A Systematic Review of Effective Hardware and Software Factors Affecting High-Throughput Plant Phenotyping
Abstract
1. Introduction
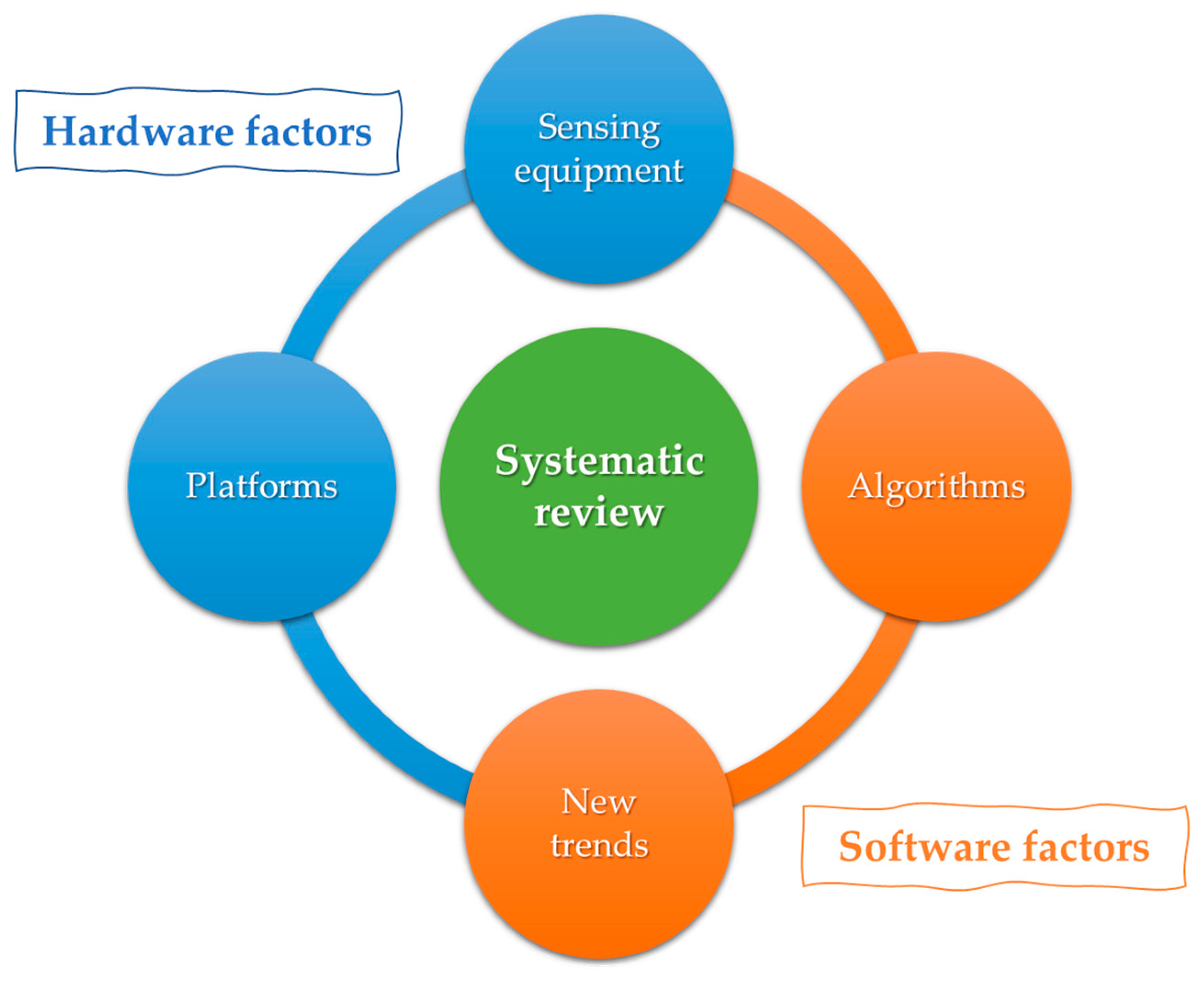
2. Background
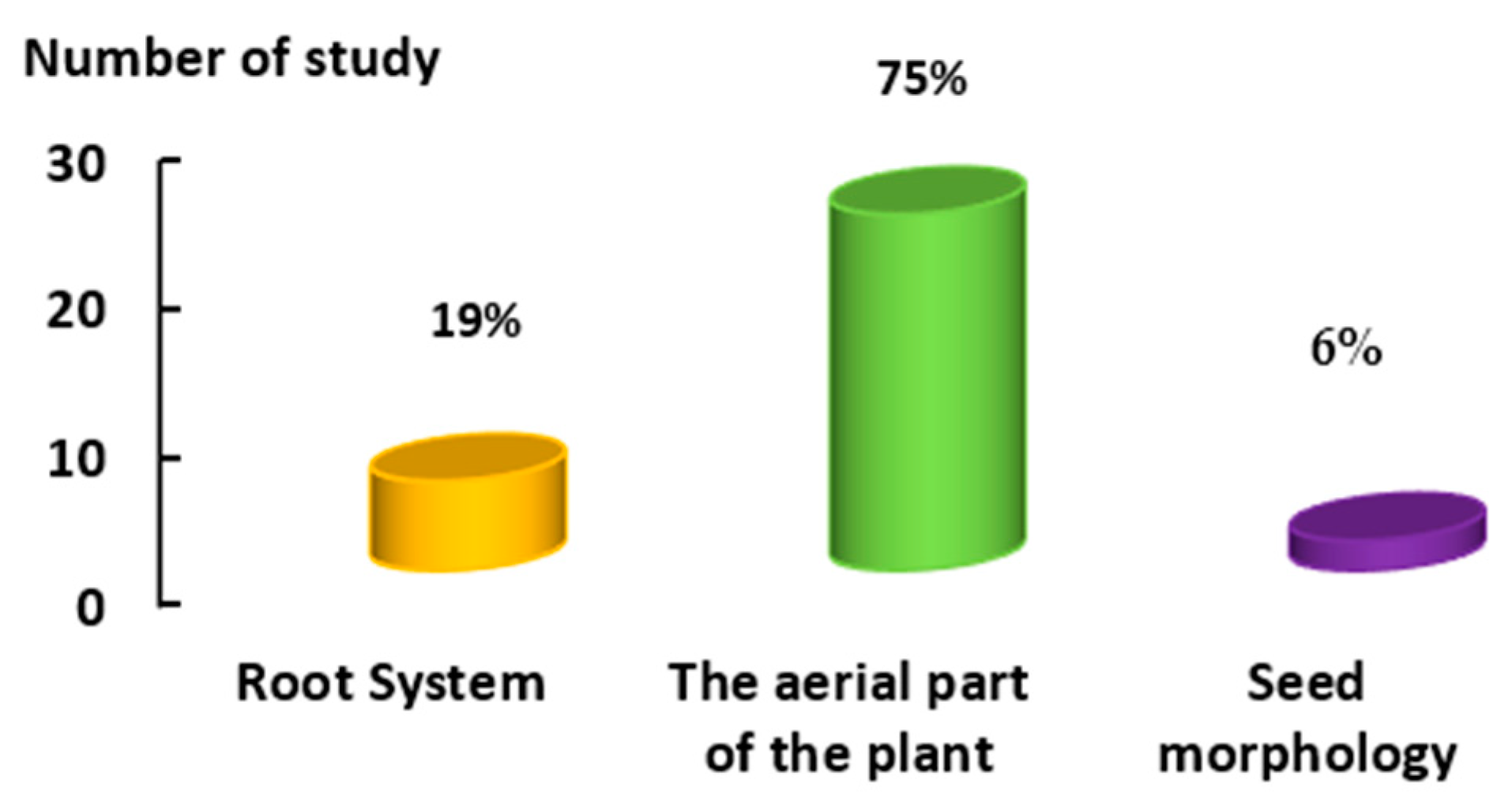
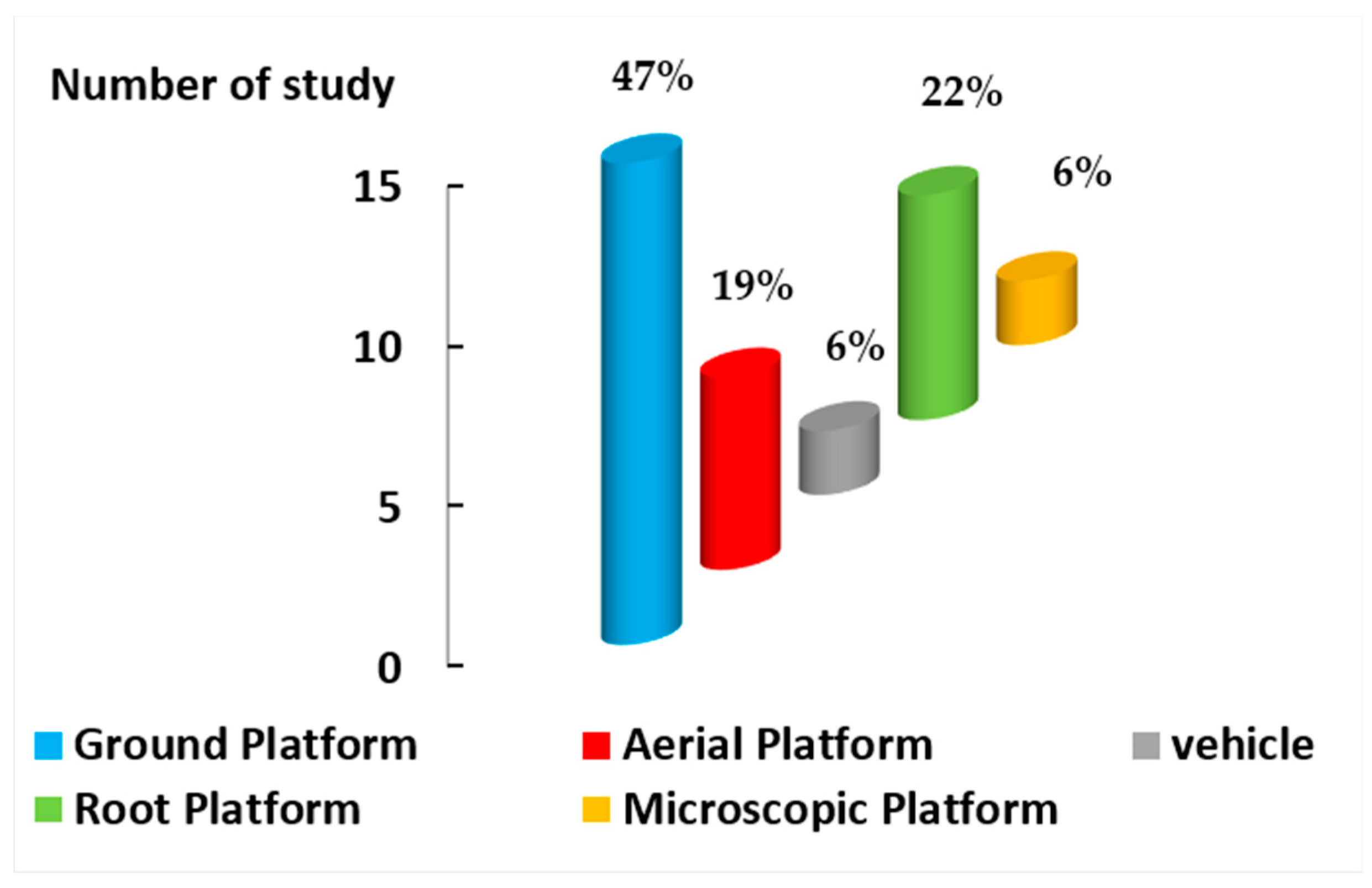
2.1. First Factor: Platforms
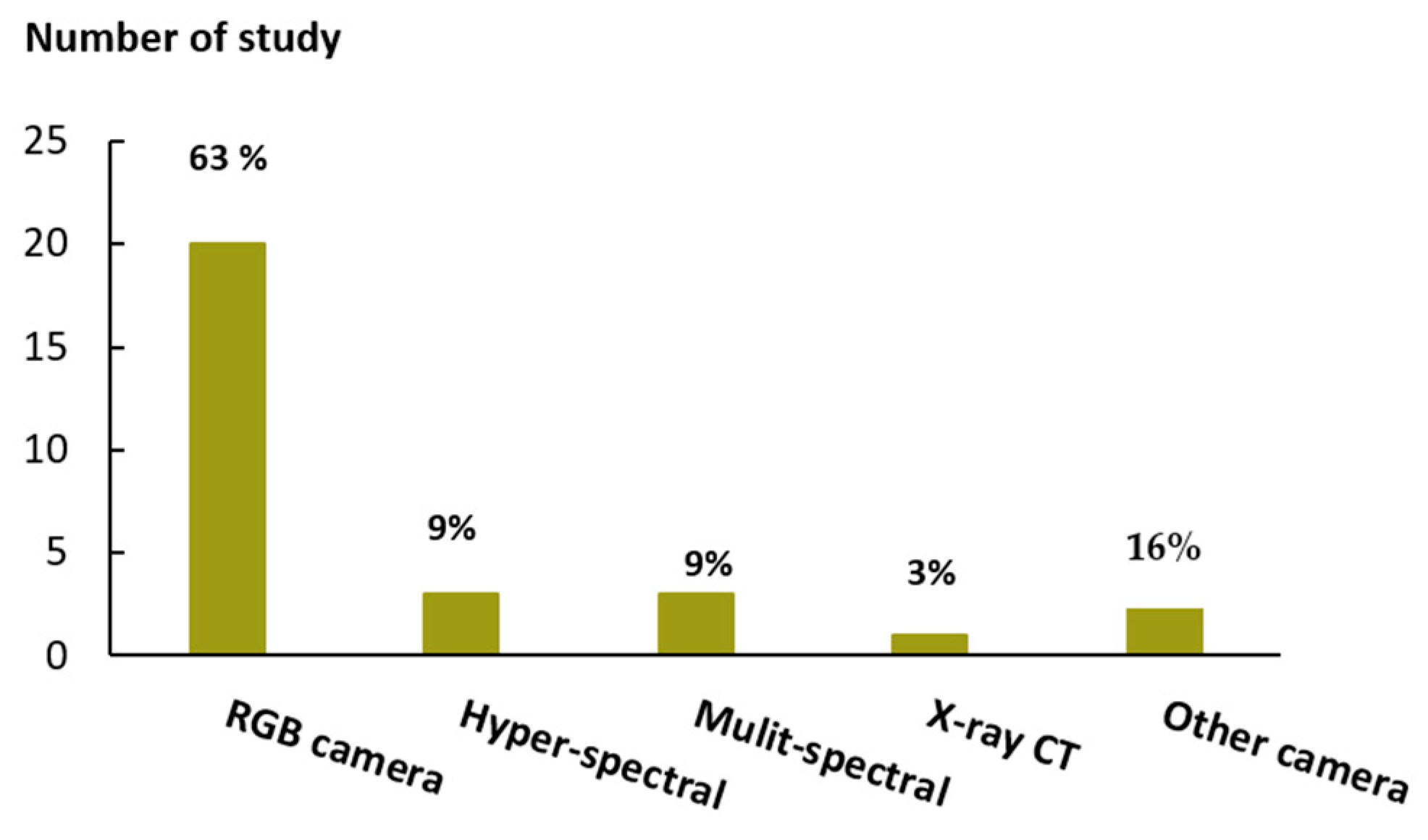
