Abstract
In Information Retrieval (IR), the semantic gap represents the mismatch between users’ queries and how retrieval models answer to these queries. In this paper, we explore how to use external knowledge resources to enhance bag-of-words representations and reduce the effect of the semantic gap between queries and documents. In this regard, we propose several simple but effective knowledge-based query expansion and reduction techniques, and we evaluate them for the medical domain. The query reformulations proposed are used to increase the probability of retrieving relevant documents through the addition to, or the removal from, the original query of highly specific terms. The experimental analyses on different test collections for Precision Medicine IR show the effectiveness of the developed techniques. In particular, a specific subset of query reformulations allow retrieval models to achieve top performing results in all the considered test collections.
1. Introduction
Searching for medical information is an activity of interest for many users with different levels of medical expertise. For example, a patient with a recently diagnosed condition would generally benefit from introductory information about the treatment of the disease, a trained physician would instead expect more detailed information when deciding the course of treatment, and a medical researcher would require references and literature relevant to their future research articles [1]. This diversity of users with different information needs and medical expertise represents a central issue in medical Information Retrieval (IR). In this paper, we are interested in the retrieval of textual medical information such as patient-specific information (i.e., electronic health records) and knowledge-based information (i.e., journal articles) [2].
A critical characteristic of the medical domain, and medical IR, is the prominence of the semantic gap [3,4,5]. The semantic gap represents the mismatch between users’ queries and how IR systems answer to these queries [6,7]. Depending on the situation, the semantic gap can hinder the retrieval of relevant documents, affect the quality of the produced ranking list, or both. For instance, traditional IR models—which compute the relevance score using heuristics defined over the lexical overlap between query and document bag-of-words representations—fall short when the user’s query and a relevant document describe the same concept but with different words, or when the query and an irrelevant document describe different concepts using the same words. Put simply, they fail to represent the semantics of queries and documents. From a user’s perspective, the semantic gap prevents users from easily finding the most relevant documents for their information need. In a medical scenario, the semantic gap leads physicians and medical researchers to read through a large amount of literature before finding the most relevant articles for their needs.
To address this issue, knowledge-enhanced query reformulation techniques have been successfully employed in medical retrieval for years [8,9,10,11]. With the advent of medical-related Text REtrieval Conference (TREC) tracks, such as the Genomics Track [12,13,14,15,16] (https://dmice.ohsu.edu/trec-gen/, accessed on 23 September 2021), the Clinical Decision Support (CDS) Track [17,18,19] (http://www.trec-cds.org/, accessed on 23 September 2021), and the recent TREC Precision Medicine (PM) Track [20,21,22] (http://www.trec-cds.org/, accessed on 23 September 2021), there have been many proposals for new techniques and systems. Among the different approaches, López-García et al. [23] relied on various domain-specific knowledge resources to perform disease and gene expansions. Given a query, the disease and gene fields were expanded using all the synonyms for the identified concepts. Then, different tuning strategies were applied to the expanded queries with the objective of diversifying the importance of the terms coming from different sources. Building on the work by López-García et al. [23], Oleynik et al. [24] developed various hand-crafted rules to mitigate the effect of detrimental information contained within either documents or queries. Along the same lines, Sondhi et al. [25] found out that combining retrieval models with selective query term weighting, based on medical thesauri and physician feedback, proves to be effective in term of retrieval performances. Similar findings were also obtained by Zhu et al. [26] and Diao et al. [27], who relied on query expansion and reweighting techniques to improve retrieval performances on medical records. Therefore, query expansion and rewriting techniques—which enhance queries by leveraging on the information contained within external knowledge resources—are relevant components in the design of effective tools for accessing and retrieving medical information.
In this paper, we present a series of studies and analyses on the TREC PM Track to investigate the effectiveness of simple query expansion and rewriting techniques on IR models. First, we perform a preliminary study [28] on the TREC PM 2018 Clinical Trials Track. Then, we deepen the analysis performed in the preliminary study and we extend it to both scientific literature and clinical trials retrieval [29] on TREC PM 2017 and 2018 Tracks. Given the outcomes of the in-depth analysis, we conduct a validation study on the TREC PM 2019 Track [30]. Based on the results achieved in the previous studies, we perform an a posteriori analysis to understand the effectiveness of the query reformulations developed for clinical trials retrieval over the three years of TREC PM [31]. Thus, this paper contributes to provide a comprehensive and coherent view of all the different studies [28,30] and analyses [29,31] we conducted over the three years of TREC PM. In particular, we set these works into a single framework, and we critically examine the results and findings obtained in a more mature perspective derived from the experience over three years of investigations [32]. Moreover, the proposed techniques are simple enough to be used by non-expert users to perform retrieval, thus helping physicians and medical researchers to perform their work in a more efficient—and effective—way.
