-3 scheme”.
In eukaryotes, the signaling system comprises many hundreds of different classes of dedicated molecular agents (receptors, ion channels, transducers, amplification cascades, second messengers, intermediate effectors, final effectors) that can be arranged differently in each tissue. Particularly throughout the very fast changes in second messenger concentrations, an integrated perspective (measurement) of the different internal and external influences at play is obtained within the cell, and is subsequently passed towards intermediate chains and the final effectors. At the end of the signaling command-chain, the gene expression machinery is waiting for appropriate signal combinations in order to change the transcriptional status of the genome—so that the well measured signals from the cytoplasmic signalome may be finally enacted as new transcription programs in relation to the advancement of the cell cycle or with the specialized function of the cell [19].
2.3. Information and the Advancement of a Life Cycle: Meaning, Value, Adaptability (Fitness), and Knowledge
The cell's life cycle is advanced by the confluence of genes permanently expressed (house-keeping systems) and genes that are expressed or inhibited after detection of specific environmental signaling coalitions or “affordances”. It is in the latter where the bulk of complexity lies, at least from the information-science point of view. The signaling system provides the cell with a generic capability to “grab” or “abduct” information from the environment and to make distinctions on the adjacent [2]. A “smart”, adaptable life-cycle is then possible.
The living cell may systematically respond to signal coalitions (“affordances”) from the environment and produce the meaning they imply by letting the signals themselves circulate throughout the signaling system pathways and meddle with the ongoing self-production “flow”. Meaning may be defined throughout molecular mining: as the (signal) induced changes in transcriptional connectivity and in constitutive enzyme-protein populations, plus associate metabolites and substrates. Nowadays the technology of microarrays routinely provides the meaning derived from gene down-regulation or up-regulation, in the form of a long list of gene expression alterations. The technology works, thus, like an alter ego of the signaling system, showing the result of selectively activating or inhibiting some particular path. Each gene transcription change, or protein concentration change, is appropriately gauged (percentage of increase or decrease) and can be incorporated into some functional cluster ontologically definable within the whole transcription list. Further components of meaning might be obtained by measuring alterations in the metabolome—and in other omes such as degradome and signalome—but usually they are of less relevance.
The value or relevance of impinging signals is established within a crescendo of occurrences: metabolic buffering, protein modifications, second messengers' switch, single gene expression, massive transcriptional rewiring. The cell cycle “checkpoints”are at the top of the value ladder. Crossing them marks the advancement through the stages of the cell cycle; this is the fundamental instance of reference. Completion of the cycle is achieved only after all the intracellular signaling instances known as checkpoints have been crossed favorably. Otherwise ad hoc inner signals will be deployed provoking arrest, latency, apoptosis, necrosis, etc. When the life cycle is successfully completed, value is tantamount to fitness. One can differentially estimate the inherent value of some signaling, metabolic or genetic variation by approaching the change in fitness it has induced. Presumably, a finer metric could be obtained by means of the ascendancy concept developed by R.E. Ulanowicz for capturing the sustainability of ecosystems [20]. Based on Information Theory, ascendancy quantifies the beneficial reserves that unused capabilities can afford a (self-producing) ecological system in its response to disturbance. In the cellular case, it is clear that one of the main concerns of signaling pathways is the governance of gene circuits in order to move the whole system towards its most sustainable configuration: sensitivity analysis with respect to each individual path would quantify the value of that link “at the margin”.
The phenomenon of knowledge may be appended too, once the DNA codes of the participating protein domains have been selected, refined, and cohered within the evolutionary succession of life cycles. As we will discuss below, domain shuffling or domain recombination—actually, knowledge recombination in the realm of the living cell—appear as the main evolutionary force propelling the development of signaling systems and achieving the adaptive coupling with the environment, either in prokaryotes or in eukaryotes.
Are cells but self-producing computers? The organization of prokaryotic genomes has been compared to computer operating systems in terms of the topology and evolution of their regulatory networks [21]; and new terms such as “paleome”, “cenome” “minimal genome” and others have also been coined in this computational direction [22]. However, the connection between transcription networks, signaling systems, and evolutionary strategies [23] must conduce to a reflection on the molecular keys of cellular intelligence. By means of systems biology, bioinformatics, synthetic biology, and network science we can start to translate the conceptual cluster around information (meaning, value, knowledge, intelligence) in rather precise molecular terms.
2.4. Cellular Intelligence and the Evolution of Protein Domains
The importance of domain recombination in the whole biomolecular evolution is not sufficiently appreciated due to the simplified conceptions generally held about biological function. Briefly returning to the molecular recognition theme, the integral function of each biomolecular agent coded in the genome implies not only the recognition of the functional “what” dictated e.g., in the active site of the enzyme, but also a series of accompanying processes distributed over different parts of the molecular structure, which might include: modulation by effectors, intracellular transportation, permanent (post-translational) modification, formation of complexes, time-frames derived from transcription and translation, and timing of the final (or partial) degradation.
Thus, in functional terms, the “what” of the primary function should be accompanied by many other functional circumstances such as: how fast, where, which way, with whom, when, and how long. In general, they are independently defined onto separate DNA sequences, constituting so to speak “addresses” dispersed within different domains of the same protein, separately coding for the specific operations of control (each one implying some specific “secondary address” in the DNA coding, irrespective that it may be functionally operative in the DNA, RNA, or in the protein stages). Changing any one of these domains might dramatically alter the deployment of the function, its timing, connectivity, cellular location, etc. Besides, every molecular stage (transcription, folding, modification, transportation, complexes, degradation) specifically coded onto DNA addresses may be used as a new functional element of control. Evolutionary solutions might be chosen, then, from an augmented set of molecular building blocks. The so called “Central Dogma” of classical molecular biology should not be taken as a closed black-box; rather the successive stages and intermediate transcripts could participate as legitimate molecular partners, each one endowed with endogenous recognition capabilities, within a whole transmolecular matrix of controlling interactions [7].