2.2. Second Factor: Sensing Equipment
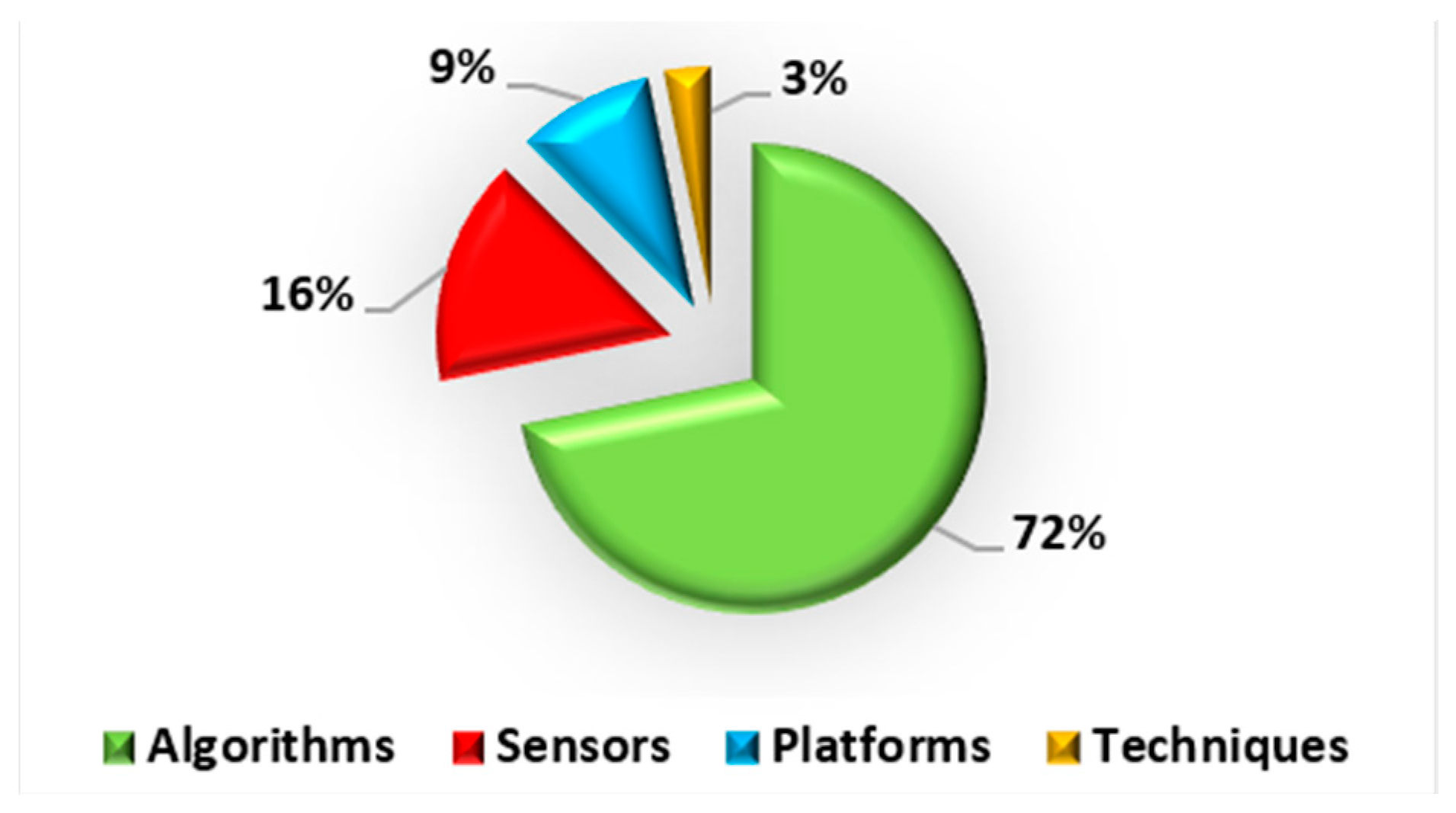
2.3. Third Factor: Algorithms
2.4. Fourth Factor: New Trends
3. Methodology
3.1. Review Questions
- -
- About platforms—RQ1: What platforms are used in HTP for aerial parts of plants and roots, and what are their strengths and challenges?
