DBA_SSD: A Novel End-to-End Object Detection Algorithm Applied to Plant Disease Detection
Abstract
:1. Introduction
- (1)
- We proposed a novel end-to-end detection algorithm for plant disease, DBA_SSD, by combining the attention mechanism and convolution kernel, which combines the attributes of the plant leaf disease pictures and pay more attention to disease details when testing plant disease.
- (2)
- We graded the health of the fruit and vegetable leaves. According to the research results of the paper, different measures can be taken according to the severity of the diseases of the fruit and vegetable leaves. Increasing the yield of plants is of great significance.
- (3)
- We implemented the classic SSD, YOLOv4, YOLOv3, Faster RCNN, and YOLOv4 tiny models and compared them with our proposed DBA_SSD. Our method is better than the classic baseline method on the vegetable and fruit leaf data set.
2. Materials and Methods
2.1. Related Work
2.2. Novel End-to-End Method for Leaf Disease Detection
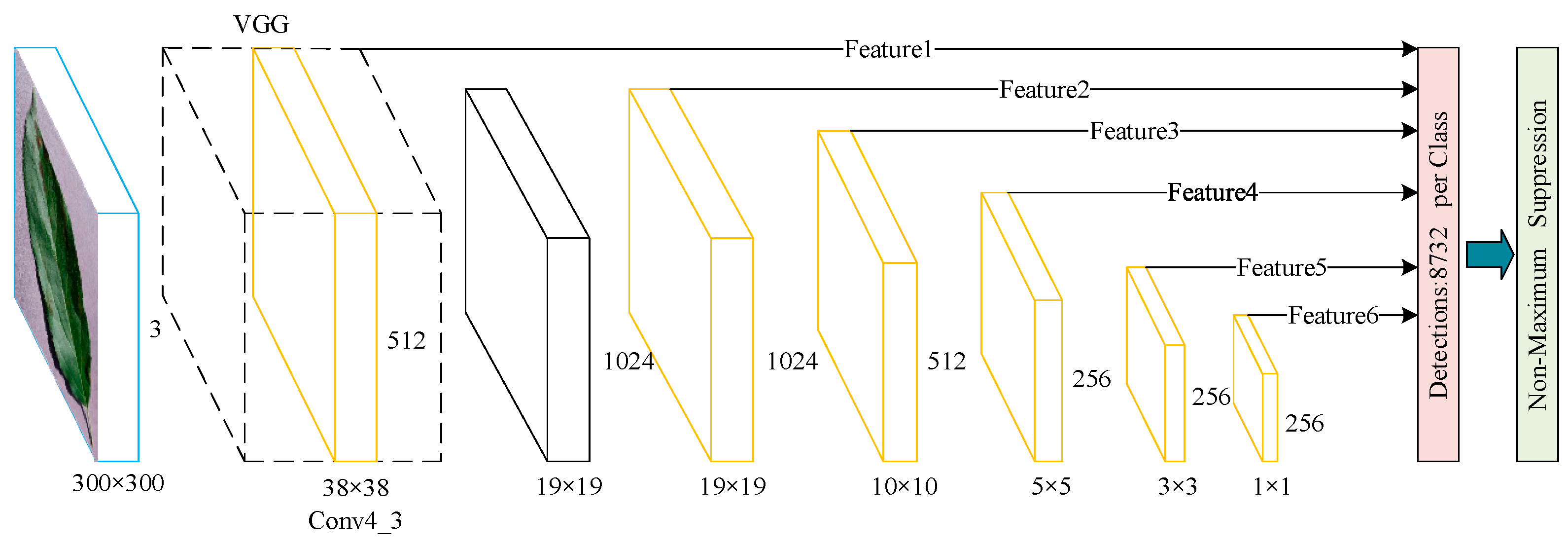
2.2.1. SSD Network
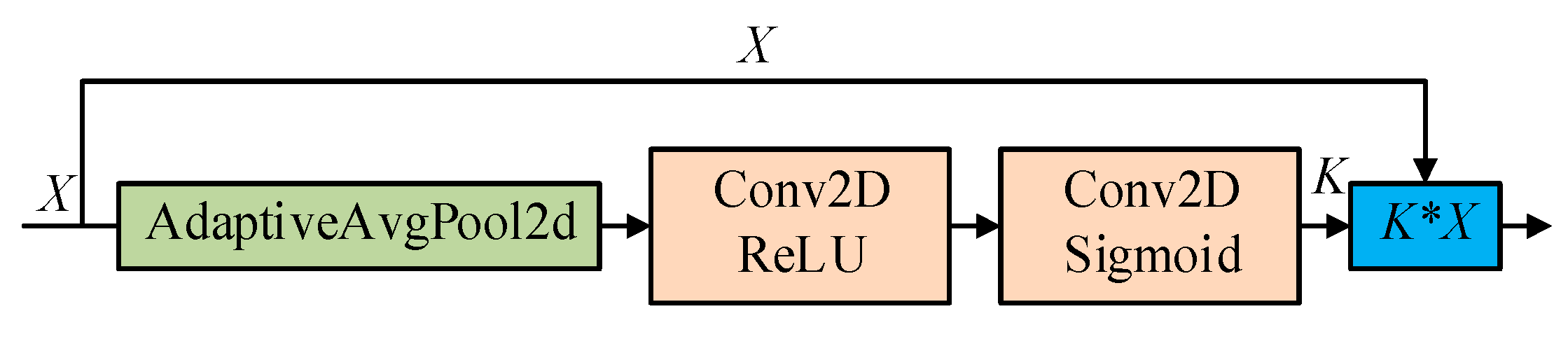
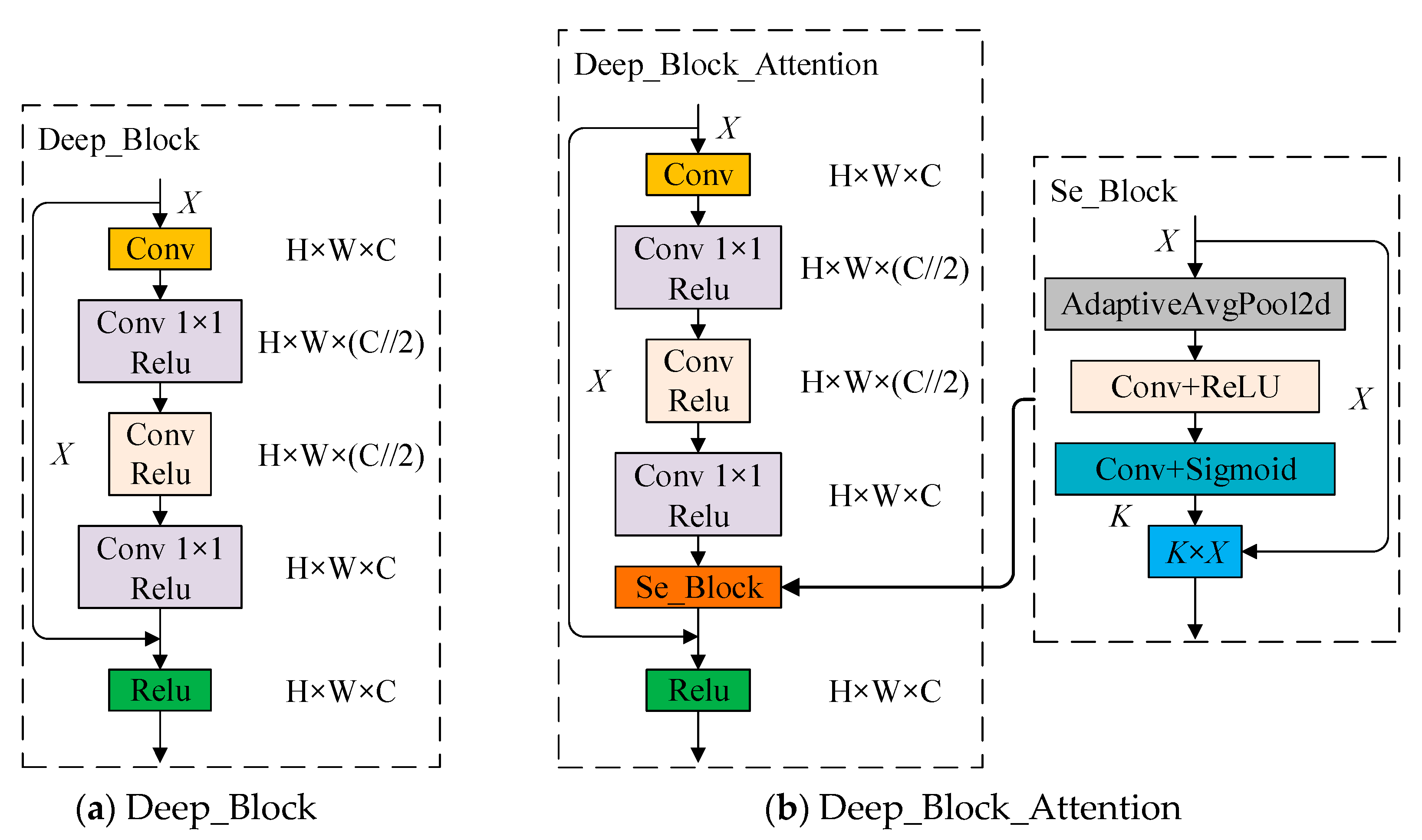
2.2.2. Squeeze-and-Excitation SSD (Se_SSD) Network
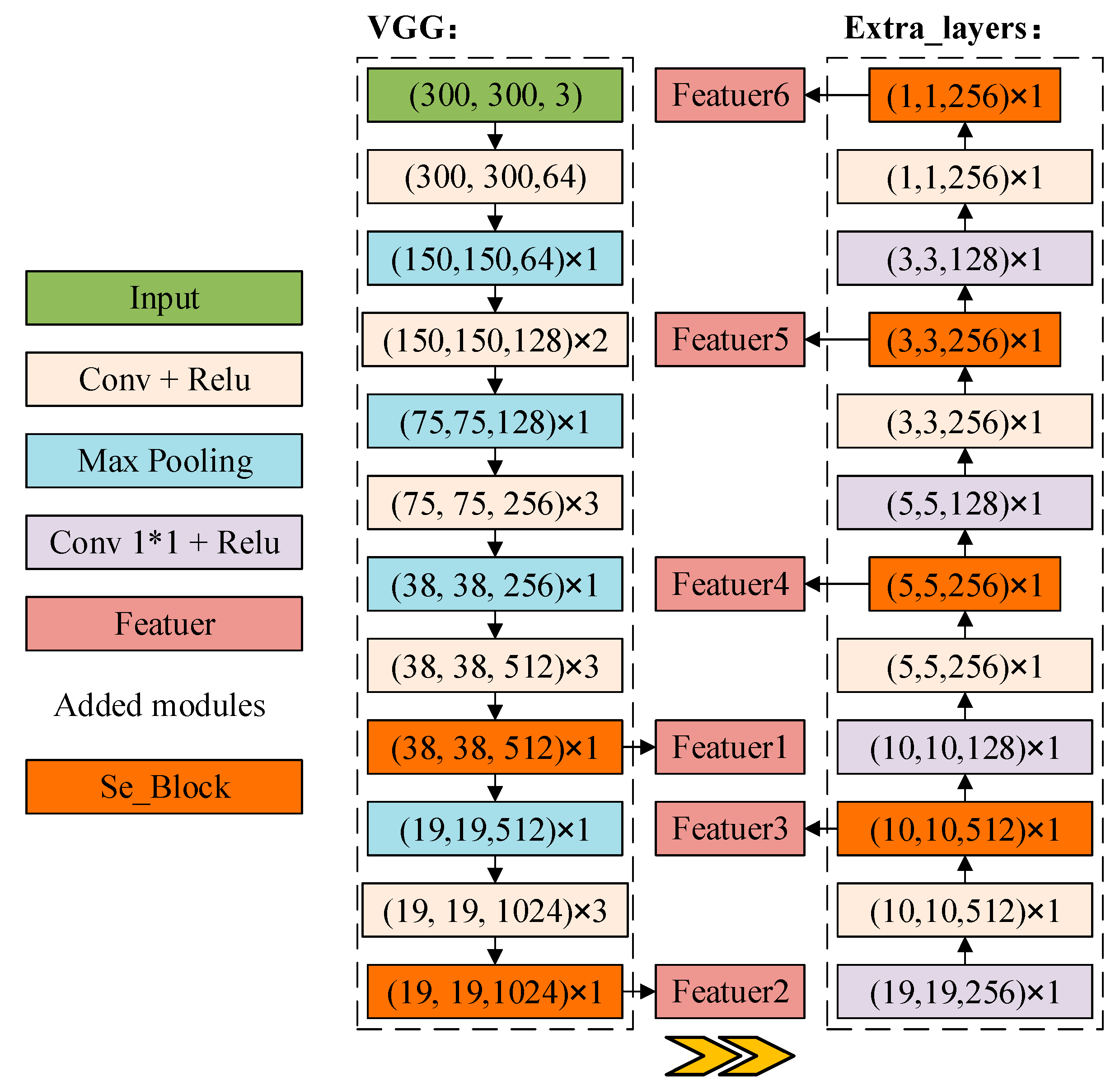
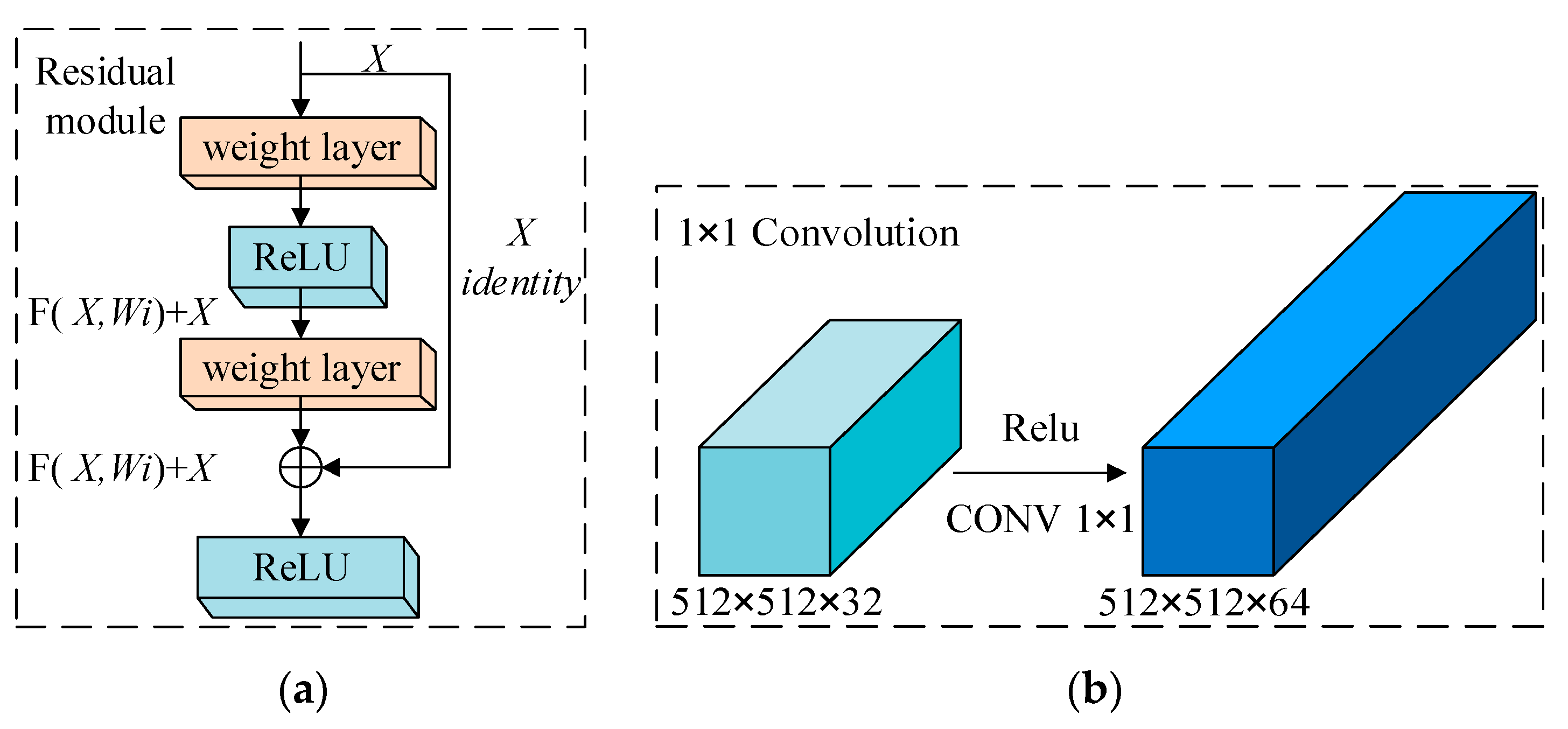
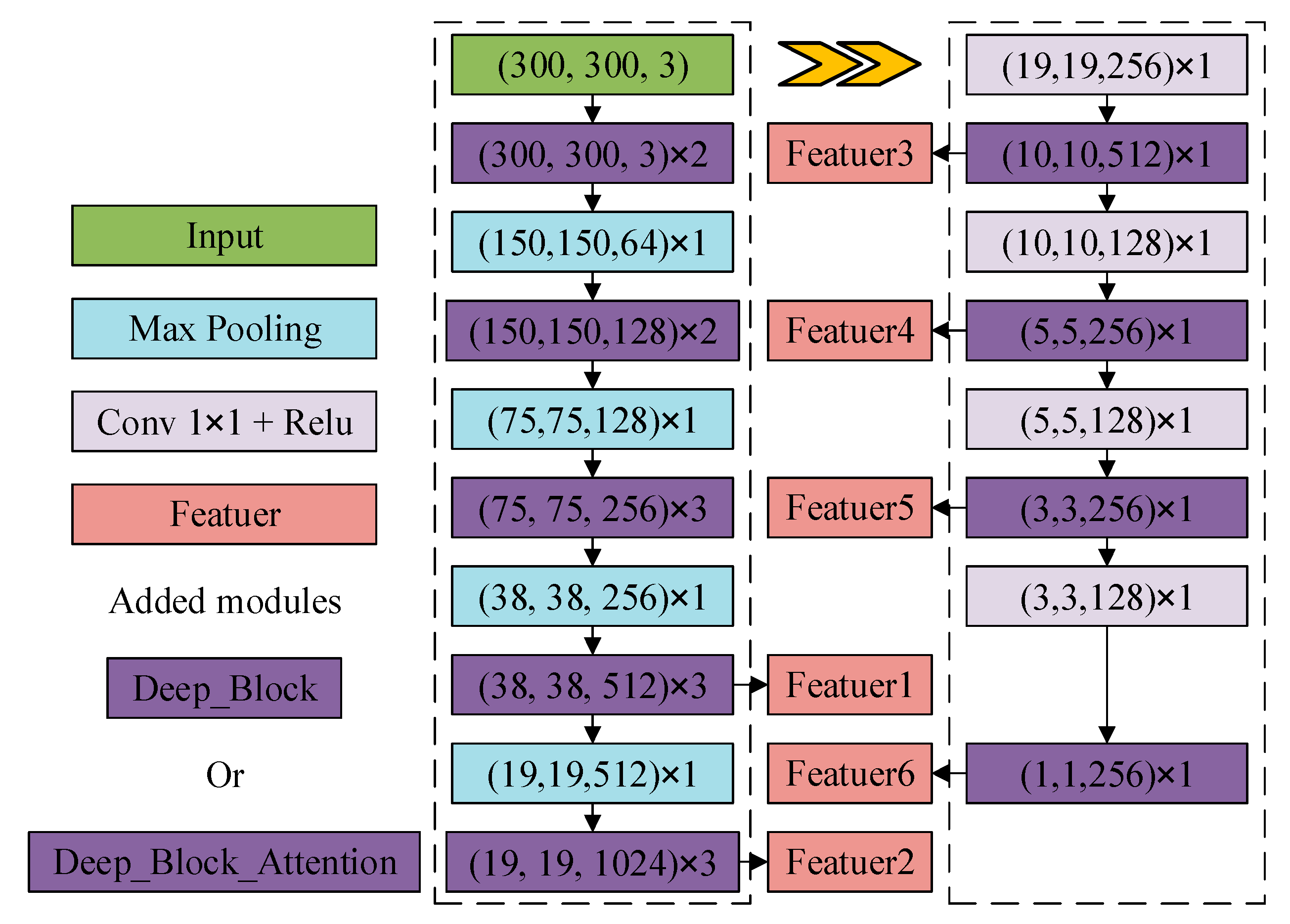
2.2.3. DB_SSD and DBA_SSD Network
3. Experimental Environment and Experimental Design
3.1. Experimental Environment
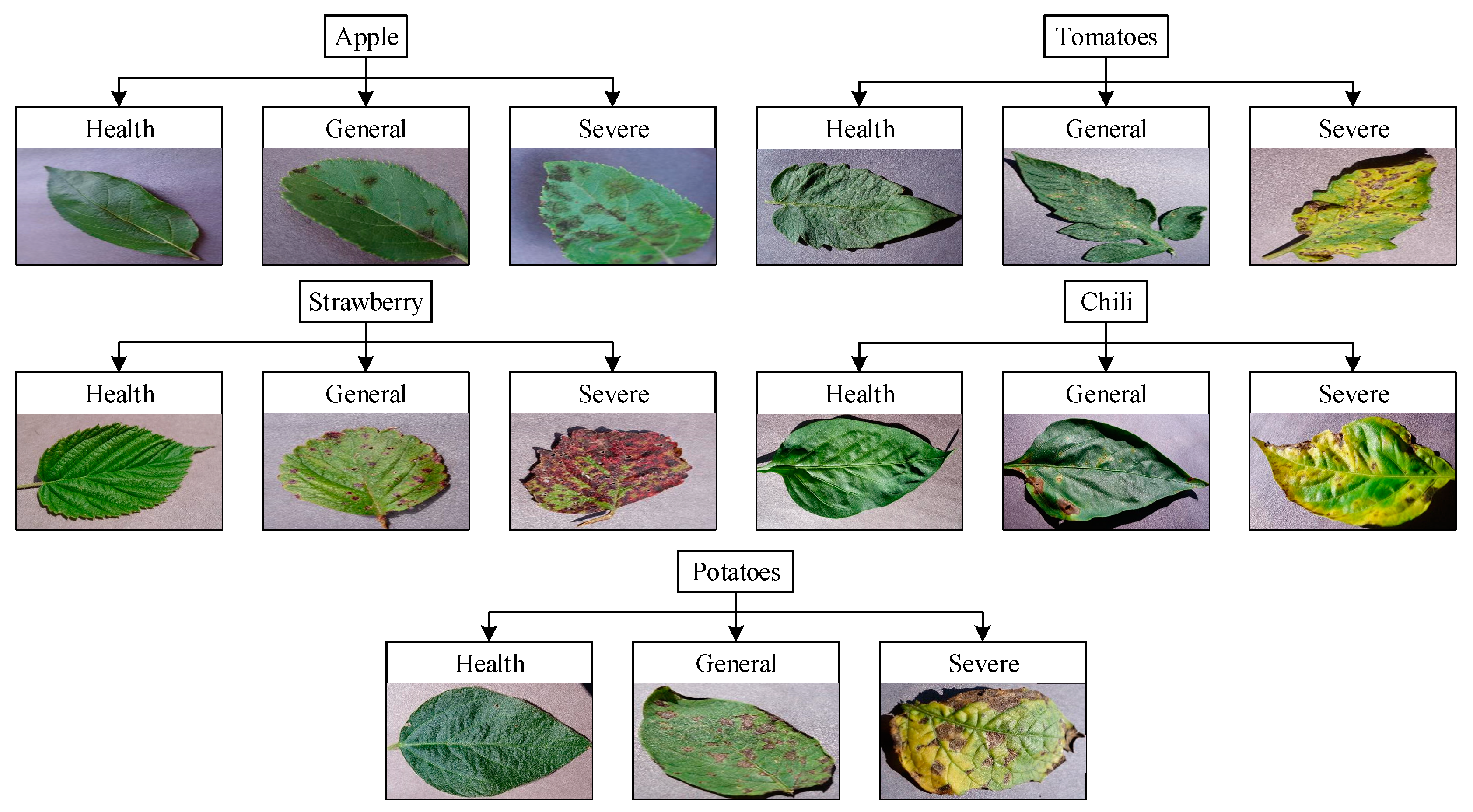
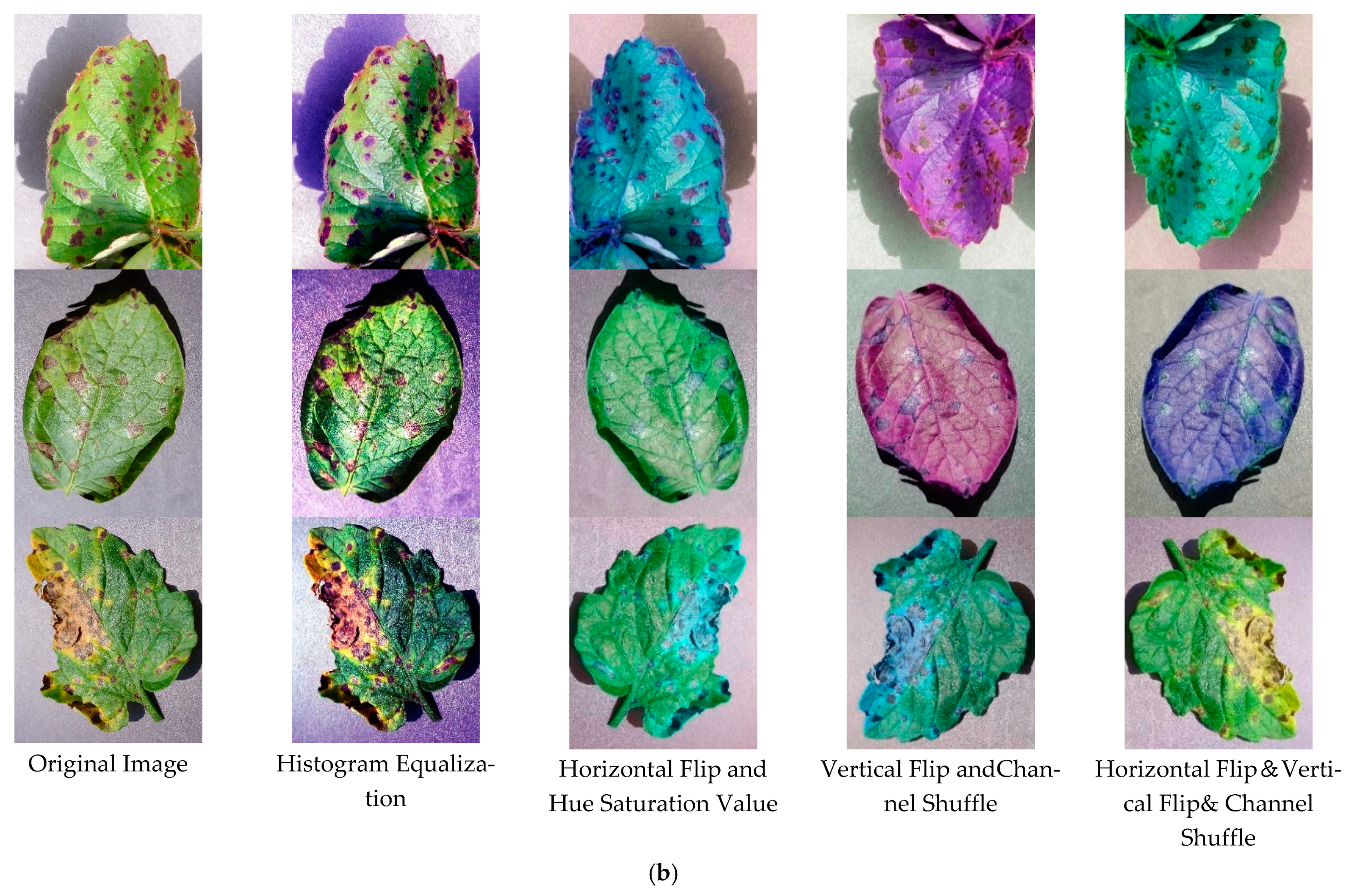
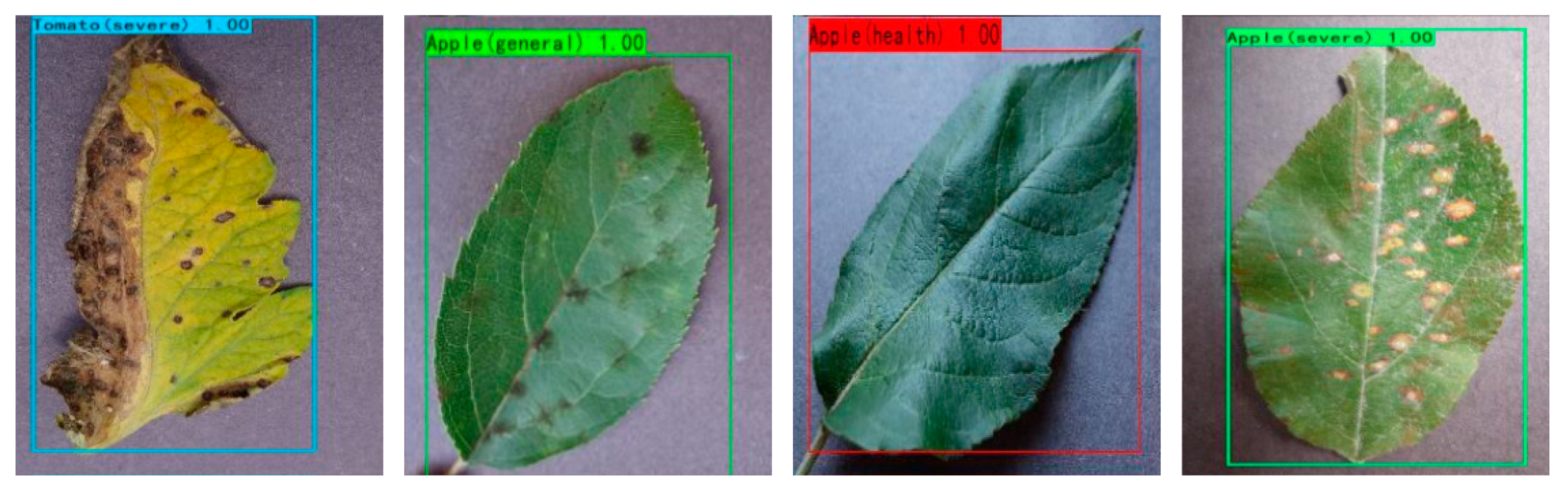
3.2. Dataset