The possibility of systematic tinkering upon multiple domains becomes one of the most distinctive evolutionary strategies of eukaryotes, the tool-box of their multicellularity. In prokaryotes, the global arrangement of embodiment processes is correspondingly simpler; their protein components are smaller and contain comparatively fewer domains. In eukaryotes, the serial-combinatoric arrangement of introns and exons (which usually constitute autonomous folding domains) allows, by differential splicing, a far bigger proteome than prokaryotes—around one or two orders of magnitude, without multiplying the number of genes involved [24]. To reiterate, by tinkering and playing combinatory games upon DNA codes containing a vast array of primary and secondary addresses, eukaryotic and prokaryotic cells may systematically explore and change the whole boundary conditions surrounding the triggering of each biomolecular function—mastering all those circumstances of when, where, how fast, which way, for how long, with whom, etc., which together co-determine functional action. It is crucial, then, that primary function addresses and secondary addresses regarding the circumstances of primary functions have been put together (often in separate domains) onto the same DNA memory bank. The parallel with the von Neumann scheme of modern computers seems unavoidable: for memory addresses and logical functions are also coded side-by-side into the CPU memory (Central Processing Unit) of computers.
The generation of variety within biological genetic algorithms is surprisingly complex in most eukaryotic genomes, potentially involving occurrences such as: genetic polymorphisms —Single Nucleotide Polymorphisms (SNPs), Restriction Fragment Length Polymorphisms (RFLPs), Variable Number Tandem Repeats (VNTRs)—, repetitive DNA, mobile elements, transposons, retrotransposons, meiotic recombination, telomere shortening, gene and segmental duplications, chromosome fissions and fusions, whole genome duplications, symbiosis, etc. The striking complexity of eukaryotic bauplans and organismic physiologies has been achieved only by the combined action of all these engines of variation—most of them implying domain recombination—that impinge in the embodiment of biomolecular functional agents. Subsequently, in the evolutionary strategies of prokaryotic and eukaryotic cells, their respective quasi-universal problem solving capabilities have been addressed directly towards the solution of molecular “assimilation” phenomena in one case, while in the other case they were addressed towards harnessing molecular “organization” phenomena (morphology and differentiation) [15].
The “big bang” of protein evolution, mainly based on domain recombination (fusion, shuffling, combination, accretion), has subtended the whole deployment of biological ingenuity and complexity—being both its evolutionary cause and consequence [25–27]. Ancient domains and ancient proteins are still diverging from each other, indicating an ongoing expansion of the protein sequence universe (Figure 2). Based on the total number of known life forms, it is estimated that there are close to 50 billion different types of proteins today, and the protein universe could hold many trillions more [28,29]. By means of bioinformatic tools, very old domains can be tracked down as they have interpenetrated and recombined with recent domains within more complex proteins, following horizontal gene transfer as well as genetic recombinations of all kinds in both prokaryotes and eukaryotes, always searching for putting into action more efficient genomes with improved sets of protein domain functionalities (primary functions plus the retinue of accompanying functional circumstances). The existing protein domains coded into the genomic DNA might be seen as the stock of knowledge of each species and of the biosphere as a whole. Through their temporary presence in the genome of each organism, protein domains continue playing their recombinatory games in the existential niches they are adapted to, and keep expanding the protein universe.
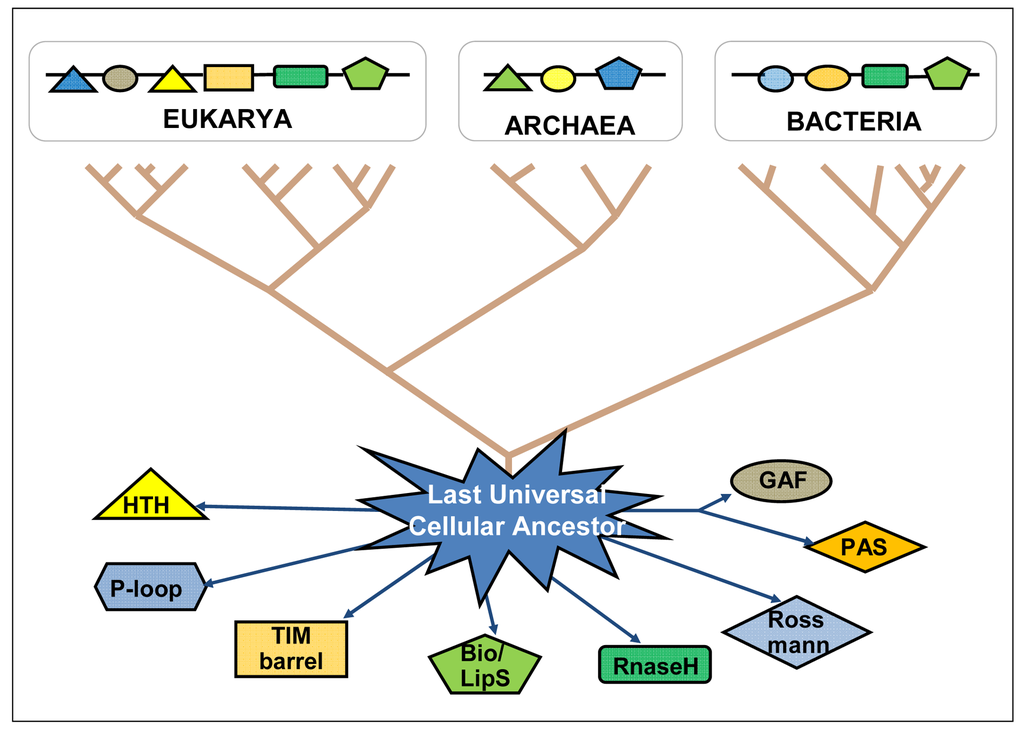
To recapitulate, are bacteria computers? [21,22]. Not exactly: in bacteria, very limited protein “agents” are capable of developing a collective processing that goes far beyond the computing bounds of each single agential entity, including self-production, multidimensional functionalities, structural self-coding, “representations” of the environment, and combinations of successful memories of past evolutionary experiences—knowledge recombination—which are continuously selected. From the point of view of artificial intelligence and natural computing there would be some lessons to learn and common goals to explore [30]. Along the emerging bioinformation paradigm we would finally have at hand a realistic definition of cellular intelligence, molecularly-based.