- -
- About sensing equipment—RQ2: What sensors do experts use to capture plant traits, and what data do these sensors collect for analysis?
- -
- About algorithms—RQ3: What algorithms can better extract and predict traits obtained from specific phenotypic data?
- -
- About trends—RQ4: What are the main trends toward which research in the HTP field is moving?
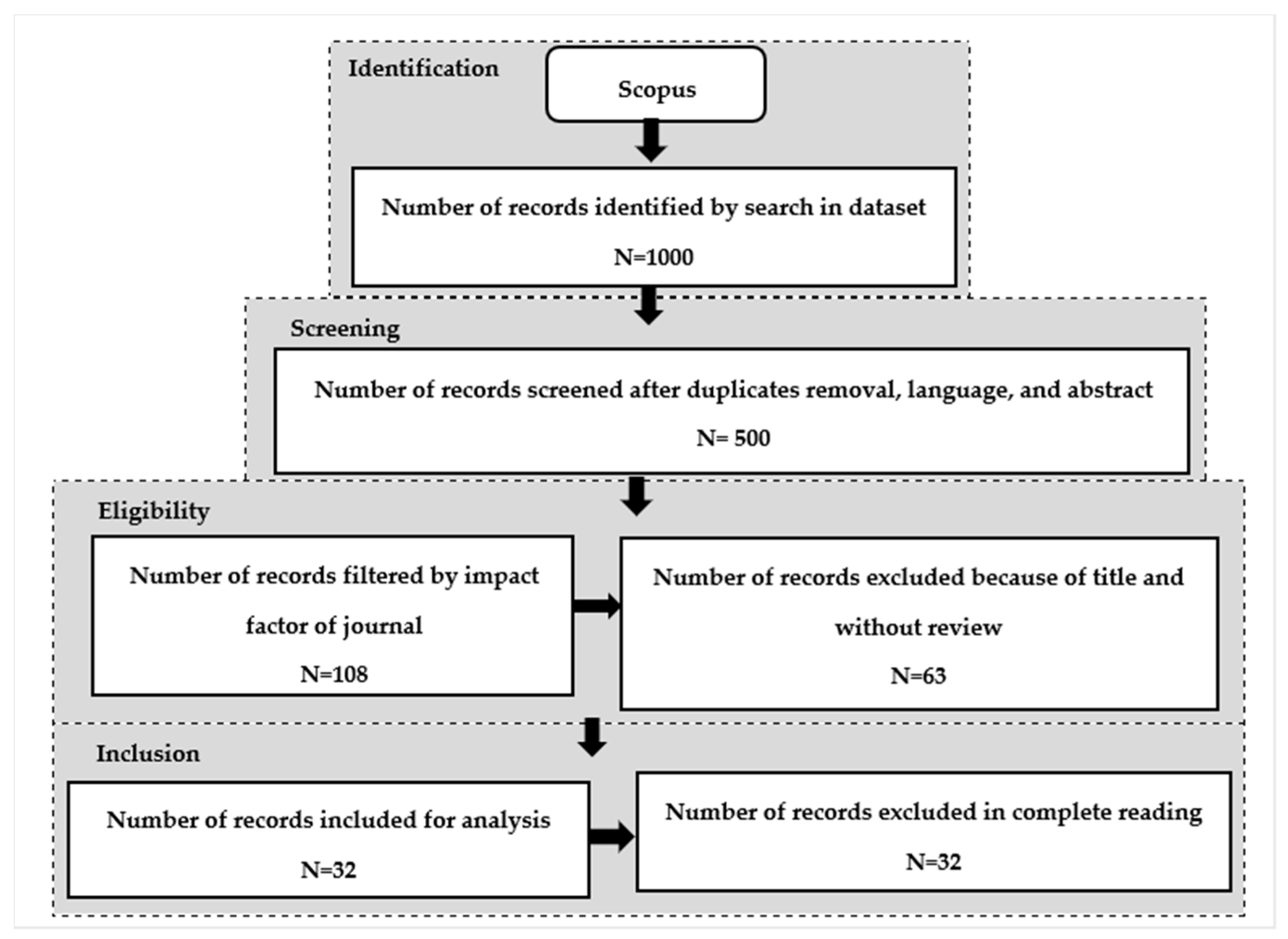
3.2. Literature Search Strategy
- According to [59], using more than one database for a literature search does not guarantee a positive impact on the research outcome.
- The high degree of reliability of Scopus guarantees the evaluation of high-quality papers published in qualified journals.
- “Sensor” AND “high-throughput plant phenotyping”.
- “Machine learning” OR “deep learning” AND “high-throughput plant phenotyping”.
- “Platform” AND “high-throughput plant phenotyping”.
- “Image acquisition technique” AND “high-throughput plant phenotyping”.