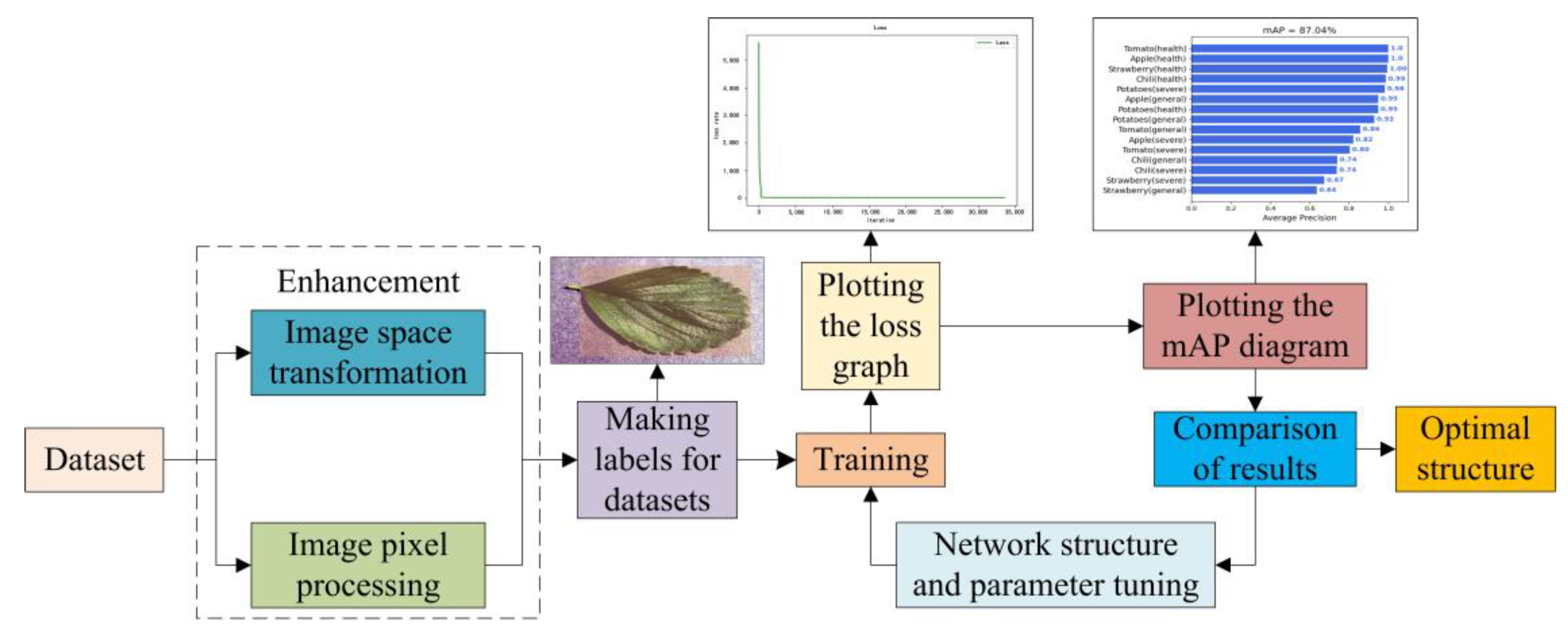
3.3. Experimental Design
3.4. Performance Evaluation Metrics
4. Analysis of Experimental Results
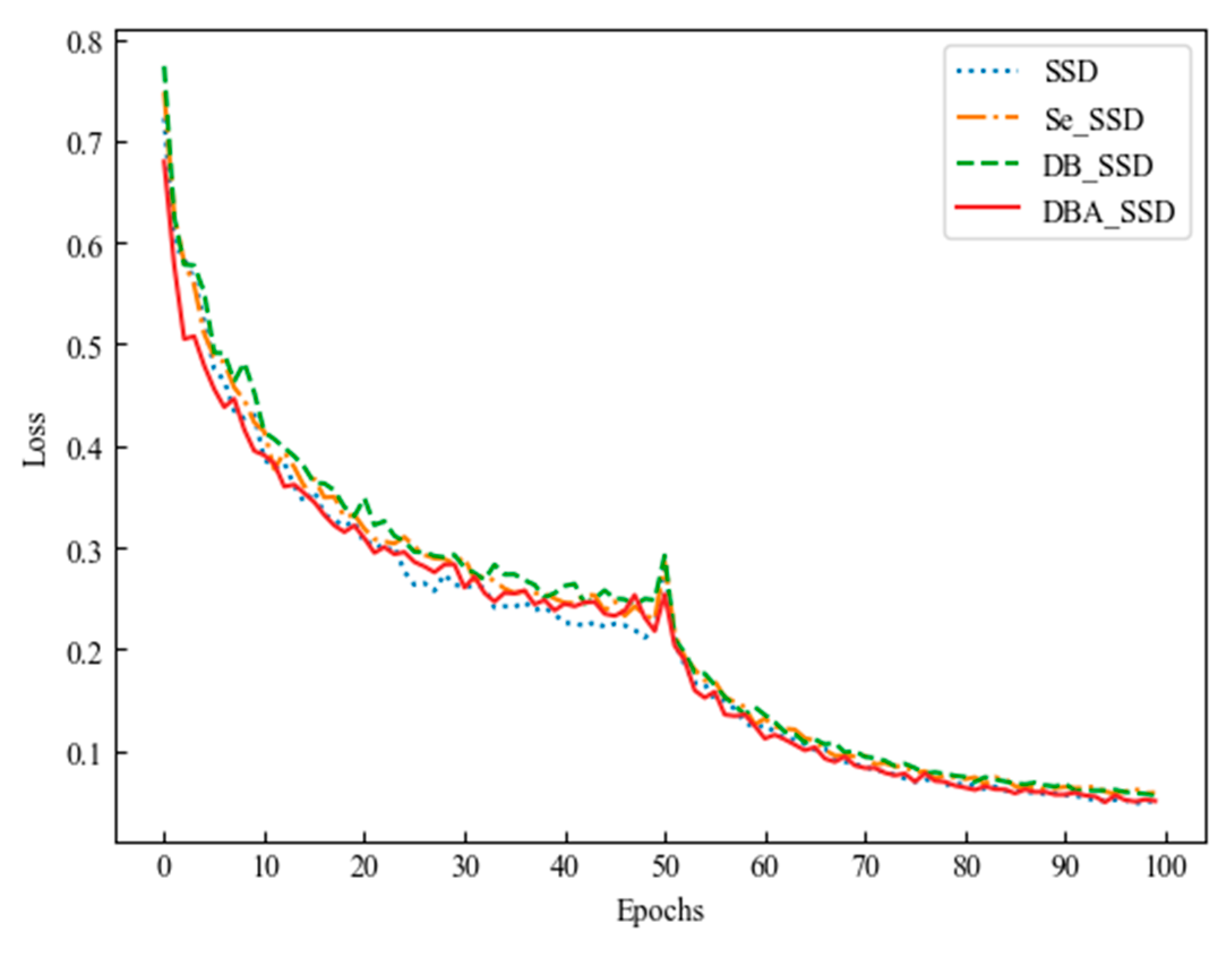
4.1. DBA_SSD Model Experimental Comparison Analysis
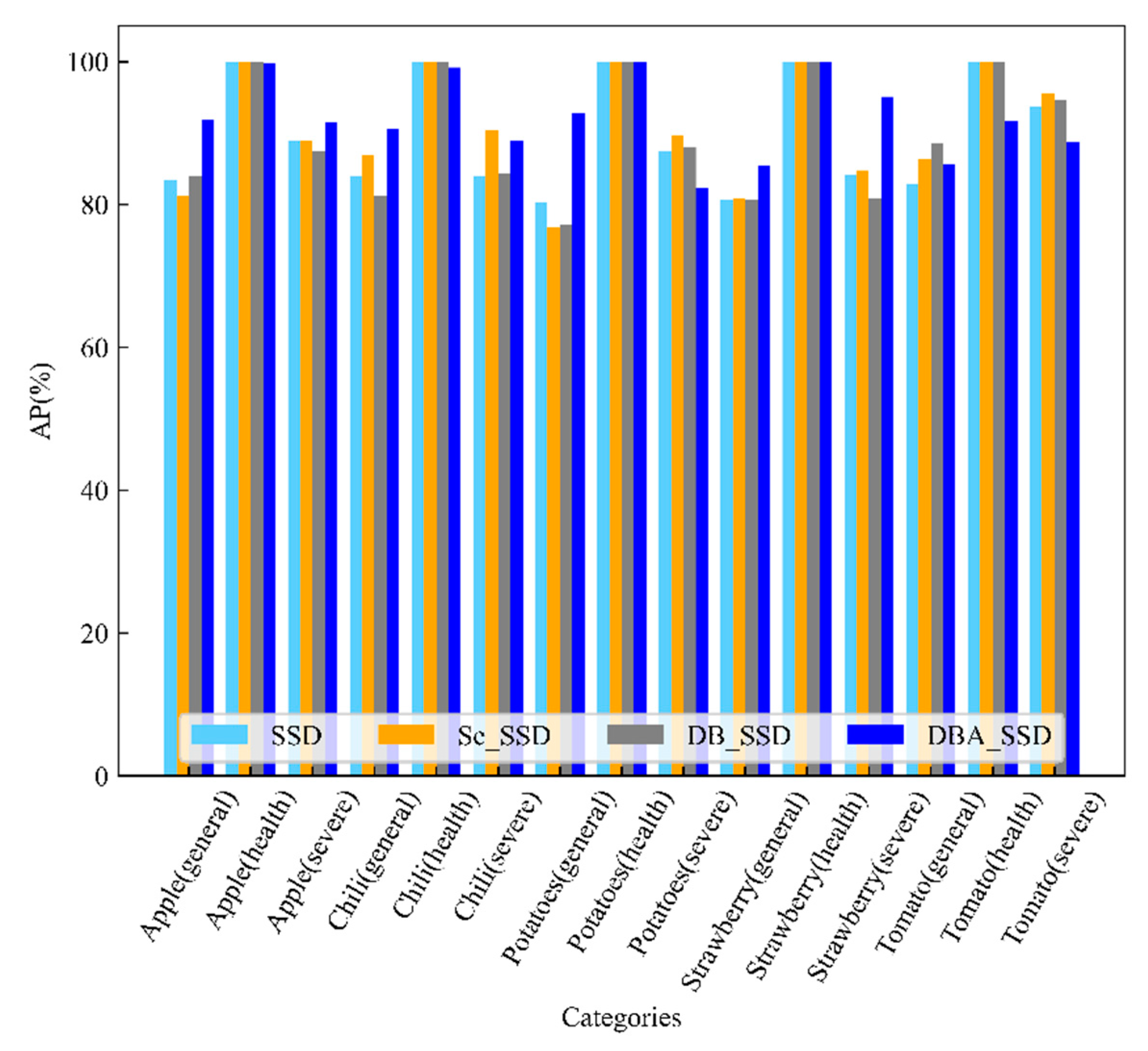
4.2. Comparative Analysis with Classical Target Detection Algorithms
5. Discussion
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Reza, N.; Na, I.S.; Baek, S.W.; Lee, K.-H. Rice yield estimation based on K-means clustering with graph-cut segmentation using low-altitude UAV images. Biosyst. Eng. 2018, 177, 109–121. [Google Scholar] [CrossRef]
- Quan, L.; Feng, H.; Lv, Y.; Wang, Q.; Zhang, C.; Liu, J.; Yuan, Z. Maize seedling detection under different growth stages and complex field environments based on an improved Faster R–CNN. Biosyst. Eng. 2019, 184, 1–23. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, X.; Yuan, M.; Ren, L.; Wang, J.; Chen, Z. Automatic in-trap pest detection using deep learning for pheromone-based Dendroctonus valens monitoring. Biosyst. Eng. 2018, 176, 140–150. [Google Scholar] [CrossRef]
- Barbedo, J.G.A. Plant disease identification from individual lesions and spots using deep learning. Biosyst. Eng. 2019, 180, 96–107. [Google Scholar] [CrossRef]
- Taheri-Garavand, A.; Nasiri, A.; Fanourakis, D.; Fatahi, S.; Omid, M.; Nikoloudakis, N. Automated In Situ Seed Variety Identification via Deep Learning: A Case Study in Chickpea. Plants 2021, 10, 1406. [Google Scholar] [CrossRef] [PubMed]