3. Knowledge Recombination in Cerebral “Workspace”
The transition from cells to brains implies an important change concerning the disciplinary backgrounds—and even more concerning the problems to be tackled. The absence of a consistent central theory in the neurosciences, similar to the Darwinian theory central role in the biological realm (or classical mechanics in physics), is a notorious problem in these sciences. The main directions of a possible theoretical development, also related with knowledge production, will be drafted in the following.
The NCT scheme (understood as a rough draft of a “Neurodynamic Central Theory”) addresses a new way of explaining the organization of brain information processes [31,32]. It establishes the correspondence between neurodynamics and behavior by means of a central theory grounded on dynamic connectivity (conectome) and on optimality (principles of brain economy). At the core of this theory, it is proposed that there is the development of an informational “behavioral-processual engine” ingraining the multidimensional operations of composition-decomposition of sensorimotor afferences and efferences with the realization of an action/perception cycle, producing adaptive behavior and associative learning (efficient knowledge) as outcomes. A number of disparate behavioral and cognitive aspects might be unified out from the development of this theory, including the recently coined brain's “dark energy” [33,34] and the global “workspace” proposed by Changeux, Dehaene, and others [5].
Very recent findings about the “conectome” need to be elaborated and generalized, both in their theoretical interpretation and in their experimental content [35,36]. Some works about the conectome have pointed at the emergence of dynamic core aggregates that fleetingly appear and disappear in milliseconds after any complex stimulus or mental process (originated either from the “outside” or from the “inside”). Such unending dynamics of fleeting aggregates has recently been dubbed as the brain's “dark energy”, and different cognitive-behavioral interpretations have been suggested, but until now they have not evolved into any sensible scheme [33,34]. Herein we suggest that the dynamic conectome has to be interpreted in terms of supersystem configurations of an information processing engine realized by cortical areas and medial nuclei, along an optimization process of local/global nature, and following symmetry-breaking/symmetry-restoration operations that make each cortically stored information unique and recoverable [31,32,37,38].
In the optimality aspect, the NCT scheme integrates the above findings with principles of maximum economy in space and time, and with symmetry-breaking and group theory concepts for distributed processes, that together configure a hierarchical-heterarchical scheme of information processing, learning and adaptive behavior [39].
In the human case (and in most advanced central nervous systems), it is the action/perception cycle that serves as the universal substratum for organizing behavior and subsequently tending the fabrication of meaning, categories and knowledge. Seemingly, we confront the world in accordance with such action/perception cycles or oscillations, regularly switching between dominant modes of behavior (motor centered versus sensory centered). The advancement of the cycle is based on a global minimization process performed upon an entropic global/local variable that cortical columns and medial nuclei cooperatively create and annihilate on a local basis, but also mediated by the organization of variable supersystem configurations, and implying formal rules of symmetry-breaking and symmetry-restoration.
The brain appears as an abstract problem-solving playground where topologically distributed variables (“tuning precision voids”) occurring at the neuronal columns of cerebral maps are processed as some overall entropy that different brain substructures and specialized modules tend to minimize. Because of the evolutionary design of nervous systems (e.g., the vertebrate phenomenon of decussation of the nerve fibers) internal and external organismic “problems” locally increase that entropy value. The subsequent blind (abstract) minimization by the nervous system's topological mechanisms and modular specialized subsystems produces as a byproduct the adequate behavioral and learning outputs. A problem-solving behavior well adapted to the advancement of the individual's life cycle emerges from all those distributed processes and minimization operations [40].
It is of particular interest in the human case that the combined system formed by the frontal and prefrontal areas with their massive increase in connectivity are breaking the brain's reliance on modular specialized subsystems and maximally expanding the combinatory possibilities. Following Dehaene [41], a “neuronal workspace” emerges whose main function is to assemble, confront, recombine, and synthesize knowledge. This system is further endowed with a fringe of spontaneous fluctuation that allows for the testing of new ideas, related to both the emergence of reflexive consciousness and the human competence for cultural invention. Although conscious brain activity fluctuates stochastically it does not wander at random. Selection mechanisms stabilize the combinations of ideas that are most interesting, useful or just contagious: privileged neuronal projections coming from the evaluation and reward circuits of orbitofrontal and cingulate cortex as well as the subcortical nuclei of amygdala and the basal ganglia are participating in this process.
Therefore, in the extent to which those premises are correct, a compact approach to knowledge automation and recombination by the central nervous system seems achievable, and further, a new kind of “theory of mind” could be contemplated. It will be close to current attempts on formulating a motor-centered epistemology, which has been deemed by relevant neuroscientists as one the best foundations for explaining our “automated cognition”. See different expostulations about the organization of action and advanced cognition [41–47].
In the perspective of the present paper, having found the knowledge recombination dynamics at the center of human cognition, facilitates the transition to discuss the same phenomenon within the highly specialized system of social knowledge we call science.
4. Social Recombination of Knowledge
Although we still lack adequate reliable “central theories” and “theories of mind” (as already said, a very unfortunate theoretical void) approaching science itself as a composite informational construction and particularly as knowledge recombination becomes feasible.
We can quote from Brian Arthur [48], in his recent approach to the nature of technological change, which is so close to the dynamics of science itself:
“Conventional thinking ascribes the invention of technologies to “thinking outside the box”, or vaguely to genius or creativity, but this book shows that such explanations are inadequate. Rather, technologies are put together from pieces—themselves technologies—that already exist. Technologies therefore share common ancestries and combine, morph, and combine again to create further technologies. Technology evolves much as a coral reef builds itself from activities of small organisms—it creates itself from itself; all technologies are descended from earlier technologies…”
The “natural” division of work within scientific communities, finally derived from the limitations of the individual scientist, seems to reflect the presence of knowledge recombination processes: the cooperative links between a number of separate, specialized disciplines (particularly after the scientific and industrial revolutions); the disciplinary reliance on accepted paradigms as well as the inevitable emergence of new explorations and interdisciplinary fields; the regular increase in the number of disciplines (at least during last two centuries); the networking structures and citation clusters highlighting the sharing of knowledge among heterogeneous scientific publications… It is quite difficult to estimate, however, what are the different social, psychological, or cognitive factors that determine the tolerable size of a field or specialization, or when the branching out of a new discipline will be produced. In order to contemplate the dynamics of the sciences, a new vantage point is needed.