3.3. Inclusion and Exclusion Criteria
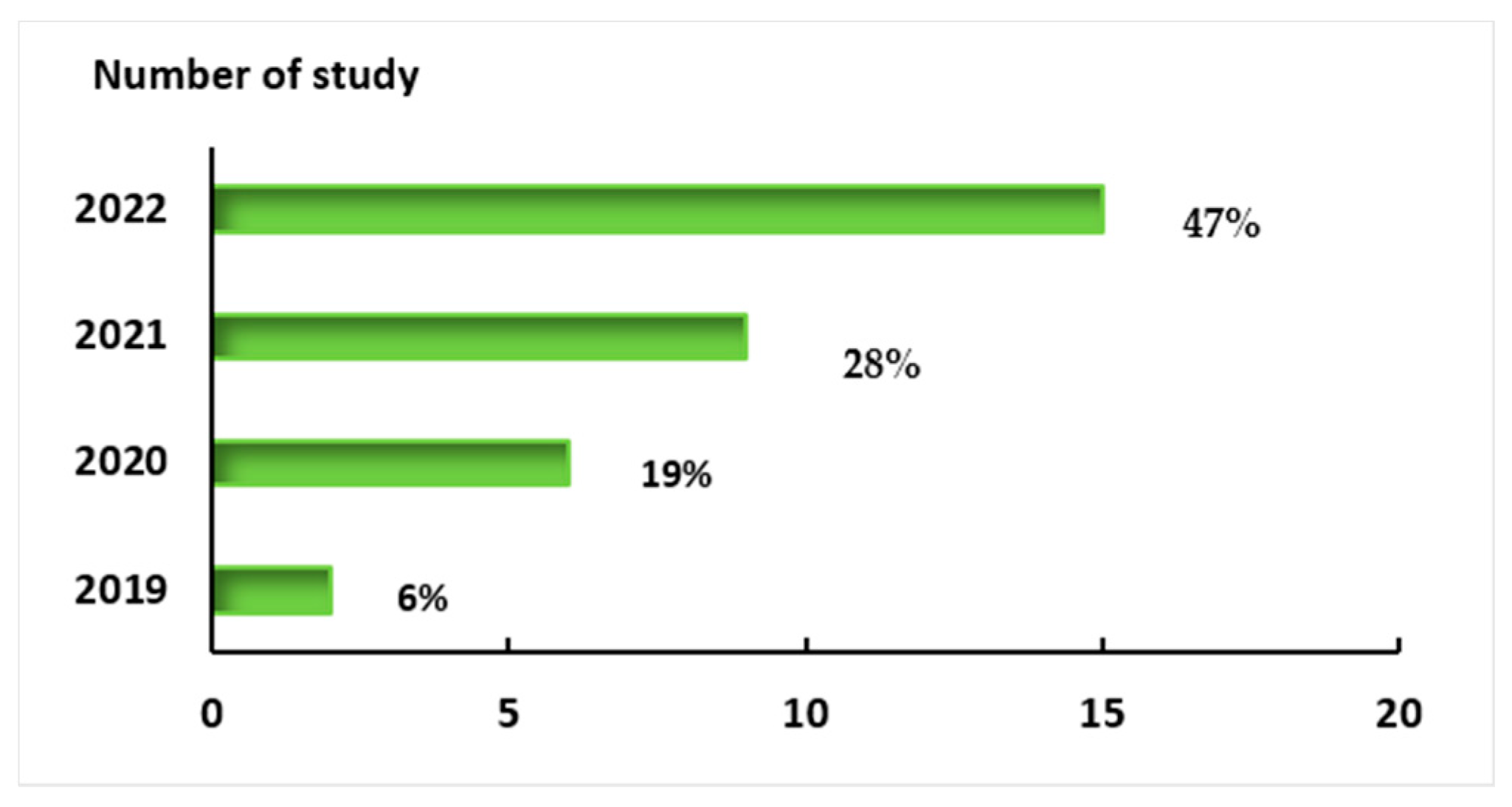
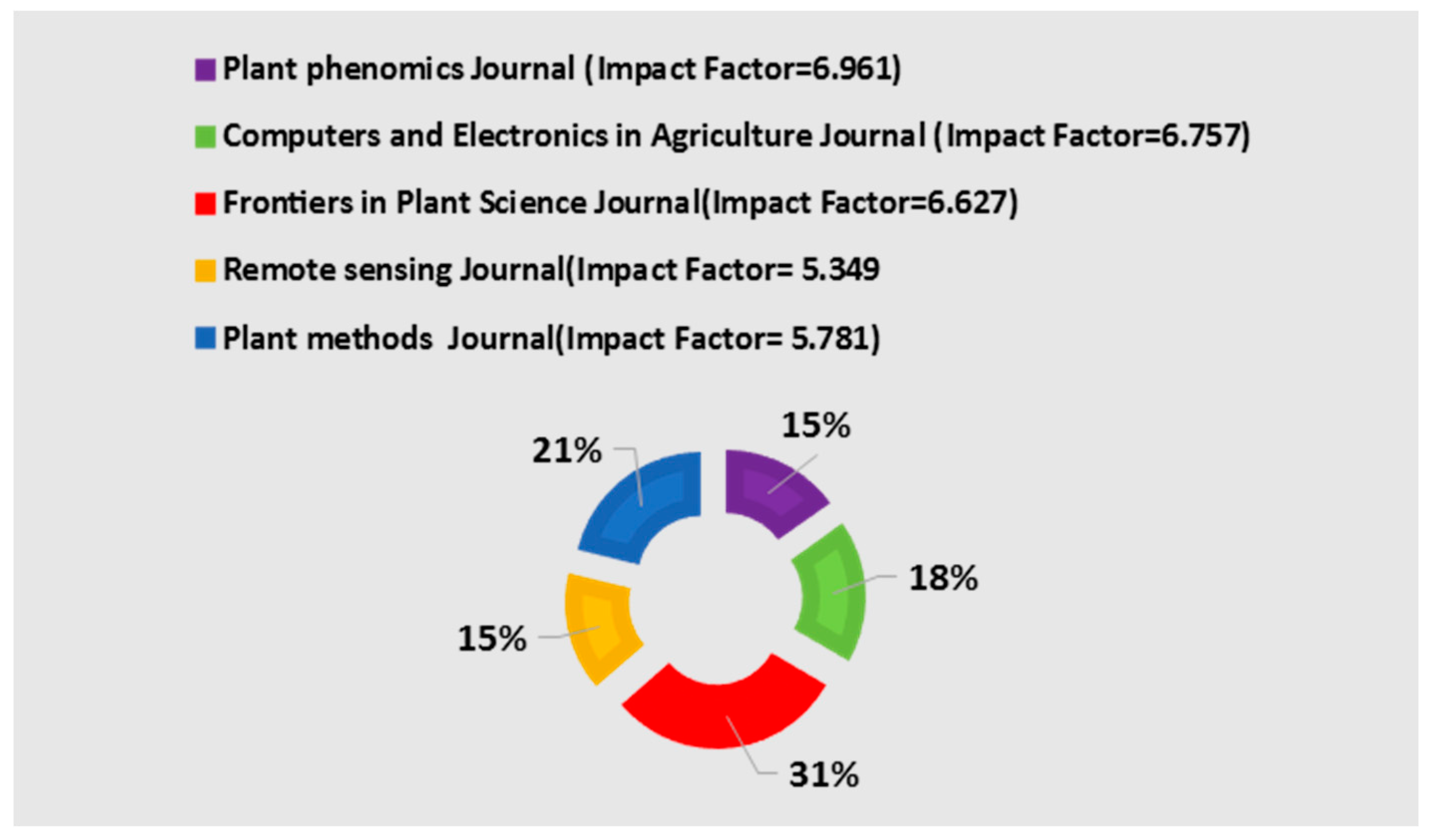
4. Results
5. Discussion
5.1. Platforms
5.2. Sensors
5.3. Algorithms
5.4. New HTP Research Ideas and Proposals
6. Conclusions and Future Work
- Ground platforms were among the most commonly used platforms for the aerial part of plants. They were used in laboratories due to their cost and time effectiveness, simplicity, and compatibility for data collection.
- Researchers widely used digital RGB cameras because of their compatibility and ease of integration with all plant phenotype platforms; moreover, using RGB cameras, it is possible to capture images of both the aerial part of the plants and the root system architecture, thus lowering the costs and achieving relatively good quality images in terms of resolution and general appearance.