- Nasiri, A.; Taheri-Garavand, A.; Fanourakis, D.; Zhang, Y.-D.; Nikoloudakis, N. Automated Grapevine Cultivar Identification via Leaf Imaging and Deep Convolutional Neural Networks: A Proof-of-Concept Study Employing Primary Iranian Varieties. Plants 2021, 10, 1628. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Feng, Y.; Wu, J.; Liu, Z.; Gao, F.; Majeed, Y.; Al-Mallahi, A.; Zhang, Q.; Li, R.; Cui, Y. Fast and accurate detection of kiwifruit in orchard using improved YOLOv3-tiny model. Precis. Agric. 2020, 22, 754–776. [Google Scholar] [CrossRef]
- Zhao, H.; Zhou, Y.; Zhang, L.; Peng, Y.; Hu, X.; Peng, H.; Cai, X. Mixed YOLOv3-LITE: A Lightweight Real-Time Object Detection Method. Sensors 2020, 20, 1861. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Nouaze, J.C.; Mbouembe, P.L.T.; Kim, J.H. YOLO-Tomato: A Robust Algorithm for Tomato Detection Based on YOLOv3. Sensors 2020, 20, 2145. [Google Scholar] [CrossRef] [Green Version]
- Redmon, J.; Farhadi, A. YOLOv3: An Incremental Improvement. arXiv 2018, arXiv:1804.02767. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 1137–1149. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Wang, X. Early recognition of tomato gray leaf spot disease based on MobileNetv2-YOLOv3 model. Plant Methods 2020, 16, 1–16. [Google Scholar] [CrossRef]
- Yi, J.; Krusenbaum, L.; Unger, P.; Hüging, H.; Seidel, S.J.; Schaaf, G.; Gall, J. Deep Learning for Non-Invasive Diagnosis of Nutrient Deficiencies in Sugar Beet Using RGB Images. Sensors 2020, 20, 5893. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. arXiv 2016, arXiv:1608.06993. [Google Scholar]