The way different disciplines “process” their specific information and create new knowledge, and keep it in record, while at the same time this knowledge is widely disseminated so that it can be put into action, and again combined and recombined with elements of all the other disciplines, generates a “swarm intelligence” that goes far beyond the perception and action capabilities of the limited individual. The success of science in this informational jumping over the individual's limitations has been rationalized as the superiority of the scientific method (leaving aside any communication and thought-collective aspects) or directly attributed to “the unreasonable effectiveness of mathematics” [49]. However, there is not much understanding of the underlying informational causes [50,51].
The creation of knowledge is always an informational process, ultimately derived from knowledge recombination processes in the cerebral “workspace” of individuals. Reliable knowledge mediates action/perception cycles of individuals and prolongs them, supra-individually, making possible a more cogent and integrated closure at the social scale. The scientific method itself appears from this perspective as the conditions to be met for a coherent decomposition of problems by communities of problem-solvers whose workings are separated in time and space. In point of fact, the strict conditions put by the scientific method are also efficient protocols that grant the social decomposability of problems [52]. Standards, measurements, mathematical operations, formalizations, and so on, become ways and means to extract mental operations out of the individual's nervous system and directly interconnect perceptions and actions at a vast institutional-social scale [53]. We really see a “collective nervous system”, a “social workspace” in action.
Disciplines, rather than being isolated fields, are continuously mixing and rearranging their contents, recombining them, for the sake of the problems they have to solve, and factually giving birth to successive generations of inter-disciplines (e.g., information-physics, physical chemistry, biophysics, biochemistry, bioenergetics, bioengineering, socio-physics, sociobiology, psycho-sociology, neuro-psychiatry, socio-information, etc.). See in Figure 3 how research in a very advanced field—biomaterial research—is contemplated by one of its leading practitioners [54]. The crowding of subdisciplines and specialties is remarkable: up to 32 different ones are listed. It could remind the domain accretion of some large protein of late eukaryotic evolution, as the figure itself suggests by representing specialties linearly in a common thread of domains. Like in the evolutionary process, it makes sense that the most advanced scientific explorations incorporate larger troves of disciplines and specializations. That is particularly true in biomedical research, which has become one of the central and most complex scientific hubs of today.
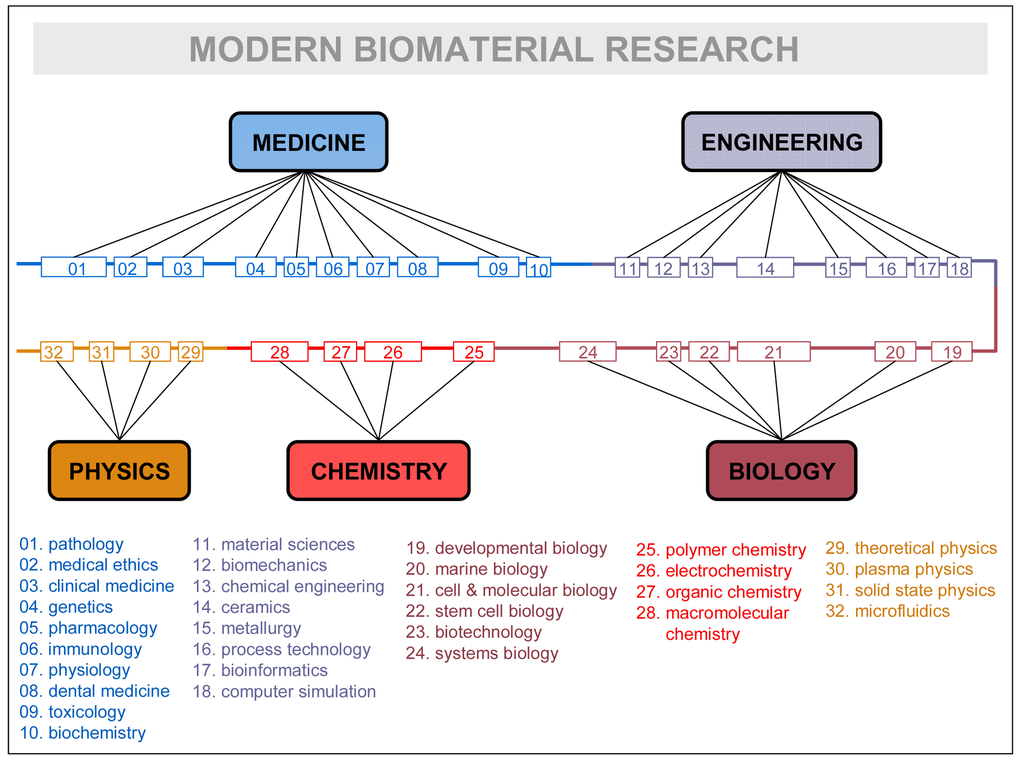
How can such a number of disciplines consistently relate within a research field? The relationship between disciplines, and in particular their overlapping or mixing, need in-depth clarification. The philosophical emphases on “reduction”, and the factual abandonment of reflection about its complementary “integration”, do not help explain the disciplinary (inter-multi-pluri-trans) game. The inconsistency of institutional discourses involving disciplines, research fields, domains, specializations, etc. does not help either.
