KEGGSum: Summarizing Genomic Pathways
Abstract
1. Introduction
- We explore seven centrality measures (betweenness centrality, degree centrality, closeness centrality, PageRank centrality, Katz centrality, eigenvector centrality, and harmonic centrality) proposed in graph theory for capturing the importance of the graph nodes, examining their applicability to KEGG graphs as well.
- Besides identifying the most important nodes of the KEGG graphs, we link those nodes in order to present to the users a summary, i.e., a subgraph out of the original graph. In order to do so, we model the problem of summary generation as a Steiner tree problem, an approach commonly adopted for semantic summaries but not yet explored for KEGG graphs.
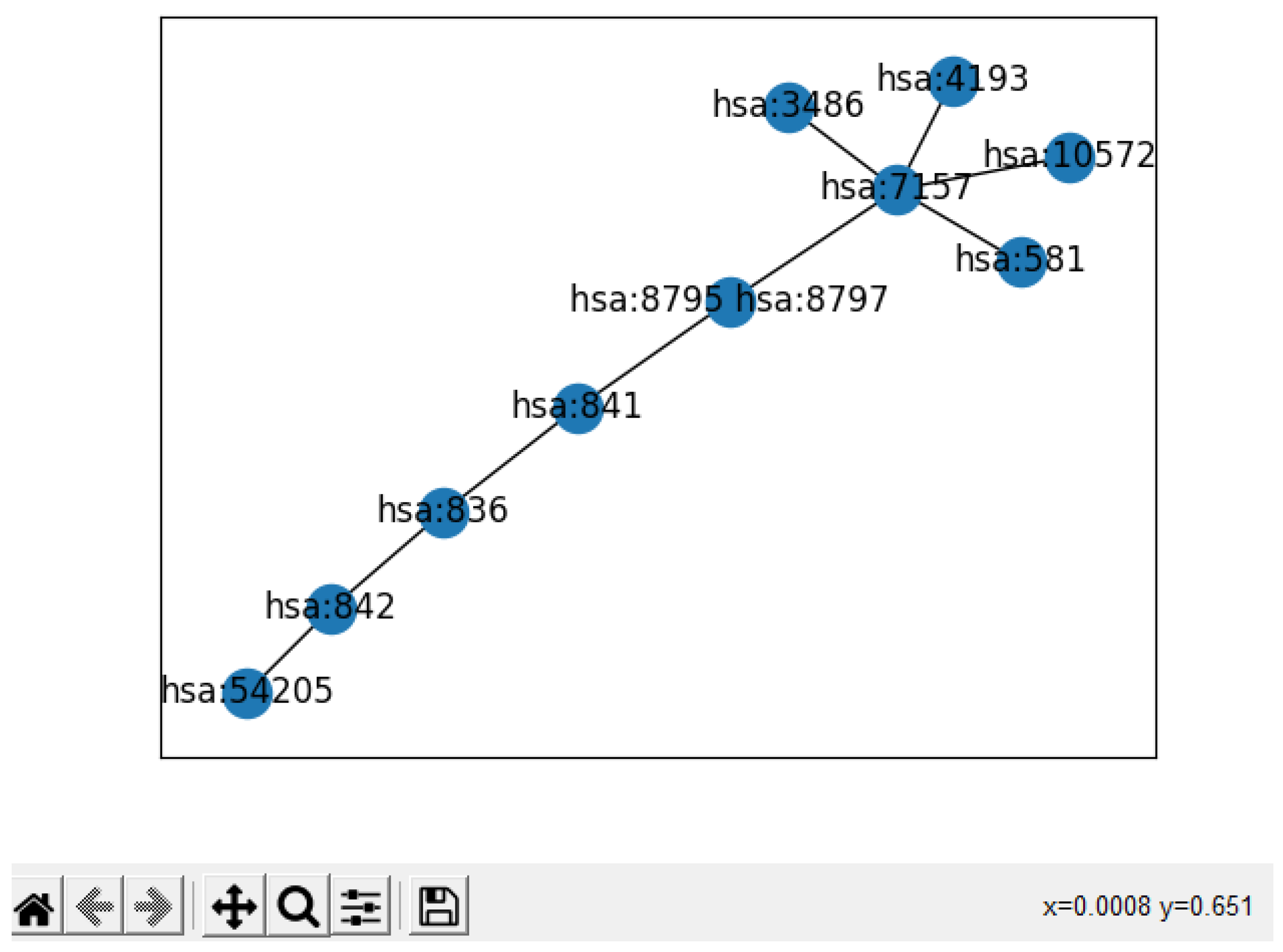
- We present a proof-of-concept visualization of the result summary, showing the researchers the most important subgraph from the original KEGG graph.
- Finally, we conduct an experimental investigation into the quality of the generated summary using three experts, identifying the usefulness of our approach. Our approach is able to select construct summaries with higher quality than using a competitive centrality measure proposed for biological networks, whereas it runs one order of magnitude faster.