- Iandola, F.N.; Han, S.; Moskewicz, M.W.; Ashraf, K.; Dally, W.J.; Keutzer, K. SqueezeNet: AlexNet-level accuracy with 50x fewer parameters and <0.5 MB model size. arXiv 2016, arXiv:1602.07360. [Google Scholar]
- Liu, W.; Anguelov, D.; Erhan, D.; Szegedy, C.; Reed, S.; Fu, C.Y.; Berg, A.C. SSD: Single Shot MultiBox Detector. arXiv 2015, arXiv:1512.02325. [Google Scholar]
- Taheri-Garavand, A.; Nejad, A.R.; Fanourakis, D.; Fatahi, S.; Majd, M.A. Employment of artificial neural networks for non-invasive estimation of leaf water status using color features: A case study in Spathiphyllum wallisii. Acta Physiol. Plant. 2021, 43, 78. [Google Scholar] [CrossRef]
- Chao, X.; Sun, G.; Zhao, H.; Li, M.; He, D. Identification of Apple Tree Leaf Diseases Based on Deep Learning Models. Symmetry 2020, 12, 1065. [Google Scholar] [CrossRef]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. arXiv 2016, arXiv:1610.02357. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Shrivastava, V.K.; Pradhan, M.K. Rice plant disease classification using color features: A machine learning paradigm. J. Plant Pathol. 2020, 103, 17–26. [Google Scholar] [CrossRef]
- Rao, A.; Kulkarni, S. A Hybrid Approach for Plant Leaf Disease Detection and Classification Using Digital Image Processing Methods. Int. J. Electr. Eng. Educ. 2020, 0020720920953126. [Google Scholar] [CrossRef]
- Abdulridha, J.; Ampatzidis, Y.; Roberts, P.; Kakarla, S.C. Detecting powdery mildew disease in squash at different stages using UAV-based hyperspectral imaging and artificial intelligence. Biosyst. Eng. 2020, 197, 135–148. [Google Scholar] [CrossRef]