As Figure 3 suggests, all major research fields have to be surrounded by a “cloud” of disciplines in order to convey the necessary scientific-technologic knowledge. We propose the term “domain of knowledge” to the particular collegiums of disciplines surrounding every major research field and potentially contributing to its knowledge recombination processes. It is clear, as in the case of Figure 3, that only some specialties or subdisciplines of each major science are actively involved in the exchange processes. Even at the level of a concrete subdiscipline, the real granularity of the exchanges concerns “modules of specialization” that incorporate theoretical and practical knowledge as embodied by some specialist, researcher, PhD student, etc. Knowledge is never disembodied, and the fundamental unit that contributes to the multidisciplinary enterprise is the individual practitioner—usually working in complex groups and providing expertise on his/her disciplinary grounds. In general, researchers will either need to develop those specialized skills or know enough about them to work with the specialist carrying out those tasks. Thus, it is the specialist who becomes the “module” supporting the different portions accreted in the knowledge recombination process, the equivalent of protein domains at the cellular level. Research fields are but niches of opportunity that attract experts of different disciplines and organize new domains of knowledge; if the research is successful and expectations are fulfilled, new disciplines of inter-multi-disciplinary nature will arise subtended by a new, ad hoc research community. In synthesis, that would be the relationship between fields of research, domains of knowledge, disciplines, subdisciplines, and specialized modules at play in the knowledge recombination process.
It has been estimated that after the industrial revolution the number of scientists and of research fields has roughly doubled with each passing generation [55]. (To be precise, “Smith terms this transformation cladistatic kinema-dynamism”, see Reference [55].) At the height of the 1990s it has been estimated that more than 8000 research topics were supported by approximately 4000 disciplines [56]. In the extent to which those estimates are cogent, the number of disciplines could have increased to 5000–6000 nowadays, supporting around 10,000 research fields.
Scientometric studies have already provided rigorous and useful “science maps”, during the last three decades; they were based on citation structures and have grown enormously—keeping pace with Moore's Law—up to impressive dimensions and multidisciplinary coverage. Representations derived from some million papers covering almost all research fields have been obtained recently [57]. Notwithstanding that, generative hypotheses on the overall science and society processes underlying the scientific growth continue to be in short supply.
From the point of view of information science, in the same way that philosophies of science, history of science, and psychology and sociology of science have already been developed, we would also need a genuine informational approach to science: scientomics. Exploring the relationship of the new approach with such big fields of research is clearly beyond the scope of this paper, but at least we can provide some hints on factual cul-de-sacs in philosophy of science that could be rediscussed along new suggestions derived from both information science and scientomics, e.g., reductionism vs. holism [58], instances of paradigm change [59], scientific methodologies and emergence of new disciplines [60], historical evolution of scientific atlases and iconic representations of the sciences [57], and so on. Whole fields of history of science might cover a new dimension if explored on informational grounds. We can realize how the major transitions in the history of Western science were related to new ways and means to externally encode human expertise, register it and preserve it: tablets, scrolls, and alphabet (Ancient science and Greek science); codices, copyists' schools, universities (Medieval Awakening); printing press, bookshops, and libraries (scientific revolution); new energy and production technologies, communication and education systems (industrial revolution); engineering, electronics, new materials, and giant corporations (scientific-technological 2nd revolution); media, computers, and internet (information revolution) [61–65].
5. The Evolutionary Adventure of Knowledge: From Genomics to Scientomics
The knowledge recombination hypothesis applied to the historical evolution of science has to be considered in scientomic terms. In its methodology, scientomics would share a genomics' inspirational and computational parallel with the recent culturomics enterprise, and also with a possible future field of “technomics”. The new approach of scientomics means an epistemic, historic, and scientometric quest on specialized modules of theoretical-practical knowledge that throughout their combinatory activity would have contributed to change the history of scientific disciplines. Thus, scientomics posits an inner structure of major recombination events along science history. Like in the protein universe, foundational modules would have traveled out from their disciplinary quarters to other disciplines or research fields and changed there the local textures of knowledge, revamping problem solving activities and opening new horizons, and even influencing the whole complexion of the world of knowledge at large.
Historically, knowledge recombination looks to be a plausible hypothesis. Influential modules such as Euclidian geometry, Newtonian mechanics, differential equations, genetics, and so on (and a multitude of other minor modules), would have generated most of the history of natural sciences, not only “developmentally” inside their own disciplines and research fields, but even more “combinatorially”, propelling the multidisciplinary evolution and cross-fertilization among scientific disciplines. Together with logics and natural philosophy, most of modern science fields were originated thanks to the migration of a very few ancient modules—fundamental ones—outside their own sphere of mathematical knowledge: arithmetic, geometry, algebra, calculus. They have been represented in Figure 4, at the bottom of contemporary mathematical subdisciplines and specialties. Obviously, important influences from nonscientific realms have also to be acknowledged in science history: religious, artistic, cultural, political, etc. However, in the cognitive melting pot of societies, those factors do not travel and mix with the same consistency, inner logical structure, and separateness as disciplinary domains.