2. Background
2.1. KEGG
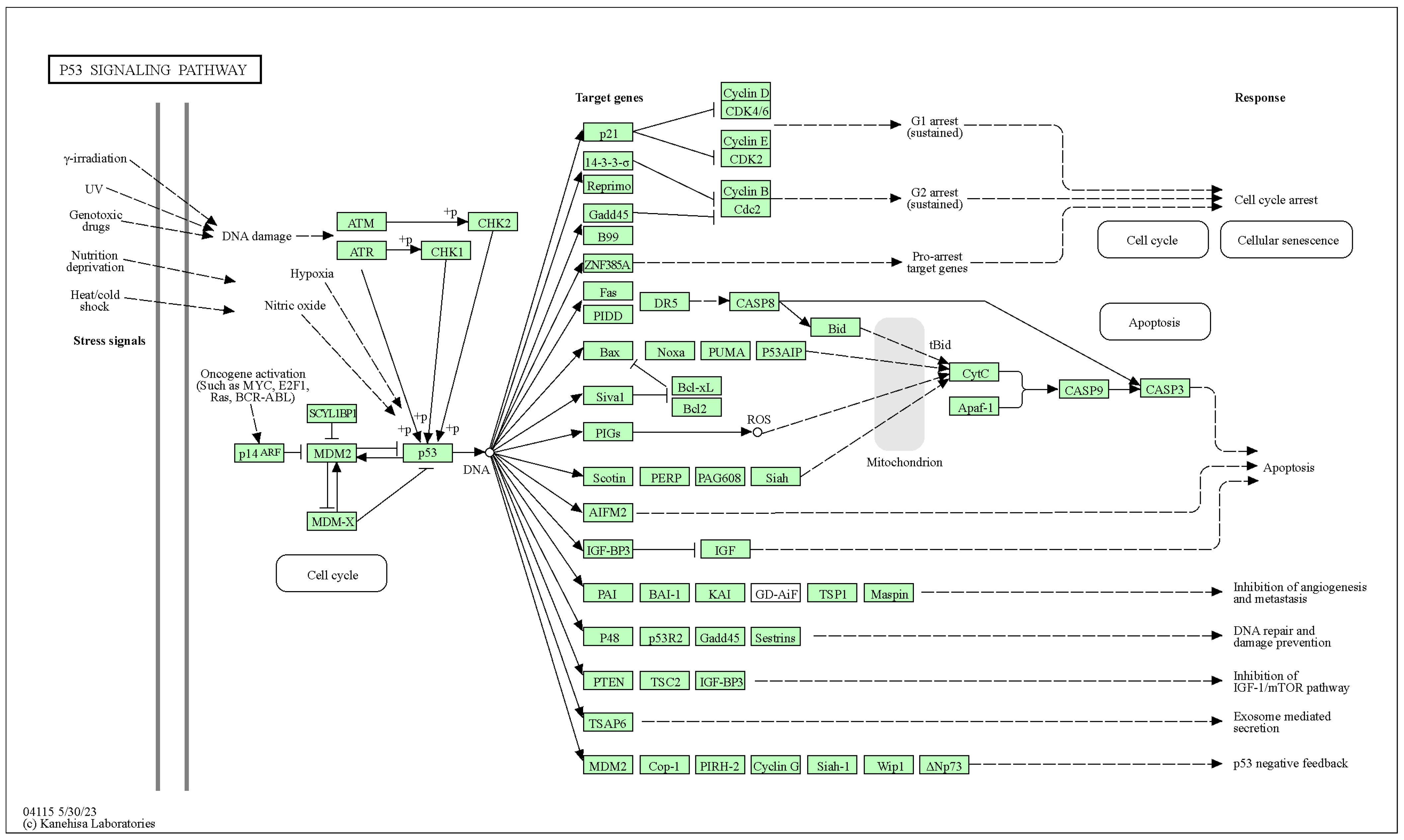
2.1.1. Pathway Maps
- Reference pathways, which are manually drawn;
- Organism-specific pathways, which are computationally generated based on reference pathways (https://www.genome.jp/, accessed on 12 December 2023).
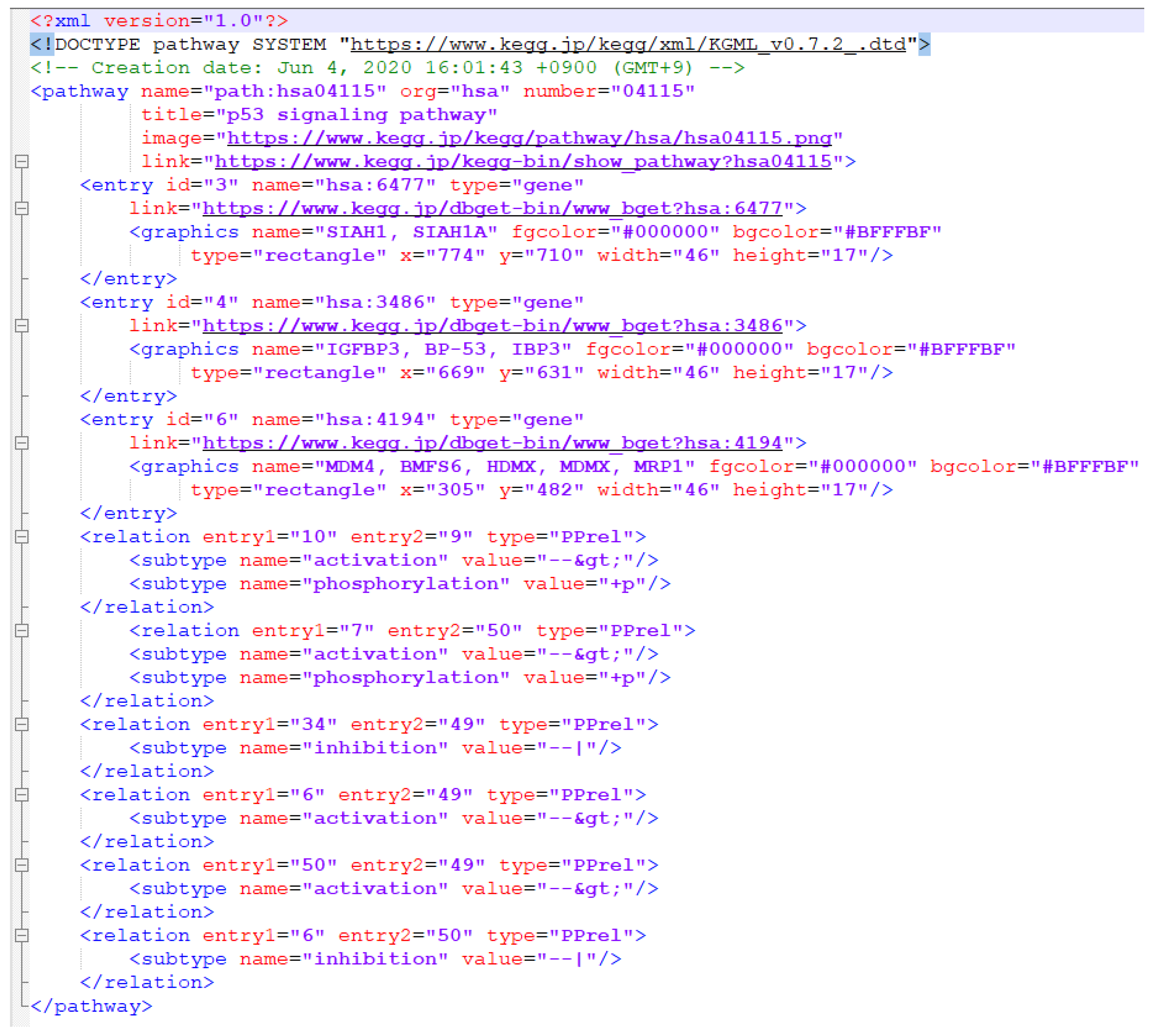
2.1.2. XML Representation of KEGG Pathway Maps
- ko—reference pathway map linked to KO entries (K numbers);
- rn—reference pathway map linked to REACTION entries (R numbers);
- ec—reference pathway map linked to ENZYME entries (EC numbers);
- org (three- or four-letter organism code)—organism-specific pathway map linked to GENES entries (gene IDs).
- Relations—relationships between boxes;
- Reactions—relationships between circles.
2.2. Graph Theory
2.3. Graph Summarization and Semantic Graph Summarization
- Structural methods take into account the graph structure first and foremost, i.e., the pathways and subgraphs encountered in the RDF graph. Because structural conditions are so important in applications and graph uses, graph structure is heavily used in summarization techniques.
- Quotient: quotient summaries use equivalence relations between the nodes and then assign a representative to each class of equivalence in the original graph. Because each graph node can only belong to one equivalence class, structural quotient methods ensure that each graph node is represented by only one summary node.