- Abdu, A.M.; Mokji, M.M.; Sheikh, U.U. Automatic vegetable disease identification approach using individual lesion features. Comput. Electron. Agric. 2020, 176, 105660. [Google Scholar] [CrossRef]
- Samiei, S.; Rasti, P.; Vu, J.L.; Buitink, J.; Rousseau, D. Deep learning-based detection of seedling development. Plant Methods 2020, 16, 103. [Google Scholar] [CrossRef] [PubMed]
- Pleșoianu, A.-I.; Stupariu, M.-S.; Șandric, I.; Pătru-Stupariu, I.; Drăguț, L. Individual Tree-Crown Detection and Species Classification in Very High-Resolution Remote Sensing Imagery Using a Deep Learning Ensemble Model. Remote Sens. 2020, 12, 2426. [Google Scholar] [CrossRef]
- Kerkech, M.; Hafiane, A.; Canals, R. Vine disease detection in UAV multispectral images using optimized image registration and deep learning segmentation approach. Comput. Electron. Agric. 2020, 174, 105446. [Google Scholar] [CrossRef]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef]
- Zhang, J.; Xie, T.; Yang, C.; Song, H.; Jiang, Z.; Zhou, G.; Zhang, D.; Feng, H.; Xie, J. Segmenting Purple Rapeseed Leaves in the Field from UAV RGB Imagery Using Deep Learning as an Auxiliary Means for Nitrogen Stress Detection. Remote Sens. 2020, 12, 1403. [Google Scholar] [CrossRef]
- Gan, H.; Lee, W.S.; Alchanatis, V.; Abd-Elrahman, A. Active thermal imaging for immature citrus fruit detection. Biosyst. Eng. 2020, 198, 291–303. [Google Scholar] [CrossRef]
- Bi, C.; Wang, J.; Duan, Y.; Fu, B.; Kang, J.-R.; Shi, Y. MobileNet Based Apple Leaf Diseases Identification. Mob. Netw. Appl. 2020, 1–9. [Google Scholar] [CrossRef]