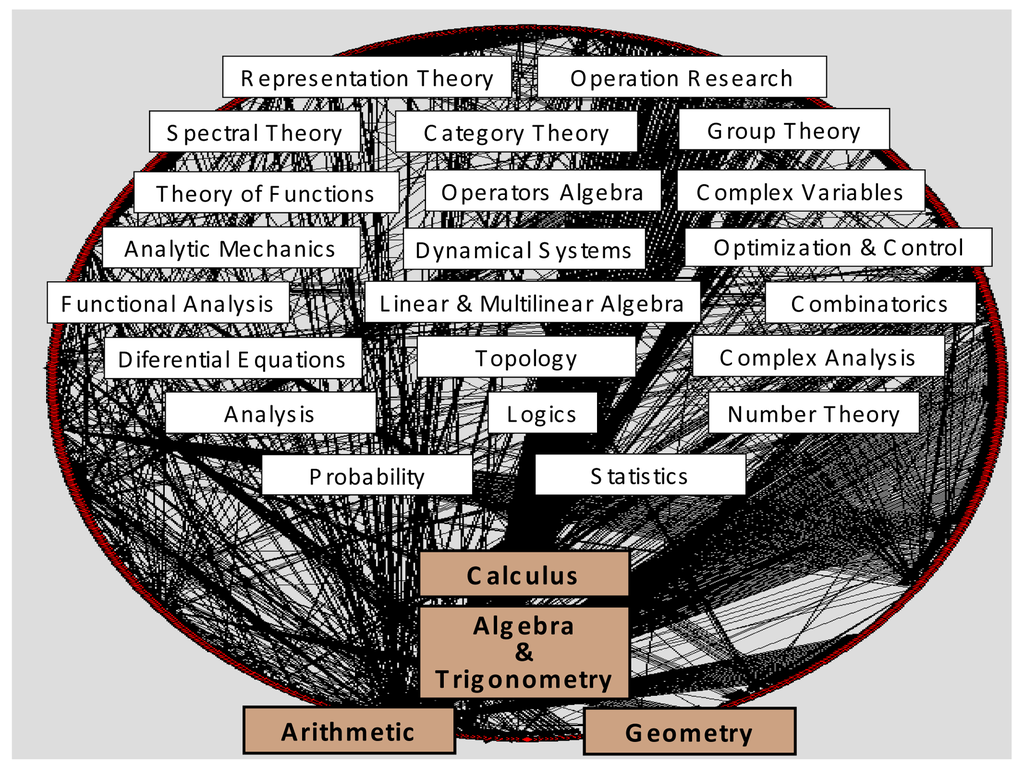
In terms of teaching and education science a comparable sequence of combinatory processes would take place. An abridged recapitulation resembling Haeckel's law seems to occur in the ontogenetic development of an individual's knowledge, which somehow recapitulates the fundamentals of the social acquisition of knowledge along history. Numbers, writing, arithmetics, geometry, etc. are progressively internalized as modules of increasing complexity along an education process that finally conduces to advanced expertise in some specialized matter.
The development of scientomics comes of age. It has already been started though in a limited way within the scientometric studies. Culturomics might have already paved part of the way too: borrowing the main concepts and techniques from evolutionary biology, J.B. Michel and E.L. Aiden were able to track the growth, change, and decline of the most meaningful published words during the last centuries [66]. The new term they have coined, culturomics, means the application of “genomic techniques” of high-throughput data collection and analysis to the study of human culture, as sampled in a vast mapping of words from a corpus of digitized books, containing about 4% of all printed books ever published. Further sources might be incorporated to the culturomic stock: newspapers, manuscripts, maps, artwork, etc. Analysis of this corpus enables a new qualitative and quantitative investigation of cultural trends, social and political influences, fashions, and all sort of cultural phenomena.
Scientomics could be an important future task for the consolidation of information science. It would appear as a multidisciplinary research-project running in parallel to current achievements of culturomics and scientometrics, though pointing at some more ambitious epistemic goals. It should carefully revise the history of knowledge in order to instantiate the changing modular structure of the major disciplines and the associated research fields, probably by means of genomic-inspired tools like culturomics and also by means of network science instruments like scientometrics; it should also revise the representations, maps of knowledge, and classifications of the sciences in the different epochs, as well as the teaching corpus of major disciplines. Means of communication and knowledge supports are also crucial elements of analysis, as they have marked the rhythms of information exchange (both oral and written) and have produced accelerations with dramatic expansion consequences (“scientific revolutions”)… Finally the central goal of scientomics is capturing the “big bang” of the science universe.
These are rough, preliminary reflections. In the extent to which a germinal project succeeds in joining these basic ideas, the creation of a proficient scientomics new field of research would help to make a more cogent sense of the historical processes of science, and of human knowledge in action. Social planning and management of the scientific effort in the “knowledge society” would undoubtedly benefit.
6. Concluding Comments
It can be argued that the growth of informational complexity of cells, nervous systems, and societies along their respective evolutionary, ontogenetic, and historical trajectories has been based on the cumulative consequences of knowledge recombination phenomena. As said, from the point of view of artificial intelligence and natural computing, there could be valuable lessons to learn and common goals to achieve [30]. Very limited “agents” are capable of developing a collective processing that goes far beyond the computing bounds of each single agential entity; and their collective processing includes the dynamics of knowledge recombination in all the different information realms. One of the common abstract traits is the existence of inner “knowledge repositories” where real physical structures are encoding the expertise of the system in its relationship with the environment. It is amazing that among the procedures to create novelty in those repositories, a central strategy becomes the swapping of knowledge “chunks” from one construct to another, so that new cognitive constructs emerge and new adaptive capabilities are deployed. We have seen how this occurs evolutionarily in cellular protein-domain recombination, and how mental constructs seem to be handled in a similar way. From a highly macroscopic point of view, the different disciplines would also partake in this common strategy of knowledge recombination, as we have discussed for the concrete case of biomaterial research.
The recognition of this cognitive commonality, and indeed the explicit definition of knowledge recombination, has been obscured among other causes, by the structural and dynamic heterogeneity of repositories in the different informational entities, and above all by being the subject of quite separate clusters of scientific disciplines: molecular and evolutionary biology, cognitive neurodynamics, philosophy of science/geography of science. Seemingly, increased epistemic distance translates into more difficult and less frequent interrelation processes.
For the time being, practically testing the recombination idea might be achieved only partially. There is as yet insufficient development in the neurosciences concerning the set of concepts mentioned (“workspace”, “behavioral-processual engine”, “dark energy”). However, there might be sufficient room to compare the biological evolution of DNA codes of protein domains and the social-historical evolution of scientific disciplinary contents. Do cognitive “modules” exist within disciplines that travel to other disciplines and generate new fields there? If so, could the combinatory processes in both realms be interrelated in a new, deeper way? That is what scientomics is about.
Tentatively applying these insights to the development of information science itself, a more open, participatory attitude would be suggested. Rather than the self-centered inward looking, vying for completion of the paradoxical or for unification of the intrinsically unbounded—is information definable at all?—a redirection of some of those energies is suggested: towards outward seeking. Information science has to actively and intensely engage in the collective participatory game of the sciences, in the generation of new knowledge throughout the recombination common play. Actually both directions of thought are complementary and have to be kept in mutual balance: otherwise it may well happen that a promising development turns into pitiful stagnation.