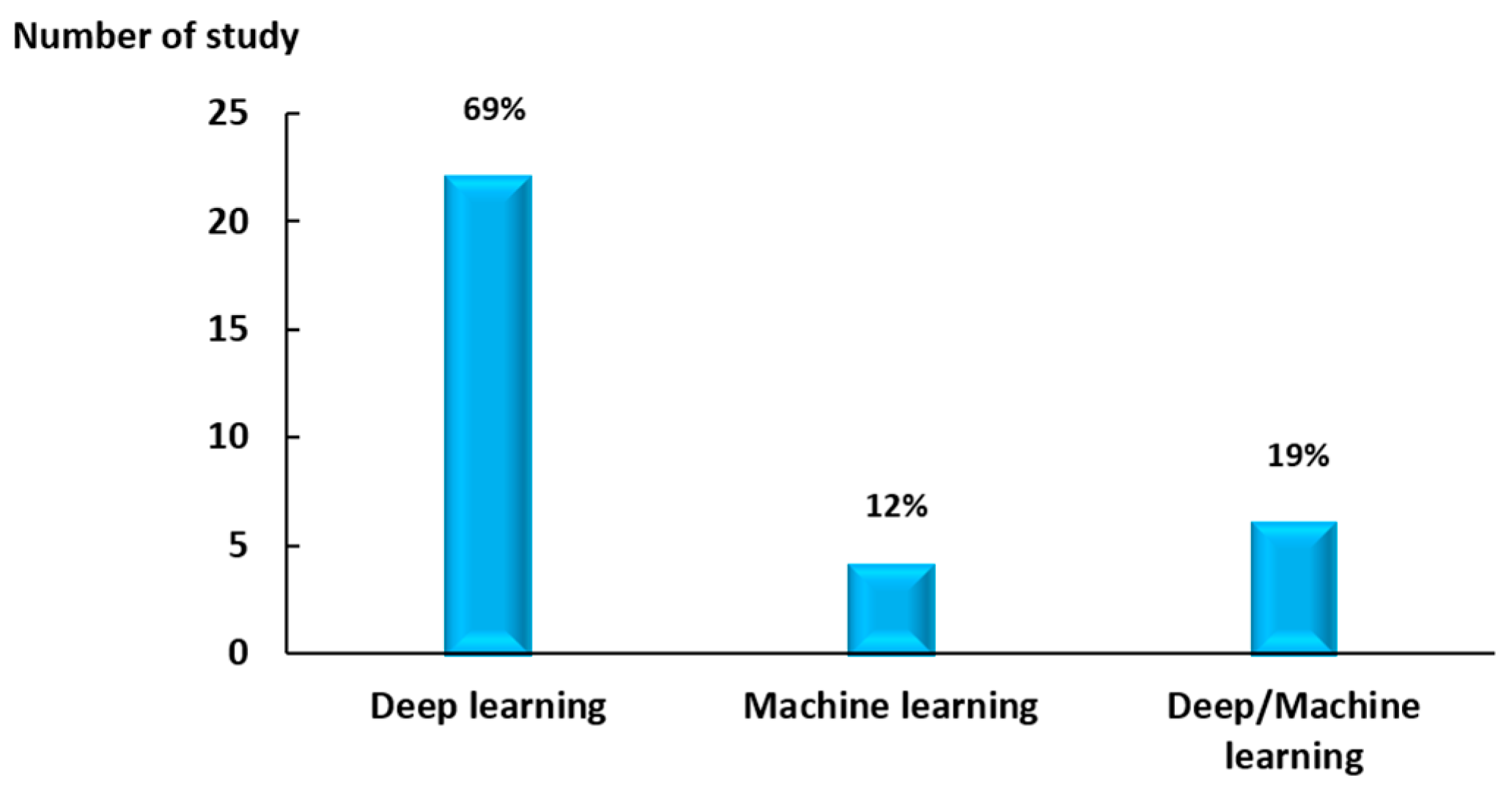
- Deep learning models were among the most widely used methods in plant phenotyping in the last few years. These models can detect and accurately measure, for example, specific parts of the plant (fruit, flowers, roots, etc.).
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Coping with Water Scarcity: An Action Framework for Agriculture and Food Security; FAO Water Reports; Steduto, P., Faurès, J.-M., Hoogeveen, J., Winpenny, J.T., Burke, J.J., Eds.; Food and Agriculture Organization of the United Nations: Rome, Italy, 2012; ISBN 978-92-5-107304-9. [Google Scholar]
- Danzi, D.; Briglia, N.; Petrozza, A.; Summerer, S.; Povero, G.; Stivaletta, A.; Cellini, F.; Pignone, D.; De Paola, D.; Janni, M. Can High Throughput Phenotyping Help Food Security in the Mediterranean Area? Front. Plant Sci. 2019, 10, 15. [Google Scholar] [CrossRef] [PubMed]
- Colla, G.; Rouphael, Y. Special Issue: Biostimulants in Horticulture. Sci. Hortic. 2015, 196, 1–134. [Google Scholar] [CrossRef]
- Rouphael, Y.; Spíchal, L.; Panzarová, K.; Casa, R.; Colla, G. High-Throughput Plant Phenotyping for Developing Novel Biostimulants: From Lab to Field or From Field to Lab? Front. Plant Sci. 2018, 9, 1197. [Google Scholar] [CrossRef] [PubMed]
- Vasconez, J.P.; Delpiano, J.; Vougioukas, S.; Auat Cheein, F. Comparison of Convolutional Neural Networks in Fruit Detection and Counting: A Comprehensive Evaluation. Comput. Electron. Agric. 2020, 173, 105348. [Google Scholar] [CrossRef]
- Paustian, M.; Theuvsen, L. Adoption of Precision Agriculture Technologies by German Crop Farmers. Precis. Agric. 2017, 18, 701–716. [Google Scholar] [CrossRef]
- Chen, D.; Neumann, K.; Friedel, S.; Kilian, B.; Chen, M.; Altmann, T.; Klukas, C. Dissecting the Phenotypic Components of Crop Plant Growth and Drought Responses Based on High-Throughput Image Analysis. Plant Cell 2014, 26, 4636–4655. [Google Scholar] [CrossRef]
- Maji, A.K.; Marwaha, S.; Kumar, S.; Arora, A.; Chinnusamy, V.; Islam, S. SlypNet: Spikelet-Based Yield Prediction of Wheat Using Advanced Plant Phenotyping and Computer Vision Techniques. Front. Plant Sci. 2022, 13, 2552. [Google Scholar] [CrossRef]
- Yang, W.; Duan, L.; Chen, G.; Xiong, L.; Liu, Q. Plant Phenomics and High-Throughput Phenotyping: Accelerating Rice Functional Genomics Using Multidisciplinary Technologies. Curr. Opin. Plant Biol. 2013, 16, 180–187. [Google Scholar] [CrossRef]
- Mazis, A.; Choudhury, S.D.; Morgan, P.B.; Stoerger, V.; Hiller, J.; Ge, Y.; Awada, T. Application of High-Throughput Plant Phenotyping for Assessing Biophysical Traits and Drought Response in Two Oak Species under Controlled Environment. For. Ecol. Manag. 2020, 465, 118101. [Google Scholar] [CrossRef]
- Fan, J.; Zhang, Y.; Wen, W.; Gu, S.; Lu, X.; Guo, X. The Future of Internet of Things in Agriculture: Plant High-Throughput Phenotypic Platform. J. Clean. Prod. 2021, 280, 123651. [Google Scholar] [CrossRef]
- Hu, P.; Chapman, S.C.; Zheng, B.; Hu, P.; Chapman, S.C.; Zheng, B. Coupling of Machine Learning Methods to Improve Estimation of Ground Coverage from Unmanned Aerial Vehicle (UAV) Imagery for High-Throughput Phenotyping of Crops. Funct. Plant Biol. 2021, 48, 766–779. [Google Scholar] [CrossRef] [PubMed]
- Atefi, A.; Ge, Y.; Pitla, S.; Schnable, J. Robotic Technologies for High-Throughput Plant Phenotyping: Contemporary Reviews and Future Perspectives. Front. Plant Sci. 2021, 12, 611940. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Marzougui, A.; Sankaran, S. High-Resolution Satellite Imagery Applications in Crop Phenotyping: An Overview. Comput. Electron. Agric. 2020, 175, 105584. [Google Scholar] [CrossRef]
- Arunachalam, A.; Andreasson, H. Real-Time Plant Phenomics under Robotic Farming Setup: A Vision-Based Platform for Complex Plant Phenotyping Tasks. Comput. Electr. Eng. 2021, 92, 107098. [Google Scholar] [CrossRef]
- Das Choudhury, S.; Samal, A.; Awada, T. Leveraging Image Analysis for High-Throughput Plant Phenotyping. Front. Plant Sci. 2019, 10, 508. [Google Scholar] [CrossRef]
- Li, D.; Li, C.; Yao, Y.; Li, M.; Liu, L. Modern Imaging Techniques in Plant Nutrition Analysis: A Review. Comput. Electron. Agric. 2020, 174, 105459. [Google Scholar] [CrossRef]
- Shibayama, M.; Sakamoto, T.; Takada, E.; Inoue, A.; Morita, K.; Takahashi, W.; Kimura, A. Continuous Monitoring of Visible and Near-Infrared Band Reflectance from a Rice Paddy for Determining Nitrogen Uptake Using Digital Cameras. Plant Prod. Sci. 2009, 12, 293–306. [Google Scholar] [CrossRef]
- Mahlein, A.-K.; Oerke, E.-C.; Steiner, U.; Dehne, H.-W. Recent Advances in Sensing Plant Diseases for Precision Crop Protection. Eur. J. Plant Pathol. 2012, 133, 197–209. [Google Scholar] [CrossRef]
- Hernández-Sánchez, N.; Hills, B.P.; Barreiro, P.; Marigheto, N. An NMR Study on Internal Browning in Pears. Postharvest Biol. Technol. 2007, 44, 260–270. [Google Scholar] [CrossRef]
- da Silva, C.B.; Bianchini, V.D.J.M.; de Medeiros, A.D.; de Moraes, M.H.D.; Marassi, A.G.; Tannús, A. A Novel Approach for Jatropha Curcas Seed Health Analysis Based on Multispectral and Resonance Imaging Techniques. Ind. Crops Prod. 2021, 161, 113186. [Google Scholar] [CrossRef]
- Köckenberger, W.; De Panfilis, C.; Santoro, D.; Dahiya, P.; Rawsthorne, S. High Resolution NMR Microscopy of Plants and Fungi. J. Microsc. 2004, 214, 182–189. [Google Scholar] [CrossRef]
- Van De Looverbosch, T.; Vandenbussche, B.; Verboven, P.; Nicolaï, B. Nondestructive High-Throughput Sugar Beet Fruit Analysis Using X-ray CT and Deep Learning. Comput. Electron. Agric. 2022, 200, 107228. [Google Scholar] [CrossRef]
- Flavel, R.J.; Guppy, C.N.; Tighe, M.; Watt, M.; McNeill, A.; Young, I.M. Non-Destructive Quantification of Cereal Roots in Soil Using High-Resolution X-ray Tomography. J. Exp. Bot. 2012, 63, 2503–2511. [Google Scholar] [CrossRef]
- Helliwell, J.R.; Sturrock, C.J.; Mairhofer, S.; Craigon, J.; Ashton, R.W.; Miller, A.J.; Whalley, W.R.; Mooney, S.J. The Emergent Rhizosphere: Imaging the Development of the Porous Architecture at the Root-Soil Interface. Sci. Rep. 2017, 7, 14875. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, J.A.; Pound, M.P.; Bennett, M.J.; Wells, D.M. Uncovering the Hidden Half of Plants Using New Advances in Root Phenotyping. Curr. Opin. Biotechnol. 2019, 55, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Bauer, F.M.; Lärm, L.; Morandage, S.; Lobet, G.; Vanderborght, J.; Vereecken, H.; Schnepf, A. Development and Validation of a Deep Learning Based Automated Minirhizotron Image Analysis Pipeline. Plant Phenomics 2022, 2022, 9758532. [Google Scholar] [CrossRef]