- Barman, U.; Choudhury, R.D.; Sahu, D.; Barman, G.G. Comparison of convolution neural networks for smartphone image based real time classification of citrus leaf disease. Comput. Electron. Agric. 2020, 177, 105661. [Google Scholar] [CrossRef]
- Hu, J.; Shen, L.; Albanie, S.; Sun, G.; Wu, E. Squeeze-and-Excitation Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2019, 42, 2011–2023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buslaev, A.; Iglovikov, V.I.; Khvedchenya, E.; Parinov, A.; Druzhinin, M.; Kalinin, A.A. Albumentations: Fast and Flexible Image Augmentations. Information 2020, 11, 125. [Google Scholar] [CrossRef] [Green Version]
- Hughes, D.; Salathé, M. An open access repository of images on plant health to enable the development of mobile disease diagnostics. arXiv 2015, arXiv:1511.08060. [Google Scholar]
- Bochkovskiy, A.; Wang, C.Y.; Liao, H.Y.M. YOLOv4: Optimal Speed and Accuracy of Object Detection. arXiv 2020, arXiv:2004.10934. [Google Scholar]
- Jiang, Z.; Zhao, L.; Li, S.; Jia, Y. Real-time object detection method based on improved YOLOv4-tiny. arXiv 2020, arXiv:2011.04244. [Google Scholar]
- Taheri-Garavand, A.; Mumivand, H.; Fanourakis, D.; Fatahi, S.; Taghipour, S. An artificial neural network approach for non-invasive estimation of essential oil content and composition through considering drying processing factors: A case study in Mentha aquatica. Ind. Crop. Prod. 2021, 171, 113985. [Google Scholar] [CrossRef]

| True Class | ||
|---|---|---|
| Predict class | TP True Positive | FP False Positive |
| FN False Negative | TN True Negative | |