- Non-quotient: additional approaches to structurally summarizing semantic graphs rely on other metrics, such as centrality, to select and connect the most essential nodes in the summary.
- Pattern-mining methods: these methods make use of data-mining techniques to find patterns in the data, which are then used to construct the summary.
- Statistical methods: these methods quantitatively summarize the contents of a graph. The emphasis is on counting occurrences, such as counting class instances or creating value histograms by class, property, and value type; other quantitative measurements include the frequency of use of specific attributes, vocabularies, average string literal length, and so on. Statistical methods can also be used to investigate (usually minor) graph patterns, but only from a quantitative, frequency-based perspective.
- Hybrid methods: hybrid approaches integrate structural, statistical, and pattern-mining techniques fall into this category.
2.4. Centralities for KEGG Graphs
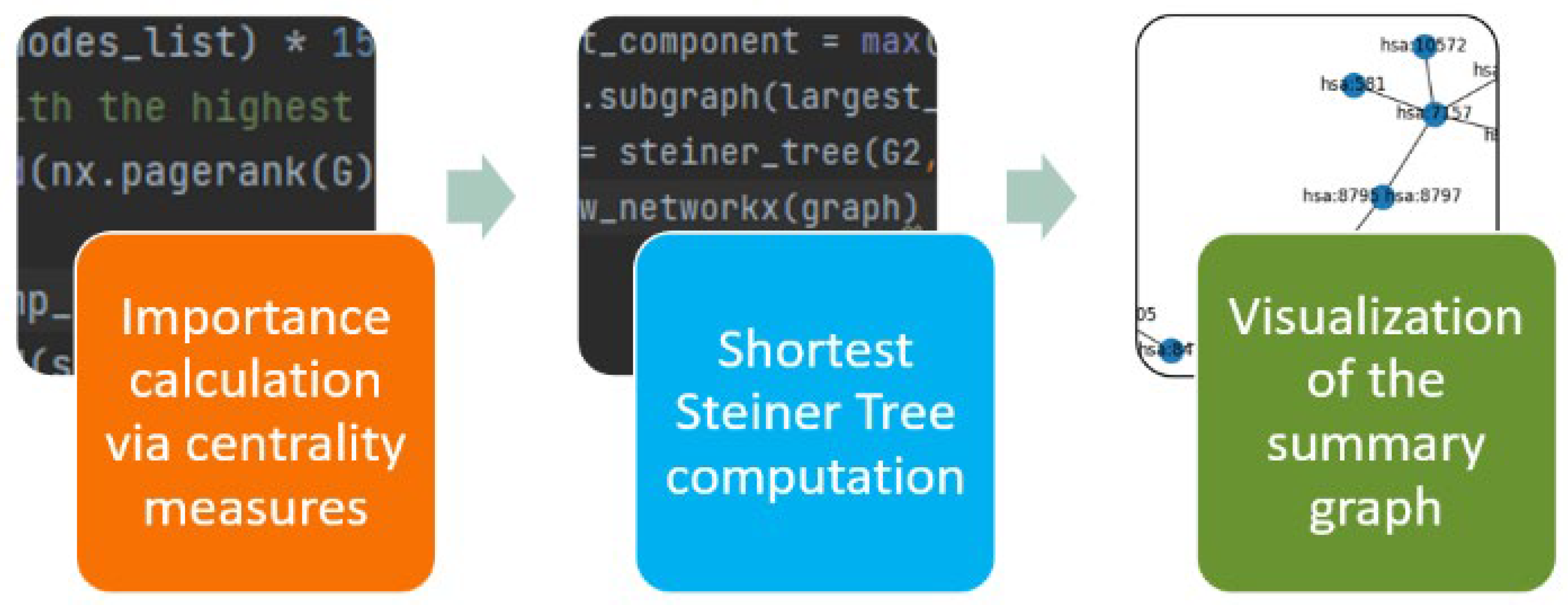
3. The KEGGSum Algorithm
3.1. Pre-Processing
- Filtering of chemical substrates and reactions. In our effort to create a more dense but rapidly comprehensible graph, we need to deal with the most significant and relevant, down to the final result, data. The main idea of the summarization process concerns the visualization of a number of significant genes. For this reason, nodes that represent chemical substrates and edges that represent reactions need to be filtered out.