The rest of this paper is organized as follows. In Section 2, we report on the two types of resources required to investigate and address the semantic gap in a precision medicine scenario. In Section 3, we present the methodology we used in the different studies and analyses we performed. In Section 4, we describe the preliminary study we conducted on TREC PM 2018. In Section 5, we present the in-depth analysis performed on the scientific literature and clinical trials retrieval tasks of TREC PM 2017 and 2018. In Section 6, we report the validation study we conducted on the tested query reformulations for TREC PM 2019. In Section 7, we present the a posteriori analysis we performed on the query reformulations developed for clinical trials retrieval. Finally, in Section 8, we conclude the paper with a discussion on main achievements and future directions.
2. Resources
In order to investigate the problem of the semantic gap in medical IR and how we can employ authoritative and formal knowledge to reduce it, we require two types of resources: test collections and knowledge resources. Test collections are reusable and standardized resources that can be used to evaluate IR systems with respect to the system. Knowledge resources provide access to authoritative relational information that can be used by IR systems to address the semantic gap. In the following sections, we report on the test collections and knowledge resources used in our work.
2.1. Test Collections
The TREC Precision Medicine (PM) track revolves around a real use case in Clinical Decision Support (CDS), which is to provide physicians with tools capable of retrieving relevant information for patients with cancer related pathologies. From 2017 to 2019, TREC PM presented a unique feature: the provided test collection shared the same set of topics—which were synthetic cases built by precision oncologists—between two documents sets, targeting two different tasks. These two tasks concerned (i) the retrieval of biomedical articles addressing relevant treatments for the given patient and (ii) the retrieval of clinical trials addressing relevant clinical trials for which the patient is eligible [20,21,22]. The idea behind this twofold dataset is the fact that relevant articles can guide physicians to the best-known treatment options for the patient’s condition, but when none of the known treatments works on patients, the physician can check the existence of undergoing treatments in clinical trials.
In the following, we detail the document collections and the topics used in these two tasks.
2.1.1. Scientific Literature
The Scientific Literature document set for TREC PM 2017 (PM17) and 2018 (PM18) is composed by the following parts:
- abstracts from MEDLINE (https://www.nlm.nih.gov/bsd/pmresources.html, accessed on 23 September 2021);
- abstracts from the American Society of Clinical Oncology (ASCO) (https://www.asco.org/, accessed on 23 September 2021),
- abstracts from proceedings of the American Association for Cancer Research (AACR) (https://www.aacr.org/, accessed on 23 September 2021).
The document set for TREC PM 2019 (PM19) consists of an updated set of the MEDLINE abstracts, for a total of documents. Unlike PM17 and PM18, the ASCO and AACR abstracts were not included in the PM19 document set.
2.1.2. Clinical Trials
The Clinical Trials document set for PM17 and PM18 consists of clinical trials, obtained from ClinicalTrials.gov, a resource provided by the U.S. National Library of Medicine (NLM) (https://clinicaltrials.gov/, accessed on 23 September 2021). In this context, precision oncology trials use specific treatments for specific diseases with specific genetic variants. The retrieval of clinical trials is a challenging task for an automated retrieval system given the complexity of the criteria that describe the inclusion or exclusion of a patient in a trial. As with Scientific Literature, the document set for PM19 is updated to clinical trials.
2.1.3. Topics
PM17, PM18, and PM19 contain 30, 50, and 40 synthetic topics compiled by oncologists in 2017, 2018, and 2019, respectively. In TREC PM 2017 [20], the topics are structured in four fields: disease, genetic variants, demographic information, and additional information that may be relevant for the treatment. In TREC PM 2018 [21] and 2019 [22], the organizers provided the topics without the fourth field. Moreover, in 2019, 30 of the 40 topics were created by precision oncologists, while the other 10 topics—unrelated to cancer—were based on the American College of Medical Genetics and Genomics (ACMG) recommendations (https://www.ncbi.nlm.nih.gov/projects/dbvar/clingen/acmg.shtml, accessed on 23 September 2021). These topics were added to assess the relative difficulty of cancer search versus other disciplines requiring precision medicine.