- Mochida, K.; Koda, S.; Inoue, K.; Hirayama, T.; Tanaka, S.; Nishii, R.; Melgani, F. Computer Vision-Based Phenotyping for Improvement of Plant Productivity: A Machine Learning Perspective. GigaScience 2019, 8, giy153. [Google Scholar] [CrossRef] [PubMed]
- Wilf, P.; Zhang, S.; Chikkerur, S.; Little, S.A.; Wing, S.L.; Serre, T. Computer Vision Cracks the Leaf Code. Proc. Natl. Acad. Sci. USA 2016, 113, 3305–3310. [Google Scholar] [CrossRef] [PubMed]
- Brichet, N.; Fournier, C.; Turc, O.; Strauss, O.; Artzet, S.; Pradal, C.; Welcker, C.; Tardieu, F.; Cabrera-Bosquet, L. A Robot-Assisted Imaging Pipeline for Tracking the Growths of Maize Ear and Silks in a High-Throughput Phenotyping Platform. Plant Methods 2017, 13, 1–12. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, C. Convolutional Neural Networks for Image-Based High-Throughput Plant Phenotyping: A Review. Plant Phenomics 2020, 2020, 4152816. [Google Scholar] [CrossRef]
- Koh, J.C.O.; Spangenberg, G.; Kant, S. Automated Machine Learning for High-Throughput Image-Based Plant Phenotyping. Remote Sens. 2021, 13, 858. [Google Scholar] [CrossRef]
- Abade, A.; Ferreira, P.A.; de Barros Vidal, F. Plant Diseases Recognition on Images Using Convolutional Neural Networks: A Systematic Review. Comput. Electron. Agric. 2021, 185, 106125. [Google Scholar] [CrossRef]
- Feng, L.; Chen, S.; Zhang, C.; Zhang, Y.; He, Y. A Comprehensive Review on Recent Applications of Unmanned Aerial Vehicle Remote Sensing with Various Sensors for High-Throughput Plant Phenotyping. Comput. Electron. Agric. 2021, 182, 106033. [Google Scholar] [CrossRef]
- Näsi, R.; Viljanen, N.; Kaivosoja, J.; Alhonoja, K.; Hakala, T.; Markelin, L.; Honkavaara, E. Estimating Biomass and Nitrogen Amount of Barley and Grass Using UAV and Aircraft Based Spectral and Photogrammetric 3D Features. Remote Sens. 2018, 10, 1082. [Google Scholar] [CrossRef]
- Xu, R.; Li, C. A Review of High-Throughput Field Phenotyping Systems: Focusing on Ground Robots. Plant Phenomics 2022, 2022, 9760269. [Google Scholar] [CrossRef] [PubMed]
- de Medeiros, A.D.; da Silva, L.J.; Ribeiro, J.P.O.; Ferreira, K.C.; Rosas, J.T.F.; Santos, A.A.; da Silva, C.B. Machine Learning for Seed Quality Classification: An Advanced Approach Using Merger Data from FT-NIR Spectroscopy and X-ray Imaging. Sensors 2020, 20, 4319. [Google Scholar] [CrossRef]
- Medeiros, A.D.D.; Silva, L.J.D.; Pereira, M.D.; Oliveira, A.M.S.; Dias, D.C.F.S. High-Throughput Phenotyping of Brachiaria Grass Seeds Using Free Access Tool for Analyzing X-ray Images. An. Acad. Bras. Ciênc. 2020, 92, e20190209. [Google Scholar] [CrossRef]
- Ahmed, M.R.; Yasmin, J.; Park, E.; Kim, G.; Kim, M.S.; Wakholi, C.; Mo, C.; Cho, B.-K. Classification of Watermelon Seeds Using Morphological Patterns of X-ray Imaging: A Comparison of Conventional Machine Learning and Deep Learning. Sensors 2020, 20, 6753. [Google Scholar] [CrossRef]
- Liu, W.; Liu, C.; Jin, J.; Li, D.; Fu, Y.; Yuan, X. High-Throughput Phenotyping of Morphological Seed and Fruit Characteristics Using X-ray Computed Tomography. Front. Plant Sci. 2020, 11, 601475. [Google Scholar] [CrossRef]
- Aich, S.; Stavness, I. Leaf Counting With Deep Convolutional and Deconvolutional Networks. In Proceedings of the IEEE International Conference on Computer Vision Workshops, Venice, Italy, 22–29 October 2017; pp. 2080–2089. [Google Scholar]
- Wang, T.; Rostamza, M.; Song, Z.; Wang, L.; McNickle, G.; Iyer-Pascuzzi, A.S.; Qiu, Z.; Jin, J. SegRoot: A High Throughput Segmentation Method for Root Image Analysis. Comput. Electron. Agric. 2019, 162, 845–854. [Google Scholar] [CrossRef]
- Gong, L.; Du, X.; Zhu, K.; Lin, C.; Lin, K.; Wang, T.; Lou, Q.; Yuan, Z.; Huang, G.; Liu, C. Pixel Level Segmentation of Early-Stage in-Bag Rice Root for Its Architecture Analysis. Comput. Electron. Agric. 2021, 186, 106197. [Google Scholar] [CrossRef]
- Chavarría-Krauser, A.; Nagel, K.A.; Palme, K.; Schurr, U.; Walter, A.; Scharr, H. Spatio-Temporal Quantification of Differential Growth Processes in Root Growth Zones Based on a Novel Combination of Image Sequence Processing and Refined Concepts Describing Curvature Production. New Phytol. 2008, 177, 811–821. [Google Scholar] [CrossRef]
- Araus, J.L.; Cairns, J.E. Field High-Throughput Phenotyping: The New Crop Breeding Frontier. Trends Plant Sci. 2014, 19, 52–61. [Google Scholar] [CrossRef]
- Minervini, M.; Giuffrida, M.V.; Perata, P.; Tsaftaris, S.A. Phenotiki: An Open Software and Hardware Platform for Affordable and Easy Image-Based Phenotyping of Rosette-Shaped Plants. Plant J. 2017, 90, 204–216. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Huang, C.; Cai, J.; Miklavcic, S.J. Root Phenotyping by Root Tip Detection and Classification through Statistical Learning. Plant Soil 2014, 380, 193–209. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2015, arXiv:1409.1556. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Lu, H.; Cao, Z.; Xiao, Y.; Zhuang, B.; Shen, C. TasselNet: Counting Maize Tassels in the Wild via Local Counts Regression Network. Plant Methods 2017, 13, 1–17. [Google Scholar] [CrossRef]
- Pound, M.P.; Atkinson, J.A.; Townsend, A.J.; Wilson, M.H.; Griffiths, M.; Jackson, A.S.; Bulat, A.; Tzimiropoulos, G.; Wells, D.M.; Murchie, E.H.; et al. Deep Machine Learning Provides State-of-the-Art Performance in Image-Based Plant Phenotyping. GigaScience 2017, 6, gix083. [Google Scholar] [CrossRef]
- Misra, T.; Arora, A.; Marwaha, S.; Chinnusamy, V.; Rao, A.R.; Jain, R.; Sahoo, R.N.; Ray, M.; Kumar, S.; Raju, D.; et al. SpikeSegNet-a Deep Learning Approach Utilizing Encoder-Decoder Network with Hourglass for Spike Segmentation and Counting in Wheat Plant from Visual Imaging. Plant Methods 2020, 16, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Wang, J.; Clark, R.; Hu, Q.; Wang, S.; Markham, A.; Trigoni, N. Learning Object Bounding Boxes for 3D Instance Segmentation on Point Clouds. In Proceedings of the 33rd International Conference on Neural Information Processing Systems, Vancouver, BC, Canada, 8–14 December 2019. [Google Scholar]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You Only Look Once: Unified, Real-Time Object Detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 779–788. [Google Scholar]
- Mosley, L.; Pham, H.; Bansal, Y.; Hare, E. Image-Based Sorghum Head Counting When You Only Look Once. In Proceedings of the Hawaii International Conference on System Sciences, Maui, HI, USA, 4–7 January 2022. [Google Scholar]