- Filtering of orthologs. The KEGG pathway files sometimes include genes orthologous to the significant ones. These nodes signify genes that have similar functions in other organisms. But, for the cause of pathway summarization, they consist of data noise, which cannot be computationally processed, so they are filtered out, too, at the pre-processing stage.
- Filtering of nodes characterized as “undefined”. Sometimes, KEGG pathways have an undefined sequence as a node. These sequences’ accession numbers are, consequently, characterized as “undefined” and are of no value to the summarization process of the pathway. Thus, they are removed.
3.2. Centrality Measures Explored
3.3. Linking the Most Important KEGG Nodes
3.4. The KEGGSum Algorithm
| Algorithm 1. KEGGSum. Creates graph summaries of KEGG graphs |
| Input: kid—a KEGG Identifier, perc—percentage of important nodes to isolate, cent—centrality measure with which to determine the importance of the nodes. |
| Output: A summary graph. |
| 1. File: = download(kid) //Download and parse the KGML file based on kid |
| 2. Nodes: =Identify_most_important_nodes(perc, cent) //identify the perc most important nodes based on the cent centrality measure |
| 3. Graph: = Steiner_Tree(File, Nodes) //connects the most important nodes using he Steiner Tree algorithm |
| 4. Visualize(Graph) //visualizes the graph summary |
4. Evaluation
4.1. Datasets
4.2. Competitors
4.3. Reference Dataset Construction
4.4. Metrics
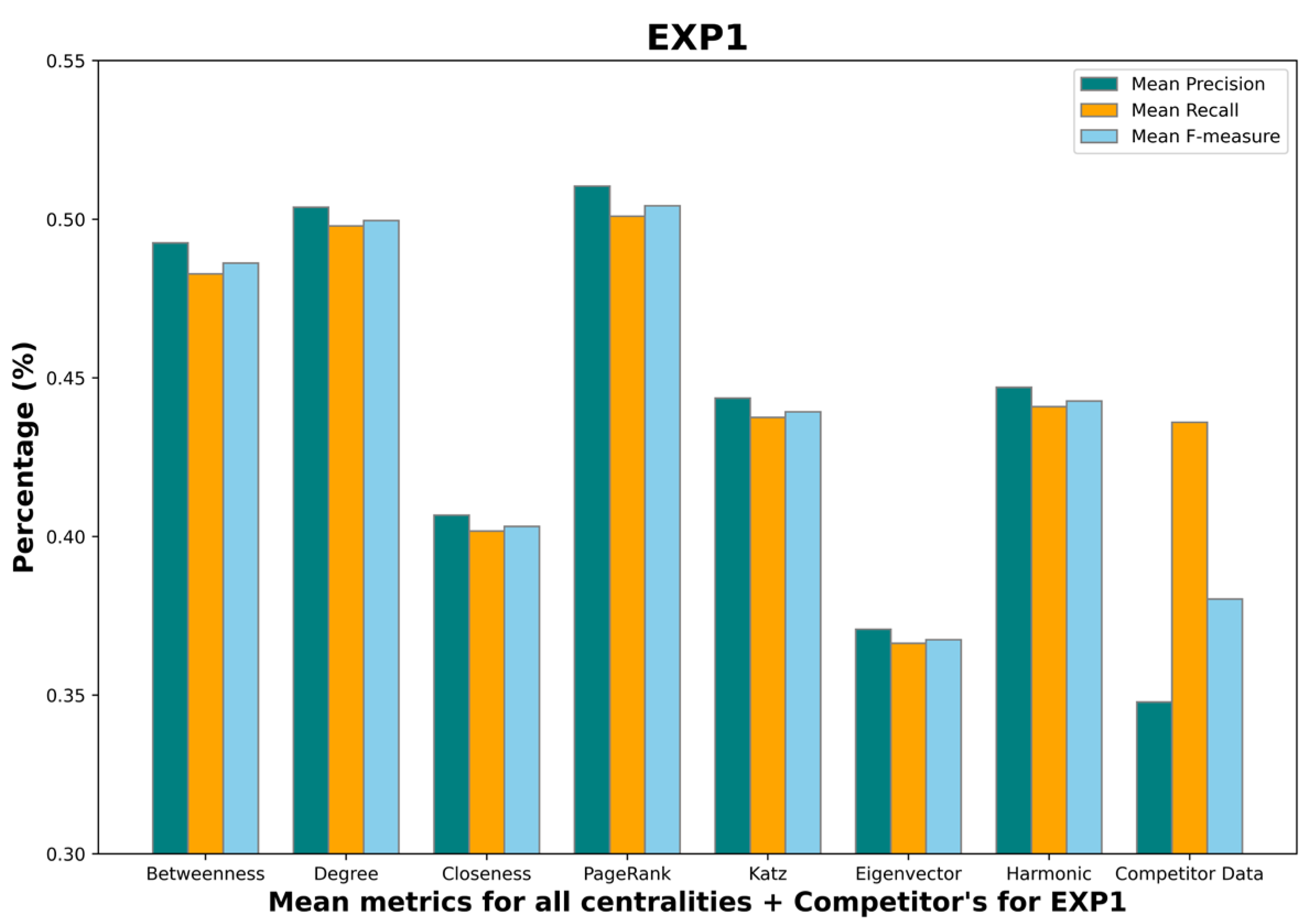
4.5. Results
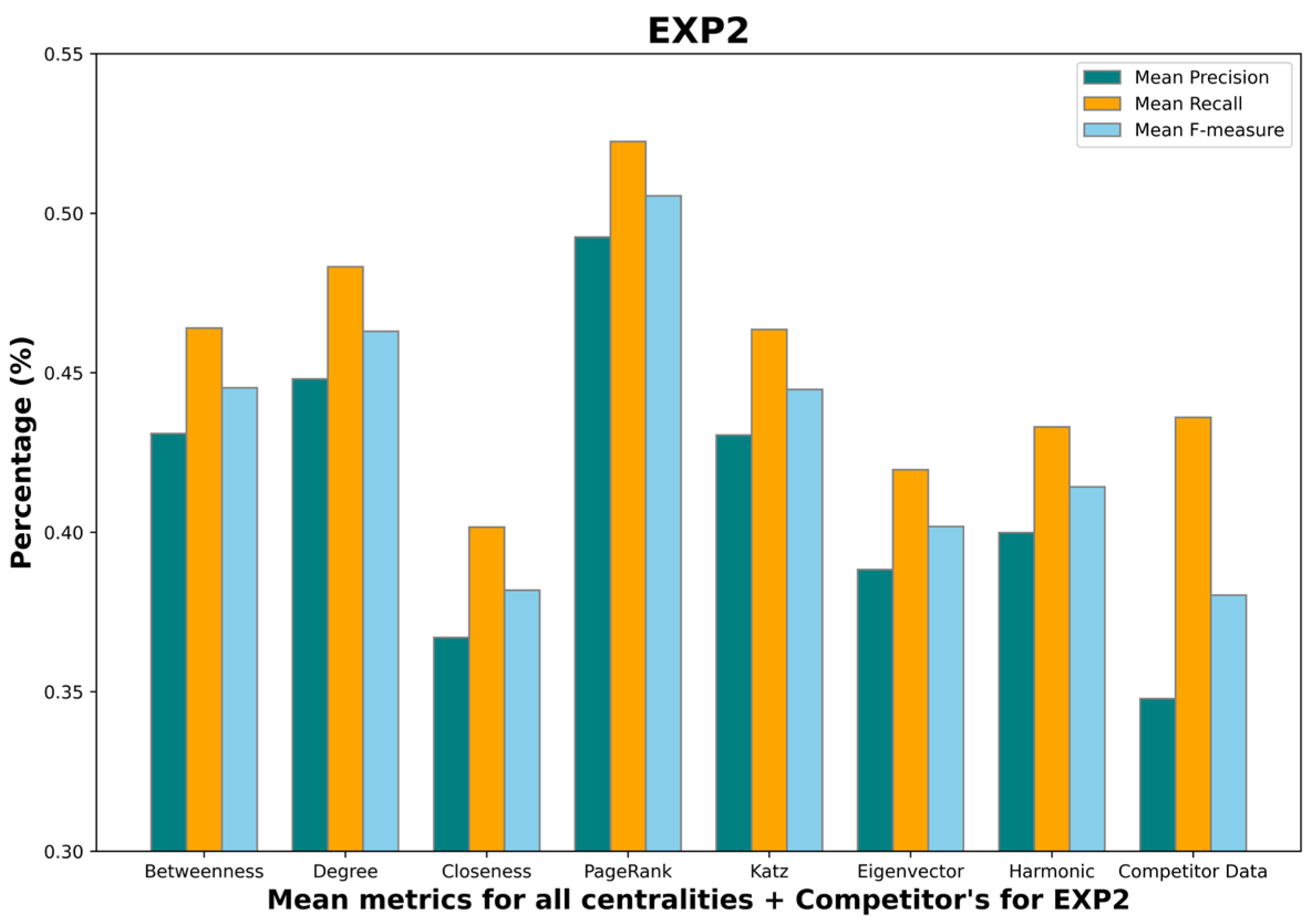
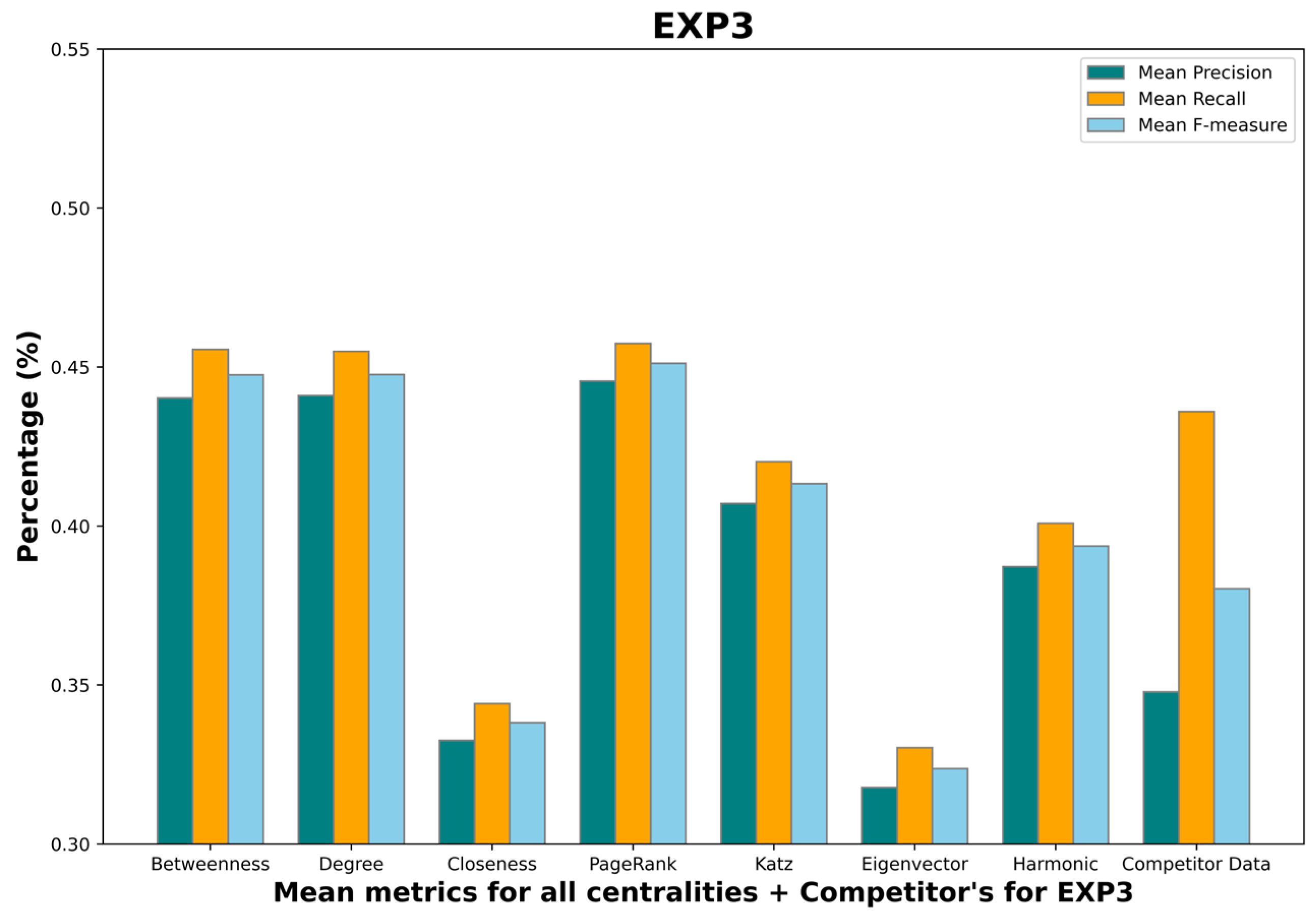
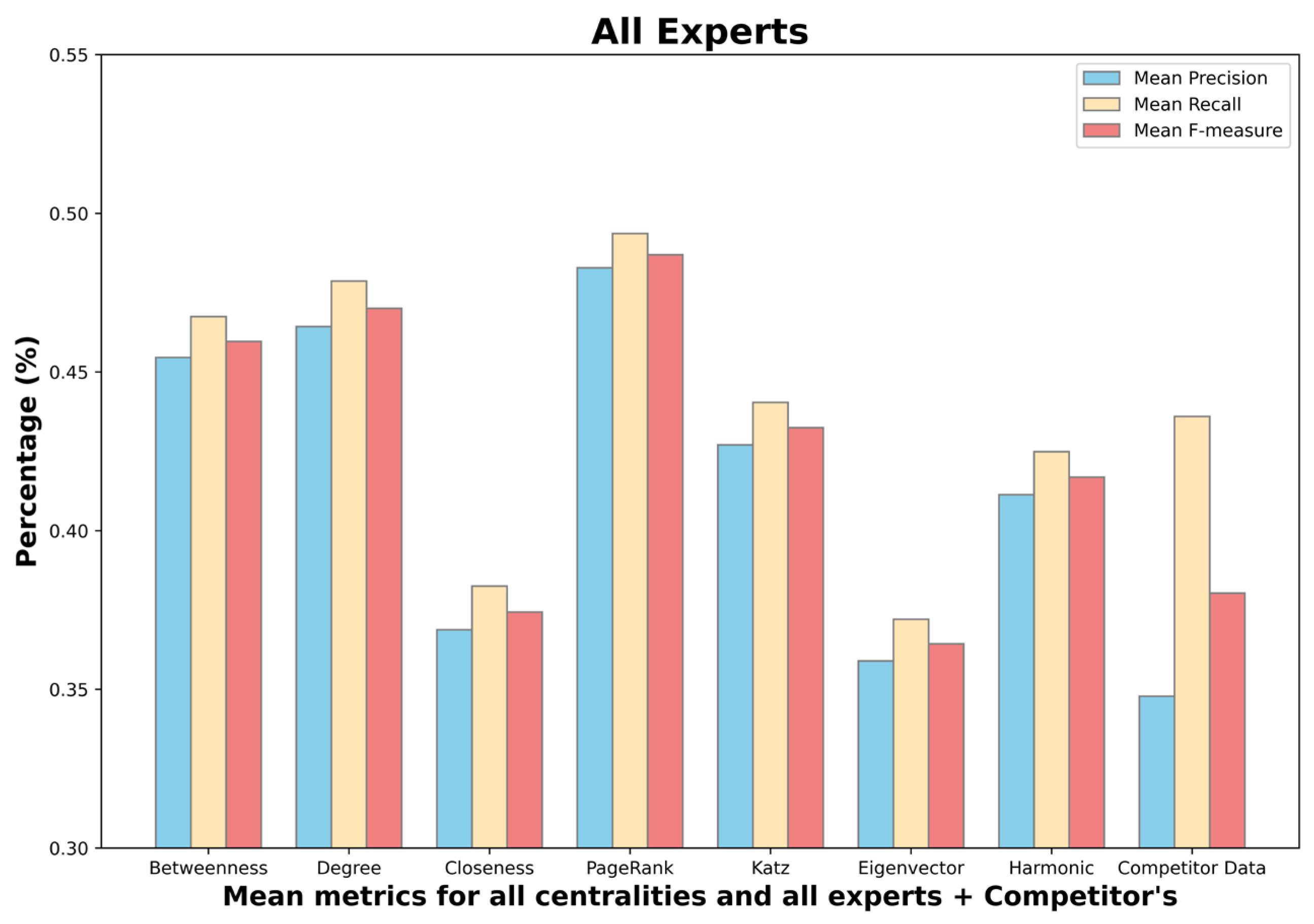
- Mean betweenness centrality F-measure: 0.45;
- Mean degree centrality F-measure: 0.48;
- Mean closeness centrality F-measure: 0.38;
- Mean PageRank centrality F-measure: 0.48;
- Mean Katz centrality F-measure: 0.43;
- Mean eigenvector centrality F-measure: 0.36;
- Mean harmonic centrality F-measure: 0.41;
- Competitor F-measure: 0.38.