Acknowledgments
Thanks to TripleC for their kind permission to base the present paper on the contribution presented in the Proceedings of FIS 2010 Beijing Conference; and for the same reason to ITHEA, in the Proceedings of GIT 2011 Varna Conference.
References
- Marijuán, P.C. First conference on foundations of information science: From computers and quantum physics, to cells, nervous systems, and societies. BioSystems 1996, 38, 87–96. [Google Scholar]
- Marijuán, P.C. The advancement of information science: Is a new way of thinking necessary? TripleC 2009, 7, 369–375. [Google Scholar]
- Lenski, W. Information: A conceptual investigation. Information 2010, 1, 74–118. [Google Scholar]
- Scott, J.C. Seeing Like a State; Yale University Press: New Haven, CT, USA, 1998. [Google Scholar]
- Dehaene, S.; Kerszberg, M.; Changeux, J.P. A neuronal model of a global workspace in effortful cognitive tasks. Ann. N. Y. Acad. Sci. 2001, 929, 152–165. [Google Scholar]
- Marijuán, P.C. Information and life: Towards a biological understanding of informational phenomena. TripleC 2004, 2, 9–19. [Google Scholar]
- Marijuán, P.C.; del Moral, R. The Informational Architectures of Biological Complexity. In Computation, Information, Cognition—The Nexus and The Liminal; Dodig-Crnkovic, G., Stuart, S., Eds.; Cambridge University Press: Cambridge, UK, 2007. [Google Scholar]
- Goodsell, D.S. Inside a living cell. TIBS 1991, 16, 203–206. [Google Scholar]
- Meggs, W.J. Biological homing: Hypothesis for a quantum effect that leads to the existence of life. Med. Hypothesis 1998, 51, 503–506. [Google Scholar]
- Lin, S.K. The nature of the chemical process. 1. Symmetry evolution—Revised information theory, similarity principle and ugly symmetry. Int. J. Mol. Sci. 2001, 2, 10–39. [Google Scholar]
- Carlton, M. The Information Paradigm, Posthumous compilation. 2002. Available online: https://webmail.unizar.es/pipermail/fis/2005-April/000956.html (accessed on 1 November 2011).
- Wichard, J.D. Binding Site Detection via Mutual Information. Proceedings of the World Congress of the Computational Intelligence (WCCI), Hong Kong, China, 1–6 June 2008; pp. 1770–1776.
- Marijuán, P.C. Bioinformation: Untangling the networks of life. BioSystems 2002, 64, 11–118. [Google Scholar]
- Balázsi, G.; Heath, A.P.; Shi, L.; Gennaro, M.L. The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. Mol. Syst. Biol. 2008, 4, 225:1–225:8. [Google Scholar]
- Navarro, J.; Goñi-Moreno, A.; Marijuán, P.C. Varieties of biological information: A molecular recognition approach to systems biology and bioinformatics. ITHEA Intern. J. Inf. Theor. Appl. 2010, 4, 56–66. [Google Scholar]
- Navarro, J. Transcriptional Regulatory Network of M. tuberculosis: Functional and Signaling Aspects. Master Thesis, Universidad de Zaragoza, Zaragoza, Spain, 2010. [Google Scholar]
- Levine, M.; Tjian, R. Transcription regulation and animal diversity. Nature 2003, 424, 147–151. [Google Scholar]
- Marijuán, P.C.; Navarro, J.; del Moral, R. On prokaryotic intelligence: Strategies for sensing the environment. BioSystems 2010, 99, 94–103. [Google Scholar]
- Janes, K.A.; Albeck, J.G.; Gaudet, S.; Sorger, P.K.; Lauffenburger, D.A.; Yaffe, M.B. A systems model of signaling identifies a molecular basis set for cytokine-induced apoptosis. Science 2005, 310, 1646–1653. [Google Scholar]
- Ulanowicz, R.E.; Goerner, S.J.; Lietaer, B.; Gomez, R. Quantifying sustainability: Resilience, efficiency and the return of information thory. Ecol. Complex. 2009, 6, 27–36. [Google Scholar]
- Yan, K.K.; Fang, G.; Bhardwaj, N.; Alexander, R.P.; Gerstein, M. Comparing genomes to computer operating systems in terms of the topology and evolution of their regulatory control networks. PNAS 2010, 107, 9186–9191. [Google Scholar]
- Danchin, A. Bacteria as computers making computers. FEMS Microbiol. Rev. 2009, 33, 3–26. [Google Scholar]
- Peisajovich, S.G.; Garbarino, J.E.; Wei, P.; Lim, W.A. Rapid diversification of cell signaling phenotypes by modular domain recombination. Science 2010, 328, 368–372. [Google Scholar]
- Claverie, J.M. What if there are only 30,000 human genes? Science 2001, 291, 1255–1257. [Google Scholar]
- Koonin, E.V.; Wolf, Y.W.; Karev, G.P. The structure of the protein universe and genome evolution. Nature 2002, 420, 218–223. [Google Scholar]
- Dokholyan, N.V.; Shakhnovich, B.; Shakhnovich, E.I. Expanding protein universe and its origin from the biological Big Bang. PNAS 2002, 99, 14132–14136. [Google Scholar]
- Chotia, C.; Gough, J.; Vogel, C.; Teichman, S.A. Evolution of the protein repertoire. Science 2003, 300, 1701–1703. [Google Scholar]
- Povolotskaya, I.S.; Kondrashov, F.A. Sequence space and the ongoing expansion of the protein universe. Nature 2010, 465, 922–926. [Google Scholar]
- Koonin, E.V.; Puigbò, P.; Wolf, Y.I. Comparison of phylogenetic trees and search for a central trend in the “forest of life”. J. Comput. Biol. 2011, 18, 917–924. [Google Scholar]
- Zhong, Y.X. On Information: An Introduction to “Principles of Information Science”. Proceedings of Fourth International Conference on the Foundations of Information Science, FIS'10, Beijing, China, August 2010.