- Cardellicchio, A.; Solimani, F.; Dimauro, G.; Petrozza, A.; Summerer, S.; Cellini, F.; Renò, V. Detection of Tomato Plant Phenotyping Traits Using YOLOv5-Based Single Stage Detectors. Comput. Electron. Agric. 2023, 207, 107757. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. Ann. Intern. Med. 2009, 151, 264–269. [Google Scholar] [CrossRef] [PubMed]
- Rice, D.B.; Kloda, L.A.; Levis, B.; Qi, B.; Kingsland, E.; Thombs, B.D. Are MEDLINE Searches Sufficient for Systematic Reviews and Meta-Analyses of the Diagnostic Accuracy of Depression Screening Tools? A Review of Meta-Analyses. J. Psychosom. Res. 2016, 87, 7–13. [Google Scholar] [CrossRef]
- Xu, Z.; York, L.M.; Seethepalli, A.; Bucciarelli, B.; Cheng, H.; Samac, D.A. Objective Phenotyping of Root System Architecture Using Image Augmentation and Machine Learning in Alfalfa (Medicago sativa L.). Plant Phenomics 2022, 2022, 9879610. [Google Scholar] [CrossRef] [PubMed]
- Islam ElManawy, A.; Sun, D.; Abdalla, A.; Zhu, Y.; Cen, H. HSI-PP: A Flexible Open-Source Software for Hyperspectral Imaging-Based Plant Phenotyping. Comput. Electron. Agric. 2022, 200, 107248. [Google Scholar] [CrossRef]
- Daviet, B.; Fernandez, R.; Cabrera-Bosquet, L.; Pradal, C.; Fournier, C. PhenoTrack3D: An Automatic High-Throughput Phenotyping Pipeline to Track Maize Organs over Time. Plant Methods 2022, 18, 130. [Google Scholar] [CrossRef]
- Yu, S.; Fan, J.; Xianju, L.; Wen, W.; Shao, S.; Guo, X.; Zhao, C. Hyperspectral Technique Combined with Deep Learning Algorithm for Prediction of Phenotyping Traits in Lettuce. Front. Plant Sci. 2022, 13, 927832. [Google Scholar] [CrossRef] [PubMed]
- Oury, V.; Leroux, T.; Turc, O.; Chapuis, R.; Palaffre, C.; Tardieu, F.; Prado, S.A.; Welcker, C.; Lacube, S. Earbox, an Open Tool for High-Throughput Measurement of the Spatial Organization of Maize Ears and Inference of Novel Traits. bioRxiv 2021. [Google Scholar] [CrossRef] [PubMed]
- Rolland, V.; Farazi, M.R.; Conaty, W.C.; Cameron, D.; Liu, S.; Petersson, L.; Stiller, W.N. HairNet: A Deep Learning Model to Score Leaf Hairiness, a Key Phenotype for Cotton Fibre Yield, Value and Insect Resistance. Plant Methods 2022, 18, 8. [Google Scholar] [CrossRef]
- Petti, D.; Li, C. Weakly-Supervised Learning to Automatically Count Cotton Flowers from Aerial Imagery. Comput. Electron. Agric. 2022, 194, 106734. [Google Scholar] [CrossRef]
- Zhao, H.; Wang, N.; Sun, H.; Zhu, L.; Zhang, K.; Zhang, Y.; Zhu, J.; Li, A.; Bai, Z.; Liu, X.; et al. RhizoPot Platform: A High-Throughput in Situ Root Phenotyping Platform with Integrated Hardware and Software. Front. Plant Sci. 2022, 13, 1004904. [Google Scholar] [CrossRef]
- Narisetti, N.; Henke, M.; Neumann, K.; Stolzenburg, F.; Altmann, T.; Gladilin, E. Deep Learning Based Greenhouse Image Segmentation and Shoot Phenotyping (DeepShoot). Front. Plant Sci. 2022, 13, 906410. [Google Scholar] [CrossRef] [PubMed]
- Zenkl, R.; Timofte, R.; Kirchgessner, N.; Roth, L.; Hund, A.; Van Gool, L.; Walter, A.; Aasen, H. Outdoor Plant Segmentation with Deep Learning for High-Throughput Field Phenotyping on a Diverse Wheat Dataset. Front. Plant Sci. 2022, 12, 774068. [Google Scholar] [CrossRef]
- Lube, V.; Noyan, M.A.; Przybysz, A.; Salama, K.; Blilou, I. MultipleXLab: A High-Throughput Portable Live-Imaging Root Phenotyping Platform Using Deep Learning and Computer Vision. Plant Methods 2022, 18, 38. [Google Scholar] [CrossRef] [PubMed]
- Jubery, T.Z.; Carley, C.N.; Singh, A.; Sarkar, S.; Ganapathysubramanian, B.; Singh, A.K. Using Machine Learning to Develop a Fully Automated Soybean Nodule Acquisition Pipeline (SNAP). Plant Phenomics 2021, 2021, 9834746. [Google Scholar] [CrossRef]
- Zhao, Y.; Zheng, B.; Chapman, S.C.; Laws, K.; George-Jaeggli, B.; Hammer, G.L.; Jordan, D.R.; Potgieter, A.B. Detecting Sorghum Plant and Head Features from Multispectral UAV Imagery. Plant Phenomics 2021, 2021, 9874650. [Google Scholar] [CrossRef]
- Guo, X.; Qiu, Y.; Nettleton, D.; Yeh, C.-T.; Zheng, Z.; Hey, S.; Schnable, P.S. KAT4IA: K-Means Assisted Training for Image Analysis of Field-Grown Plant Phenotypes. Plant Phenomics 2021, 2021, 9805489. [Google Scholar] [CrossRef]
- Chang, S.; Lee, U.; Hong, M.J.; Jo, Y.D.; Kim, J.-B. Time-Series Growth Prediction Model Based on U-Net and Machine Learning in Arabidopsis. Front. Plant Sci. 2021, 12, 721512. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Hu, Y.; Mao, H.; Li, S.; Li, F.; Zhao, C.; Luo, L.; Liu, W.; Yuan, X. A Deep Learning-Based Method for Automatic Assessment of Stomatal Index in Wheat Microscopic Images of Leaf Epidermis. Front. Plant Sci. 2021, 12, 716784. [Google Scholar] [CrossRef]
- Zhou, S.; Chai, X.; Yang, Z.; Wang, H.; Yang, C.; Sun, T. Maize-IAS: A Maize Image Analysis Software Using Deep Learning for High-Throughput Plant Phenotyping. Plant Methods 2021, 17, 48. [Google Scholar] [CrossRef]
- Pranga, J.; Borra-Serrano, I.; Aper, J.; De Swaef, T.; Ghesquiere, A.; Quataert, P.; Roldán-Ruiz, I.; Janssens, I.A.; Ruysschaert, G.; Lootens, P. Improving Accuracy of Herbage Yield Predictions in Perennial Ryegrass with UAV-Based Structural and Spectral Data Fusion and Machine Learning. Remote Sens. 2021, 13, 3459. [Google Scholar] [CrossRef]
- Banerjee, B.P.; Sharma, V.; Spangenberg, G.; Kant, S. Machine Learning Regression Analysis for Estimation of Crop Emergence Using Multispectral UAV Imagery. Remote Sens. 2021, 13, 2918. [Google Scholar] [CrossRef]
- Rehman, T.U.; Ma, D.; Wang, L.; Zhang, L.; Jin, J. Predictive Spectral Analysis Using an End-to-End Deep Model from Hyperspectral Images for High-Throughput Plant Phenotyping. Comput. Electron. Agric. 2020, 177, 105713. [Google Scholar] [CrossRef]
- Du, J.; Lu, X.; Fan, J.; Qin, Y.; Yang, X.; Guo, X. Image-Based High-Throughput Detection and Phenotype Evaluation Method for Multiple Lettuce Varieties. Front. Plant Sci. 2020, 11, 563386. [Google Scholar] [CrossRef]
- Lin, Z.; Guo, W. Sorghum Panicle Detection and Counting Using Unmanned Aerial System Images and Deep Learning. Front. Plant Sci. 2020, 11, 534853. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Li, C.; Xu, R.; Sun, S.; Robertson, J.S.; Paterson, A.H. DeepFlower: A Deep Learning-Based Approach to Characterize Flowering Patterns of Cotton Plants in the Field. Plant Methods 2020, 16, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Falk, K.G.; Jubery, T.Z.; Mirnezami, S.V.; Parmley, K.A.; Sarkar, S.; Singh, A.; Ganapathysubramanian, B.; Singh, A.K. Computer Vision and Machine Learning Enabled Soybean Root Phenotyping Pipeline. Plant Methods 2020, 16, 1–19. [Google Scholar] [CrossRef]