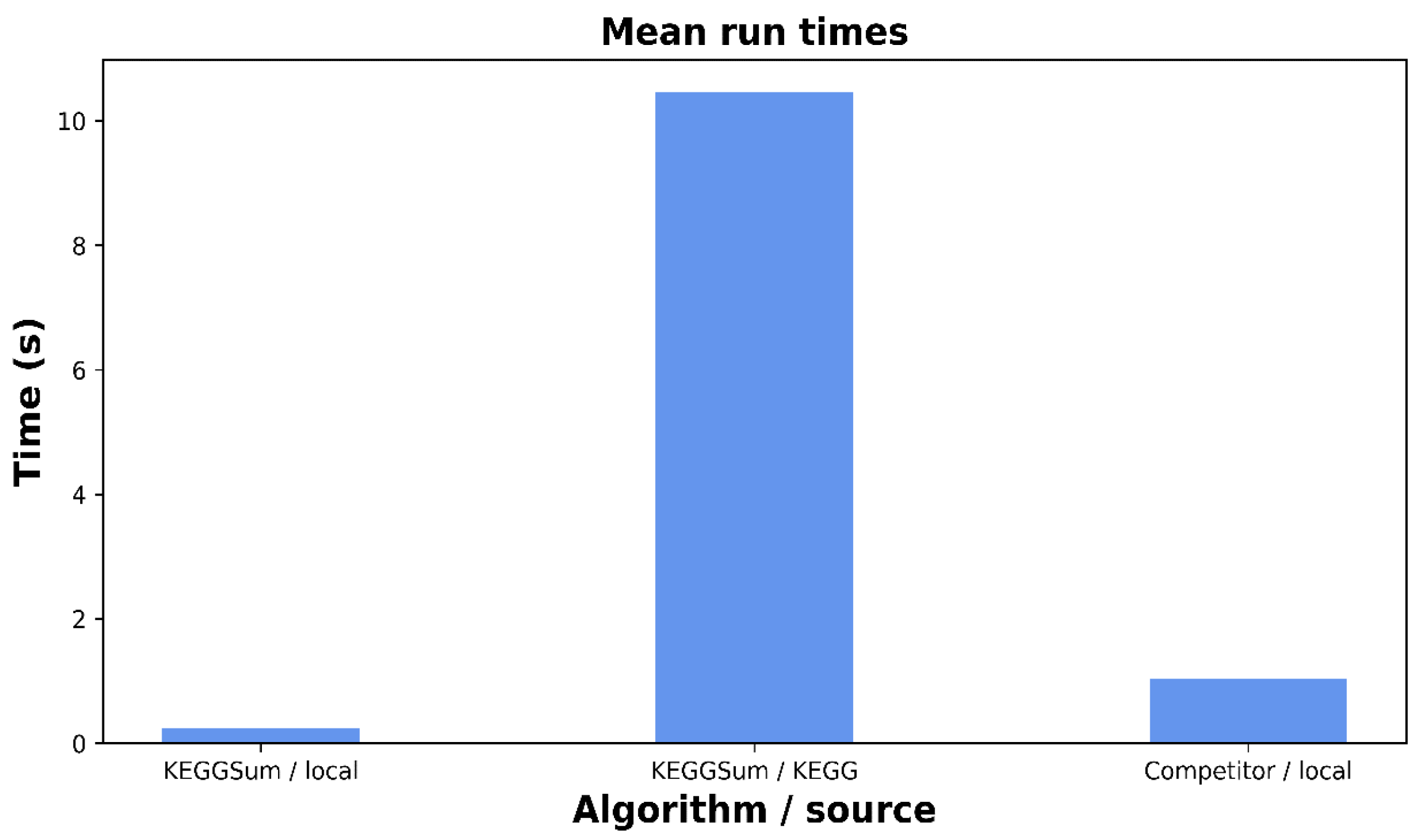
4.6. Execution Time
4.7. Use Case and Evaluator Comments
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023, 51, D587–D592. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Fu, M.; Li, J.; Chen, L.; Feng, K.; Huang, T.; Cai, Y.D. Analysis and prediction of protein stability based on interaction network, gene ontology, and kegg pathway enrichment scores. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2023, 1871, 140889. [Google Scholar] [CrossRef] [PubMed]
- Yousef, M.; Ozdemir, F.; Jaber, A.; Allmer, J.; Bakir-Gungor, B. PriPath: Identifying dysregulated pathways from differential gene expression via grouping, scoring, and modeling with an embedded feature selection approach. BMC Bioinform. 2023, 24, 60. [Google Scholar] [CrossRef] [PubMed]
- Thippana, M.; Dwivedi, A.; Das, A.; Palanisamy, M.; Vindal, V. Identification of key molecular players and associated pathways in cervical squamous cell carcinoma progression through network analysis. Proteins Struct. Funct. Bioinform. 2023, 91, 1173–1187. [Google Scholar] [CrossRef] [PubMed]
- Erciyes, K. Discrete Mathematics and Graph Theory, A Concise Study Companion and Guide; Springer Nature Switzerland: Cham, Switzerland, 2021. [Google Scholar] [CrossRef]
- Liu, Y.; Safavi, T.; Dighe, A.; Koutra, D. Graph summarization methods and applications: A survey. ACM Comput. Surv. 2018, 51, 1–34. [Google Scholar] [CrossRef]
- Cebiric, Š.; Goasdoué, F.; Kondylakis, H.; Kotzinos, D.; Manolescu, I.; Troullinou, G.; Zneika, M. Summarizing semantic graphs: A survey. VLDB J. 2018, 28, 295–327. [Google Scholar] [CrossRef]
- Naderi Yeganeh, P.; Mostafavi, M.T.; Richardson, C.; Saule, E.; Loraine, A. Revisiting the use of graph centrality models in biological pathway analysis. BioData Mining 2020, 13, 5. [Google Scholar] [CrossRef] [PubMed]
- Freeman, L.C. A Set of Measures of Centrality Based on Betweenness. Sociometry 1977, 40, 35–41. [Google Scholar] [CrossRef]
- Bloch, F.; Jackson, M. Centrality Measures in Networks. SSRN Electron. J. 2016. [Google Scholar] [CrossRef]
- Bavelas, A. Communication patterns in task-oriented groups. J. Acoust. Soc. Am. 1950, 22, 725–730. [Google Scholar] [CrossRef]
- Sabidussi, G. The centrality index of a graph. Psychometrika 1966, 31, 581–603. [Google Scholar] [CrossRef]
- Garg, M. Axiomatic Foundations of Centrality in Networks. SSRN Electron. J. 2009. [Google Scholar] [CrossRef]
- Rochat, Y. Closeness Centrality Extended to Unconnected Graphs: The Harmonic Centrality Index; ASNA: Zurich, Switzerland, 2009. [Google Scholar]