| Target Identification Methods | Inserted Modules | mAP |
|---|---|---|
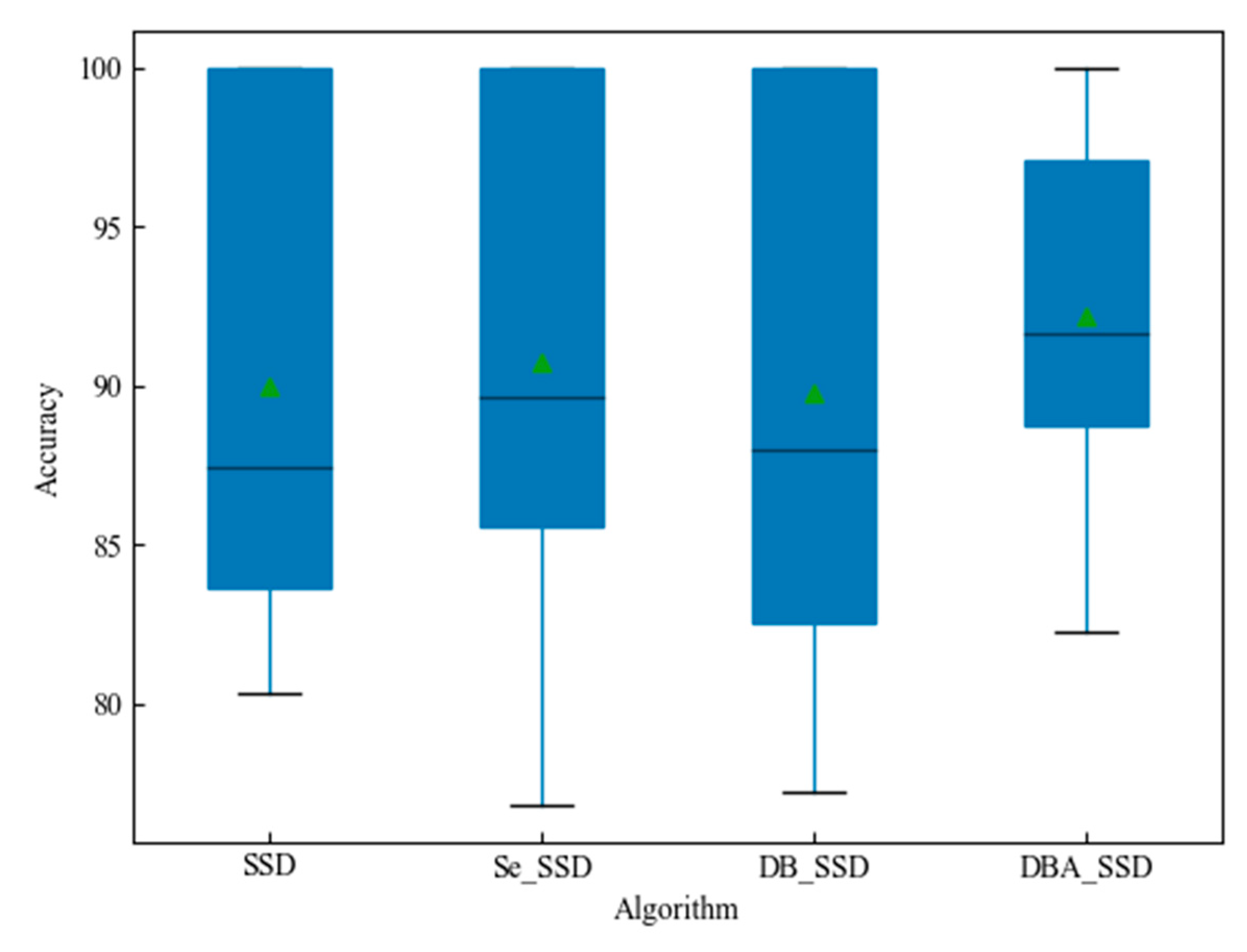
| SSD | \ | 89.96% |
| Se_SSD | Se_Block | 90.77% |
| DB_SSD | Deep_Block | 89.93% |
| DBA_SSD | Deep_Block_Attention | 92.20% |
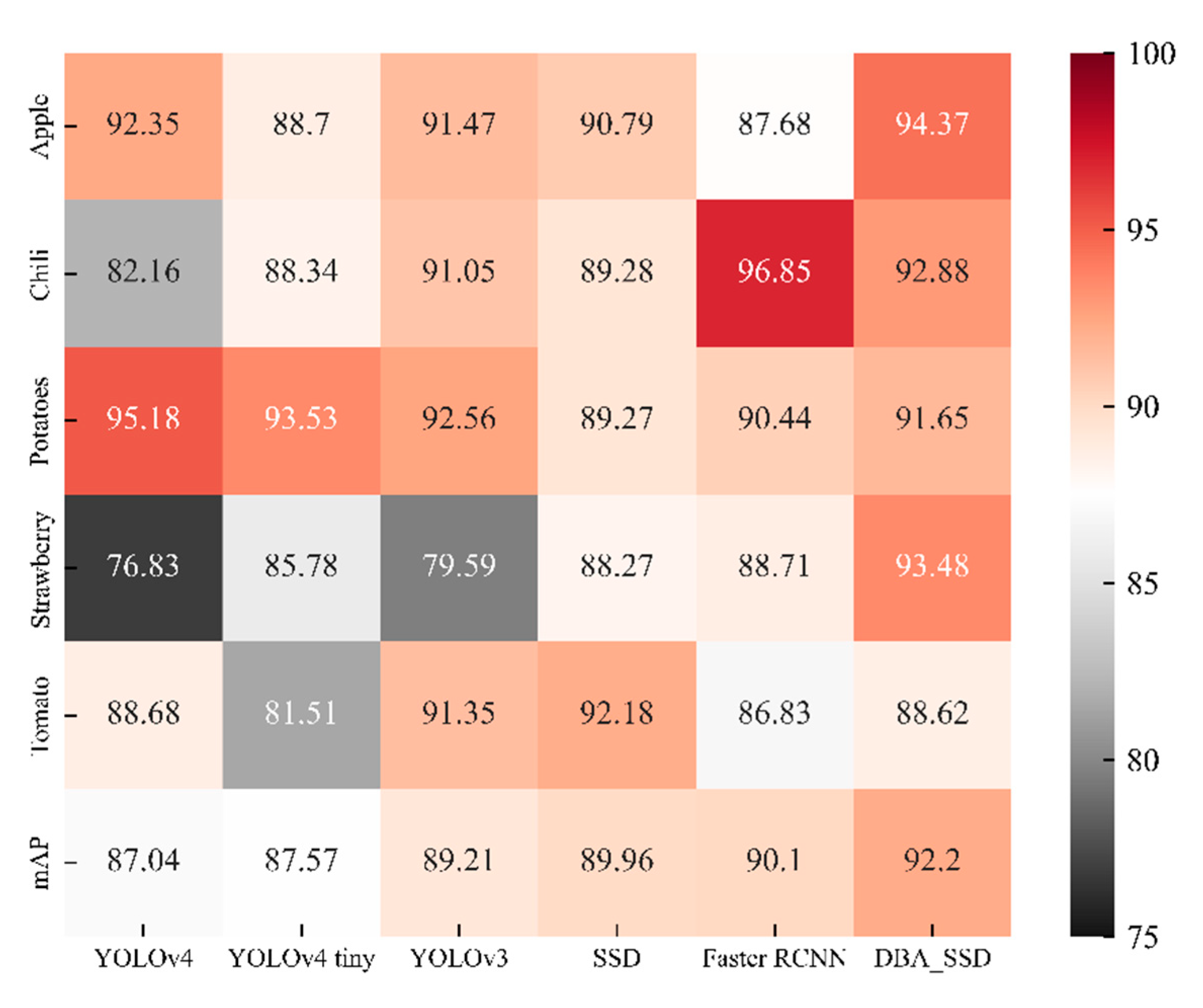
| Category | YOLOv4 | YOLOv4 Tiny | YOLOv3 | SSD | Faster RCNN | DBA_SSD | |
|---|---|---|---|---|---|---|---|
| Algorithm | |||||||
| Apple(general) | 94.79% | 78.32% | 88.87% | 83.45% | 74.85% | 91.83% | |
| Apple(health) | 100.00% | 99.78% | 94.83% | 100.00% | 100.00% | 99.73% | |
| Apple(severe) | 82.27% | 88.01% | 90.71% | 88.93% | 88.20% | 91.56% | |
| Chili(general) | 73.99% | 92.32% | 81.60% | 83.89% | 90.54% | 90.65% | |
| Chili(health) | 98.75% | 100.00% | 100.00% | 100.00% | 100.00% | 99.12% | |
| Chili(severe) | 73.74% | 72.70% | 91.55% | 83.94% | 100.00% | 88.86% | |
| Potatoes(general) | 92.71% | 89.41% | 88.37% | 80.35% | 88.65% | 92.72% | |
| Potatoes(health) | 94.74% | 100.00% | 100.00% | 100.00% | 99.80% | 100.00% | |
| Potatoes(severe) | 98.08% | 91.18% | 89.32% | 87.46% | 82.88% | 82.24% | |
| Strawberry(general) | 63.52% | 80.89% | 59.83% | 80.69% | 73.48% | 85.37% | |
| Strawberry(health) | 99.52% | 100.00% | 100.00% | 100.00% | 100.00% | 100.00% | |
| Strawberry(severe) | 67.46% | 76.44% | 78.95% | 84.11% | 92.64% | 95.07% | |
| Tomato(general) | 85.70% | 85.64% | 95.81% | 82.85% | 85.02% | 85.54% | |
| Tomato(health) | 100.00% | 94.58% | 100.00% | 100.00% | 89.14% | 91.67% | |
| Tomato(severe) | 80.35% | 64.31% | 78.24% | 93.69% | 86.33% | 88.65% | |
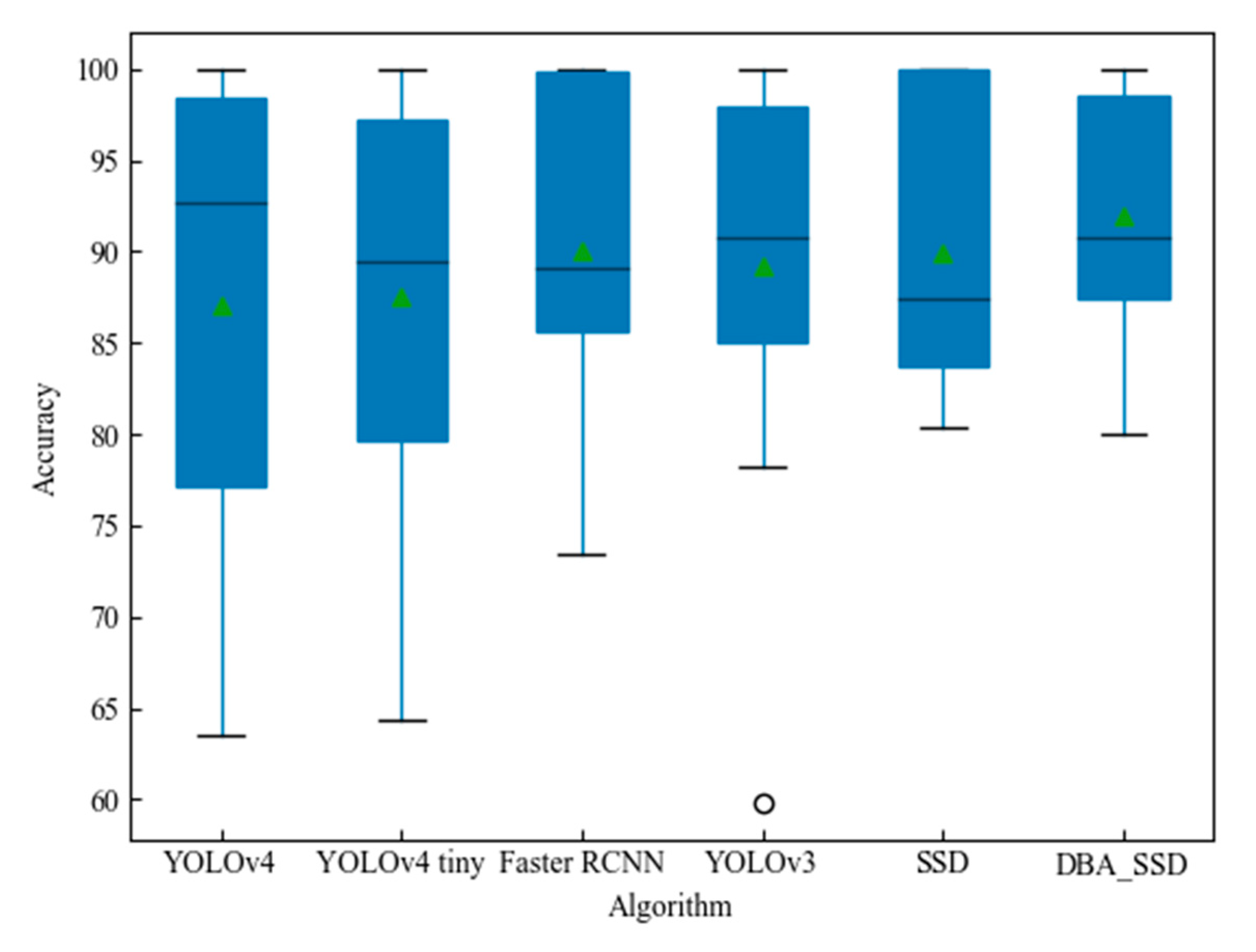