2.2. Knowledge Resources
In this section, we detail the knowledge bases that were used in our experiments.
2.2.1. Systematized Nomenclature of Medicine—Clinical Terms
The Systematized Nomenclature of Medicine - Clinical Terms (SNOMED CT) [33] is a logic-based healthcare terminology, which originated from the Systematized Nomenclature of Pathology (SNOP) (http://www.snomed.org/, accessed on 23 September 2021). It is the most comprehensive, multilingual clinical healthcare terminology worldwide. The main objective of SNOMED CT is to enable users to encode different kinds of health information in a standardized way, thus improving patient care. SNOMED CT presents a multi-hierarchical and multi-axial structure (i.e., concepts can have more than one superordinate concept) and includes three components: concepts, terms, and relations. Concepts are organized from the most general to the most specific through hierarchical, is-a relationships. Then, associative relationships connect concepts whose meaning is related in non-hierarchical ways. These relationships provide formal definitions and properties, such as causative agent, finding site, pathological process, etc. Each concept has a unique concept code (or ID) that identifies the clinical terms used to designate that concept. The terms describing concepts can be divided into fully specified names, preferred terms, and synonyms.
2.2.2. Medical Subject Headings
The Medical Subject Headings (MeSH) thesaurus [34] is a controlled and hierarchically organized vocabulary used for indexing, cataloging, and searching biomedical and health-related information (https://www.ncbi.nlm.nih.gov/mesh/, accessed on 23 September 2021) MeSH contains the subject headings appearing in MEDLINE, the NLM Catalog, and other NLM databases. MeSH is available in several languages and presents a tree structure, from the most general concept to the most specific. The terms describing concepts can be divided into preferred terms and synonyms. MeSH is constantly updated by domain specialists in various areas. Each year, hundreds of new concepts are added, and thousands of modifications are made.
2.2.3. National Cancer Institute Thesaurus
The National Cancer Institute (NCI) thesaurus [35] is the NCI’s reference terminology, covering areas of basic and clinical science and built to facilitate translational research in cancer (https://ncithesaurus.nci.nih.gov/ncitbrowser/, accessed on 23 September 2021). It contains terms, concepts, and relations. The concepts are partitioned in subdomains, which includes, among others, diseases, drugs, genes, anatomy, and biological processes—all with a cancer-centric focus in content. Each concept presents a preferred name and a list of synonyms, along with annotations like textual definitions and (optional) references to external sources. Besides, concepts are defined by their relationships to other concepts.
2.2.4. Unified Medical Language Systems Metathesaurus
The Unified Medical Language System (UMLS) metathesaurus [36] is a large, multipurpose, and multilingual vocabulary database containing information about biomedical and health related concepts, their name variants, and the relationships occurring between them (https://www.nlm.nih.gov/research/umls/knowledge_sources/metathesaurus/, accessed on 23 September 2021). The metathesaurus is composed of different thesauri, classifications, ontologies, code sets, and lists of controlled terms used in patient care, health services, biomedical literature, etc. All the concepts in the metathesaurus have a unique and permanent Concept Unique Identifier (CUI), as well as a preferred name and at least one semantic type from the UMLS Semantic Network (https://semanticnetwork.nlm.nih.gov/, accessed on 23 September 2021). Semantic types provide a consistent categorization of all concepts in the metathesaurus at the high level presented in the Semantic Network. The metathesaurus contains equivalence, hierarchical, and associative relationships between concepts. Most of these relationships derive from individual source vocabularies, but some are introduced by NLM during metathesaurus construction. The UMLS metathesaurus is updated twice per year.
2.2.5. Cancer Biomarkers Database
The Cancer Biomarkers database [37] is an extension of a previous collection of genomic biomarkers of anticancer drug response [38], which contains information on genomic biomarkers of response (sensitivity, resistance, or toxicity) to different drugs across different types of cancer (https://www.cancergenomeinterpreter.org/biomarkers/, accessed on 23 September 2021). Negative results of clinical trials are also included in the database. Biomarkers are organized according to the level of clinical evidence supporting each one, ranging from results of pre-clinical data, case reports, and clinical trials in early and late phases to standard-of-care guidelines. The Cancer Biomarkers database is updated by medical oncologists and cancer genomics experts.