- Collins, K.P. On the Automation of Knowledge within Central Nervous Systems. Proceedings of the American Association for the Advancement of Science, Washington, DC, USA, February 1991.
- Collins, K.P.; Marijuán, P.C. El Cerebro Dual: Un Acercamiento Interdisciplinar a la Naturaleza del Conocimiento Humano y Biológico; Editorial Hacer: Barcelona, Spain, 1997. [Google Scholar]
- Raichle, M.E. The brain's dark energy. Science 2006, 324, 1249–1250. [Google Scholar]
- Raichle, M.E. The Brain's Dark Energy. Sci. Am. 2010, 302, 28–33. [Google Scholar]
- Zamora-López, G.; Zhou, Ch.; Kurths, J. Cortical hubs form a module for multisensory integration on top of hierarchy of cortical networks. Front. Comput. Neurosci. 2010, 4, 1–13. [Google Scholar]
- Sporns, O. The non-random brain: Efficiency, economy, and complex dynamics. Front. Comput. Neurosci. 2011, 5, 1–13. [Google Scholar]
- Leyton, M. Symmetry, Causality, Mind; The MIT Press: Cambridge, MA, USA, 1992. [Google Scholar]
- Turvey, M.T. Impredicativity, Dynamics, and the Perception-Action Divide; Jirsa, V.K., Kelso, S., Eds.; Springer-Verlag: Berlin, Heidelberg, Germany, 2004. [Google Scholar]
- Marijuán, P.C. Cajal and consciousness: An introduction. Ann. N. Y. Acad. Sci. 2001, 929, 1–10. [Google Scholar]
- Marijuán, P.C. Information and symmetry in the biological and social realm: New avenues of inquiry. Symmetry Cult. Sci. 1996, 7, 281–294. [Google Scholar]
- Dehaene, S. Reading in the Brain; Penguin: New York, NY, USA, 2009. [Google Scholar]
- Allman, J.M. Evolving Brains; Scientific American Library: New York, NY, USA, 1999. [Google Scholar]
- Berthoz, A. The Brain's Sense of Movement; Harvard University Press: Cambridge, MA, USA, 2000. [Google Scholar]
- Edelman, G.M.; Tononi, G. A Universe of Consciousness: How Matter Becomes Imagination; Basic Books: New York, NY, USA, 2000. [Google Scholar]
- Fuster, J. Cortex and Mind: Unifying Cognition; Oxford University Press: New York, NY, USA, 2003. [Google Scholar]
- Changeux, J.P. The Physiology of Truth; Harvard University Press: Cambridge, MA, USA, 2004. [Google Scholar]
- Buzsáki, G. Rhythms of the Brain; Oxford University Press: New York, NY, USA, 2006. [Google Scholar]
- Arthur, B.W. The Nature of Technology: What it Is and How it Evolves; The Free Press and Penguin Books: New York, NY, USA, 2009. [Google Scholar]
- Wigner, E. The Unreasonable Effectiveness of Mathematics in the Natural Sciences. In Communications in Pure and Applied Mathematics; John Wiley & Sons: New York, NY, USA, 1960; Volume 13, I. [Google Scholar]
- Lanham, R.A. The Economics of Attention; The University of Chicago Press: Chicago, IL, USA, 2006. [Google Scholar]
- Wright, A. Glut: Mastering Information through the Ages; Joseph Henry Press: Washington, DC, USA, 2007. [Google Scholar]
- Rosen, R. Essays on Life Itself; Columbia University Press: New York, NY, USA, 2000. [Google Scholar]
- Hobart, M.E.; Schiffman, Z.S. Information Ages; The John Hopkins University Press: Baltimore, MD, USA, 1998. [Google Scholar]
- Kirkpatrick, J. Multi- & Interdisciplinarity in Bioengineering, Biomaterials & Nanomedicine. Proceedings of the CIBER-BBN: Networking Biomedical Research Centre for Bioengineering, Biomaterials & Nanomedicine, Zaragoza, Spain, 17–18 February 2009.
- Landes, D.S. The Wealth and Poverty of Nations: Why Some Are So Rich and Some So Poor; W.W.Norton: Brooklyn, NY, USA, 1998. [Google Scholar]
- Klein, J.T. Interdisciplinary and Complexity: An evolving relationship. Emergence 2004, 6, 2–10. [Google Scholar]
- Börner, K. Atlas of Science: Visualizing What We Know; The MIT Press: Cambridge, MA, USA, 2010. [Google Scholar]
- Dupré, J. The Disorder of Things: Metaphysical Foundations of the Disunity of Science; Harvard University Press: Cambridge, MA, USA, 1993. [Google Scholar]
- Kuhn, S.T. The Structure of Scientific Revolutions; The University of Chicago Press: Chicago, IL, USA, 1996. [Google Scholar]
- Feyerabend, P. Against Method: Outline of an Anarchistic Theory of Knowledge; Verso: London, UK; Brooklyn, NY, USA, 1993. [Google Scholar]
- Scott, J.C. Seeing Like a State; Yale University Press: New Haven, CT, USA, 1998. [Google Scholar]
- O'Donell, J.J. Avatars of the Word; Harvard University Press: Cambridge, MA, USA, 1998. [Google Scholar]
- Hobart, M.E.; Schiffman, Z.S. Information Ages; The Jonh Hopkins University: Baltimore, MD, USA, 1998. [Google Scholar]
- Lanham, R.A. The Economics of Atention; The University Chicago Press: Chicago, IL, USA, 2006. [Google Scholar]
- Wright, A. Glut: Mastering Information through the Ages; Joseph Henry Press: Washington, DC, USA, 2007. [Google Scholar]
- Michel, J.B.; Shen, Y.K.; Aiden, A.P.; Veres, A.; Gray, M.K.; Pickett, J.P.; Hoiberg, D.; Clancy, D.; Norvig, P.; Orwant, J.; Pinker, S.; Nowak, M.A.; Aiden, E.L. Quantitative analysis of culture using millions of digitized books. Science 2011, 331, 176–182. [Google Scholar]
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).