- Henni, K.; Mesghani, N.; Gouin-Vallerand, C. Unsupervised graph-based feature selection via subspace and pagerank centrality. Expert Syst. Appl. 2018, 114, 46–53. [Google Scholar] [CrossRef]
- Zhan, J.; Gurung, S.; Parsa, S. Identification of top-K nodes in large networks using Katz centrality. Big Data 2017, 4, 16. [Google Scholar] [CrossRef]
- Zaki, M.J.; Meira, W. Data Mining and Analysis: Fundamental Concepts and Algorithms; Cambridge University Press: New York, NY, USA, 2014. [Google Scholar]
- Bolid, P.; Vigna, S. Axioms for Centrality. Internet Math. 2014, 10, 222–262. [Google Scholar] [CrossRef]
- Pappas, A.; Troullinou, G.; Roussakis, G.; Kondylakis, H.; Plexousakis, D. Exploring Importance Measures for Summarizing RDF/S KBs. In Proceedings of the European Semantic Web Conference, Portorož, Slovenia, 28 May–1 June 2017; pp. 387–403. [Google Scholar] [CrossRef]
- Dreyfus, S.E.; Wagner, R.A. The steiner problem in graphs. Networks 1971, 1, 195–207. [Google Scholar] [CrossRef]
- Gwet, K. Handbook of Inter-Rater Reliability: The Definitive Guide to Measuring the Extent of Agreement among Raters; Advanced Analytics, LLC: Gaithersburg, MD, USA, 2014. [Google Scholar]
- Landis, R.J.; Koch, G.G. The Measurement of Observer Agreement for Categorical Data. Biometrics 1977, 33, 159–174. [Google Scholar] [CrossRef] [PubMed]
- McHugh, M. Interrater reliability: The kappa statistic. Biochem. Medica 2012, 22, 276–282. [Google Scholar] [CrossRef]
- Trouli, G.E.; Pappas, A.; Troullinou, G.; Koumakis, L.; Papadakis, N.; Kondylakis, H. Summer: Structural summarization for RDF/S KGs. Algorithms 2022, 16, 18. [Google Scholar] [CrossRef]
- Vassiliou, G.; Alevizakis, F.; Papadakis, N.; Kondylakis, H. iSummary: Workload-Based, Personalized Summaries for Knowledge Graphs. In European Semantic Web Conference; Springer Nature Switzerland: Cham, Switzerland, 2023; pp. 192–208. [Google Scholar]
| Measure | Complexity |
|---|---|
| Betweenness | O (V × (V × E)) |
| Degree | O (V+E) |
| Closeness | O (V × E) |
| PageRank | O (k × E) |
| Katz | O (V3) |
| Eigenvector | O (V × logV |
| Harmonic | O (N × (N+E)) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
David, C.; Kondylakis, H. KEGGSum: Summarizing Genomic Pathways. Information 2024, 15, 56. https://doi.org/10.3390/info15010056
David C, Kondylakis H. KEGGSum: Summarizing Genomic Pathways. Information. 2024; 15(1):56. https://doi.org/10.3390/info15010056
Chicago/Turabian StyleDavid, Chaim, and Haridimos Kondylakis. 2024. "KEGGSum: Summarizing Genomic Pathways" Information 15, no. 1: 56. https://doi.org/10.3390/info15010056
APA StyleDavid, C., & Kondylakis, H. (2024). KEGGSum: Summarizing Genomic Pathways. Information, 15(1), 56. https://doi.org/10.3390/info15010056