3. Methodology
The proposed methodology is composed of six steps: indexing, pre-retrieval query reformulation, retrieval, post-retrieval query reformulation, filtering, and rank fusion. Depending on the experiment, a subset of these steps is performed and evaluated.
3.1. Indexing
Indexing consists of two phases. First, for both clinical trials and scientific literature collections, we employed MetaMap [39]—a biomedical concept mapper developed by NLM—to extract from each document all the concepts belonging to the following UMLS semantic types: Neoplastic Process (neop); Gene or Genome (gngm); and Amino Acid, Peptide, or Protein (aapp) (https://metamap.nlm.nih.gov/SemanticTypesAndGroups.shtml, accessed on 23 September 2021).
Then, for clinical trials, we indexed documents using the following fields: <docid>, <text>, <max_age>, <min_age>, <gender>, and <concepts>. The <max_age>, <min_age>, and <gender> fields are obtained from the eligibility section of clinical trials and are required to perform filtering. The <text> field stores the whole content of each clinical trial, including the information contained in <docid>, <max_age>, <min_age>, and <gender> fields. The <concepts> field contains the list of UMLS CUIs extracted by MetaMap.
On the other hand, for scientific literature, we indexed documents using <docid>, <text>, and <concepts>. As with clinical trials, the <text> field stores the whole content of each document.
3.2. Pre-Retrieval Query Reformulation
We used two types of query reformulation: query expansion and query reduction.
3.2.1. Query Expansion
In this phase, a knowledge-based query expansion is performed by extracting the UMLS concepts in the queries with MetaMap. These concepts belong to Neoplastic Process (neop), Gene or Genome (gngm); Cell or Molecular Dysfunction (comd); and Amino Acid, Peptide, or Protein (aapp) UMLS semantic types. Specifically, the concepts belonging to gngm, comd, and aapp semantic types are extracted from the <gene> query field, while the concepts belonging to neop are extracted from the <disease> query field. Furthermore, when the <other> query field is present, MetaMap extracts concepts without restrictions on semantic types.
In addition, we retrieved all the name variants of the extracted concepts from the following knowledge resources within UMLS: NCI thesaurus [35], MeSH thesaurus [34], SNOMED CT [33], and UMLS metathesaurus [36]. All the knowledge resources are authoritative and manually curated by professionals. An optional step in this process is to use the term “solid” when a query is not related to blood-related cancers—such as “lymphoma” or “leukemia”—because, as suggested by Goodwin et al. [40], there are several relevant trials for a query that do not mention specific disease but use a generic term like “solid tumor”.
At the end of this process, the expanded query consists in the union of the original query terms with all the name variants associated to the extracted concepts.
3.2.2. Query Reduction
We performed this type of query reformulation to diminish the over-specificity of topics, a situation which may affect the performance of the retrieval when the query is (or contains terms) too specific compared to the documents [24]. For this reason, we removed gene mutations from the <gene> field (whenever they are present). We relied on the Cancer Biomarkers database [37] to identify gene mutations. To exemplify, if a topic contains the value “BRAF (V600E)” in the <gene> field, first, we verified whether “BRAF (V600E)” is in the Cancer Biomarkers database, then, we removed the mutation “(V600E)” from the query. After the reduction process, the <gene> field becomes “BRAF”. In addition, we did not consider the <other> query field, when this field is present, as it contains information that could be a source of noise for retrieving relevant information for patients.
3.3. Retrieval
We used BM25 [41] to perform retrieval. Given a collection D of N documents and a query q expressed as a bag-of-words , where is a word, V is the vocabulary, and is the query length, BM25 computes the score between the query q and a document d as follows:
where represents the Inverse Document Frequency (IDF) weight of the query term , represents the term frequency of within the document d, represents the document length, and b are two hyperparameters (The values for and b will be defined later on as part of the experimental procedures of the following sections.), and represents the average length of documents in the document collection. Given a query term, the formula is
where N is the size of the document collection and is the number of documents containing the query term . The value of the score computed in Equation 1 increases with the frequency of the query term in the document and it is inversely proportional to the frequency of the same term in the collection; has a minimum equal to zero if all the terms in the query are absent in the document.
Furthermore, we weighted the terms added in the expansion step with either a value of or—to limit noise injection in the retrieval process [42]—with a value of . In other words, we assigned a weight to expansion terms so that the impact of such terms when computing the score between the query q and the document d is either equal to the impact of terms in the original query or scaled by a factor of .