- Lu, H.; Cao, Z. TasselNetV2+: A Fast Implementation for High-Throughput Plant Counting From High-Resolution RGB Imagery. Front. Plant Sci. 2020, 11, 541960. [Google Scholar] [CrossRef] [PubMed]
- Milella, A.; Marani, R.; Petitti, A.; Reina, G. In-Field High Throughput Grapevine Phenotyping with a Consumer-Grade Depth Camera. Comput. Electron. Agric. 2019, 156, 293–306. [Google Scholar] [CrossRef]
- Zhou, J.; Fu, X.; Zhou, S.; Zhou, J.; Ye, H.; Nguyen, H.T. Automated Segmentation of Soybean Plants from 3D Point Cloud Using Machine Learning. Comput. Electron. Agric. 2019, 162, 143–153. [Google Scholar] [CrossRef]
- Tracy, S.R.; Nagel, K.A.; Postma, J.A.; Fassbender, H.; Wasson, A.; Watt, M. Crop Improvement from Phenotyping Roots: Highlights Reveal Expanding Opportunities. Trends Plant Sci. 2020, 25, 105–118. [Google Scholar] [CrossRef]
- Girshick, R. Fast R-CNN. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 7–13 December 2015; pp. 1440–1448. [Google Scholar]
- Wang, X.; Liu, J. Tomato Anomalies Detection in Greenhouse Scenarios Based on YOLO-Dense. Front. Plant Sci. 2021, 12, 634103. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Cai, R.; Lin, P.; Cheng, S. A Detection Approach for Bundled Log Ends Using K-Median Clustering and Improved YOLOv4-Tiny Network. Comput. Electron. Agric. 2022, 194, 106700. [Google Scholar] [CrossRef]
- Das Choudhury, S.; Guha, S.; Das, A.; Das, A.K.; Samal, A.; Awada, T. FlowerPhenoNet: Automated Flower Detection from Multi-View Image Sequences Using Deep Neural Networks for Temporal Plant Phenotyping Analysis. Remote Sens. 2022, 14, 6252. [Google Scholar] [CrossRef]
| # | Exclusion Criterion |
|---|---|
| 1 | Articles not written in English |
| 2 | Articles that do not refer to high-throughput plant phenotyping |
| 3 | Articles that relate to phenotyping traits but are unrelated to the discussion |
| 4 | Articles that do not use DL or ML |
| 5 | Articles that appear in invalid journals or those with very low-impact factors |
| 6 | Articles that are reviews |
| 7 | Articles for which only abstracts are available |
| # | Inclusion Criterion |
|---|---|
| 1 | Articles written in English |
| 2 | Articles that refer to the high-throughput plant phenotyping |
| 3 | Articles that use DL or ML |
| 4 | Articles that appear with high-impact factors |
| 5 | Research articles (non-review papers) |
| 6 | Articles that are fully available |
| 7 | Articles that relate to selected research questions |
| Extraction | Element Contents | Type |
|---|---|---|
| 1 | Title | Yes/no |
| 2 | Research questions | The clear description of the research question |
| 3 | Type of article | Problem identification |
| 4 | Study outcomes | Short description of study outcomes |
| 5 | Year | The year of publication |
| 6 | Journal | Impact factor (Q1) |
| # | Reference | Year | Plant | Platform | Sensor | Algorithm |
|---|---|---|---|---|---|---|
| [27] | Bauer et al. | 2022 | Wheat | Minirhizotron | Camera | DL |
| [60] | Xu et al. | 2022 | Alfalfa | Rhizotron | RGB | ML/DL |
| [61] | Islam Elmanawy et al. | 2022 | Oilseed rape | Hyper platform | Hyperspectral | DL |
| [23] | Van De Looverbosch et al. | 2022 | Sugar beet | Polystyrene sheets | X-ray CT | DL |
| [16] | Das Choudhury et al. | 2022 | Flowers | LemnaTec Scanalyzer | RGB, infrared | DL |
| [62] | Daviet et al. | 2022 | Maize | PhenoArch | RGB | DL |
| [63] | Yu et al. | 2022 | Lettuce | LQ-FieldPheno | Hyperspectral | DL |
| [64] | Oury et al. | 2022 | Maize | Earbox | RGB/IR | DL |
| [65] | Rolland et al. | 2022 | Cotton | Microscope | Microscope | DL |
| [66] | Petti and Li | 2022 | Cotton | UAV | RGB | DL |
| [67] | Zhao et al. | 2022 | Cotton | Rhizo-Pot platform | Scanner | DL |
| [8] | Maji et al. | 2022 | Wheat | Chamber platform | RGB | DL |
| [68] | Narisetti et al. | 2022 | Maize/wheat | Scanalyzer3D | RGB | DL |
| [69] | Zenkl et al. | 2022 | Wheat | Field platform | RGB | DL |
| [70] | Lubi et al. | 2022 | Arabidopsis | MultipleXLab | RGB | DL |
| [71] | Jubery et al. | 2021 | Soybean | SNAP platform | RGB | ML/DL |
| [72] | Zhao et al. | 2021 | Sorghum | UAV | Multispectral | ML |
| [73] | Guo et al. | 2021 | Maize | KAT4IA field | RGB | ML/DL |
| [74] | Chang et al. | 2021 | Arabidopsis | Controlled Platform | RGB | ML/DL |
| [75] | Zhu et al. | 2021 | Wheat | DP72 microscope | DP72 microscope | DL |
| [76] | Zhou et al. | 2021 | Maize | Chamber | RGB | DL |
| [77] | Pranga et al. | 2021 | Ryegrass | UAV | Multispectral/RGB | ML |
| [78] | Banerjee et al. | 2021 | Wheat | UAV | Multispectral | ML |
| [32] | Koh et al. | 2021 | Wheat | UAV | RGB | ML/DL |
| [79] | Rehman et al. | 2020 | Maize | Greenhouse platform | Hyperspectral | DL |
| [80] | Du et al. | 2020 | Lettuce | Greenhouse platform | Industrial camera | DL |
| [81] | Lin and Guo | 2020 | Sorghum | UAV | RGB | DL |
| [82] | Jiang et al. | 2020 | Cotton | GPhenoVision | RGB | DL |
| [83] | Falk et al. | 2020 | Soybean | Root platform | RGB | ML/DL |
| [84] | Lu and Cao | 2020 | Wheat/maize | Field platform | RGB | DL |
| [85] | Milella et al. | 2019 | Grapevine | Caterpillar vehicle | RGB-D | DL |
| [86] | Zhou et al. | 2019 | Soybean | Greenhouse | RGB | ML |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Solimani, F.; Cardellicchio, A.; Nitti, M.; Lako, A.; Dimauro, G.; Renò, V. A Systematic Review of Effective Hardware and Software Factors Affecting High-Throughput Plant Phenotyping. Information 2023, 14, 214. https://doi.org/10.3390/info14040214
Solimani F, Cardellicchio A, Nitti M, Lako A, Dimauro G, Renò V. A Systematic Review of Effective Hardware and Software Factors Affecting High-Throughput Plant Phenotyping. Information. 2023; 14(4):214. https://doi.org/10.3390/info14040214
Chicago/Turabian StyleSolimani, Firozeh, Angelo Cardellicchio, Massimiliano Nitti, Alfred Lako, Giovanni Dimauro, and Vito Renò. 2023. "A Systematic Review of Effective Hardware and Software Factors Affecting High-Throughput Plant Phenotyping" Information 14, no. 4: 214. https://doi.org/10.3390/info14040214
APA StyleSolimani, F., Cardellicchio, A., Nitti, M., Lako, A., Dimauro, G., & Renò, V. (2023). A Systematic Review of Effective Hardware and Software Factors Affecting High-Throughput Plant Phenotyping. Information, 14(4), 214. https://doi.org/10.3390/info14040214