| mAP | 87.04% | 87.57% | 89.21% | 89.96% | 90.10% | 92.20% | |
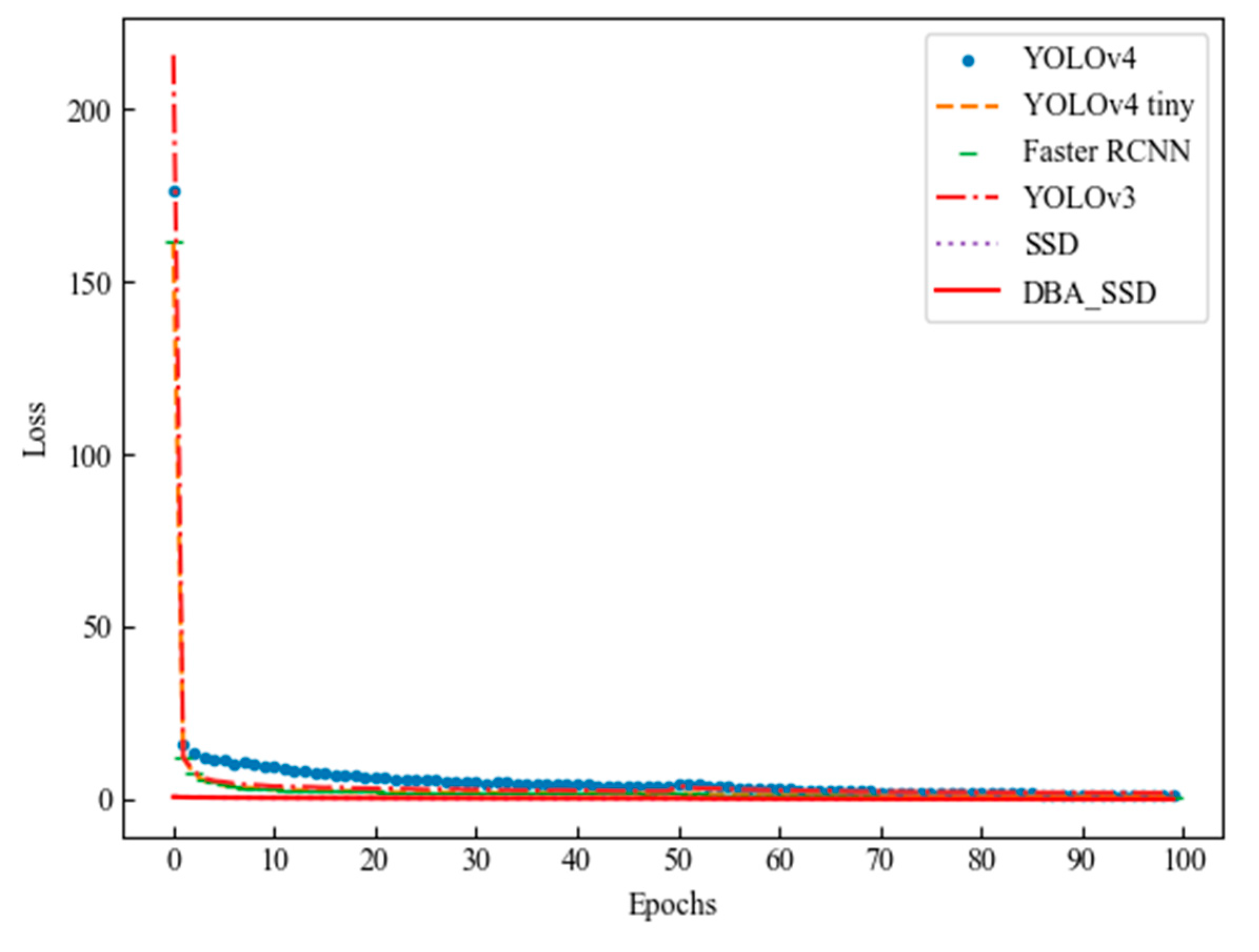
| Algorithm | Backbone Model | Image Size | Parameters | FPS | GFLOPs |
|---|---|---|---|---|---|
| YOLOv4 | CSPDarkNet53 | 512 × 512 | 64.62 M | 62 | 45.96GMac |
| YOLOv4-tiny | CSPDarknet53-tiny | 512 × 512 | 5.91 M | 75 | 5.19 GMac |
| Faster RCNN | VGG16 | 512 × 512 | 136.98 M | 9 | 86.0 GMac |
| YOLOv3 | darknet53 | 512 × 512 | 61.6 M | 34 | 49.7 GMac |
| SSD | VGG16 | 512 × 512 | 25.48 M | 45 | 85.6 GMac |
| SE_SSD | VGG16_SE | 512 × 512 | 25.60M | 43 | 85.62 GMac |
| DB_SSD | VGG_DB | 512 × 512 | 30.55 M | 40 | 86.6 GMac |
| DBA_SSD | VGG_DBA | 512 × 512 | 30.57M | 40 | 86.6GMac |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Yu, L.; Yang, J.; Dong, H. DBA_SSD: A Novel End-to-End Object Detection Algorithm Applied to Plant Disease Detection. Information 2021, 12, 474. https://doi.org/10.3390/info12110474
Wang J, Yu L, Yang J, Dong H. DBA_SSD: A Novel End-to-End Object Detection Algorithm Applied to Plant Disease Detection. Information. 2021; 12(11):474. https://doi.org/10.3390/info12110474
Chicago/Turabian StyleWang, Jun, Liya Yu, Jing Yang, and Hao Dong. 2021. "DBA_SSD: A Novel End-to-End Object Detection Algorithm Applied to Plant Disease Detection" Information 12, no. 11: 474. https://doi.org/10.3390/info12110474
APA StyleWang, J., Yu, L., Yang, J., & Dong, H. (2021). DBA_SSD: A Novel End-to-End Object Detection Algorithm Applied to Plant Disease Detection. Information, 12(11), 474. https://doi.org/10.3390/info12110474