3.4. Post-Retrieval Query Reformulation
We performed a Pseudo Relevance Feedback (PRF)-based query expansion. The set of documents retrieved by BM25 using the pre-retrieval reformulated query is used to select expansion terms for the second round of retrieval. Given the top k retrieved documents, we selected the document concepts—identified by MetaMap during the indexing step—that match the concepts associated to the query terms. Then, for each matched concept, we considered the name variants of its neighbor concepts, that is, concepts that present a hierarchical or associative relation within UMLS with the matched concept (https://www.nlm.nih.gov/research/umls/knowledge_sources/metathesaurus/release/abbreviations.html, accessed on 23 September 2021). We limited neighbor concepts to those concepts belonging to the same semantic types used in the pre-retrieval query reformulation step. In this way, we avoided introducing information that is not strictly related to the contents of the query. Finally, the name variants identified with the PRF based query expansion are used to further reformulate the query.
3.5. Filtering
Within clinical trials, the eligibility section contains some detailed information about the patient (such as age or gender) that are necessary for the eligibility of the patient for that trial. In particular:
- minimum age represents the minimum age required for a patient to be considered eligible for the trial. In case of absence of the attribute, we avoided setting a lower threshold on patient’s age.
- maximum age represents the maximum age required for a patient to be considered eligible for the trial. In case of absence of the attribute, we avoided setting an upper threshold on patient’s age.
- gender represents the gender required for a patient to be considered eligible for the trial. In case of absence of the attribute, we kept the trial regardless of patient’s gender.
In our experiments, we performed a filtering step—based on the three demographic aspects described above—to filter out clinical trials for which a patient is not eligible. Given a patient’s case (i.e., a topic), we removed trials from the list of candidate trials when the topic field <demographic>—which contains age and gender information—does not match the eligibility criteria. When part of the demographic data is missing, we kept or discarded a clinical trial according to the remaining demographic information. For example, when a clinical trial does not report a minimum age in its eligibility section, then it is kept or discarded depending on the maximum age and gender values.
3.6. Rank Fusion
We performed a rank fusion step to combine the rankings obtained with different query reformulations. We adopted CombSUM [43] to perform rank fusion and we normalized scores using min-max normalization.
4. Preliminary Study: TREC Precision Medicine 2018
In this section, we describe the preliminary study we performed on the TREC PM 2018 Clinical Trials Track. The study served to identify what features are required to build effective query expansions, and what instead should be avoided. The proposed approach consists of a procedure to (1) expand queries iteratively—relying on medical knowledge resources—to increase the probability of finding relevant trials by adding neoplastic, genetic, and proteic term variants to the original query, and (2) filter out trials, based on demographic data, for which the patient is not eligible.
The objective of the study is twofold: (i) evaluate the effectiveness of a recall-oriented approach based on iterative—and increasingly aggressive—query expansions, and (ii) investigate the correlation between the effectiveness of the retrieval approach and the quality of the relational information stored within the knowledge resource(s) used in the expansion process.
4.1. Experimental Setup
4.1.1. Test Collection and Knowledge Resource
We considered the Clinical Trials collection of the TREC PM 2018 (PM18) Track [21]. We used all the query fields and we performed experiments on the 50 topics provided. As for the knowledge resource, we relied on the 2018AA release of the UMLS metathesaurus [36].
4.1.2. Evaluation Measures
In order to compare our results with the ones provided by the organizers of the TREC PM 2018 Track, we used the following measures: inferred nDCG (infNDCG), R-precision (Rprec), and Precision at rank 10 (P@10).
4.1.3. Experimental Procedure
The implementation of (part of) the methodology described in Section 3 is provided hereby.
Indexing:
- Use MetaMap to select the UMLS concepts belonging to neop and gngm semantic types in the clinical trials and create a new field <concepts>.
- Index the fields <docid>, <text>, <max_age>, <min_age>, <gender>, and <concepts>.
Pre-Retrieval Query Reformulation:
- Use MetaMap to select the UMLS concepts belonging to neop for <disease> and gngm, comd for <gene>.
- Get name variants of selected concepts from all the knowledge sources contained within UMLS and expand the original query with them.
First Round of Retrieval:
- Weight expansion terms with a value .
- Perform retrieval with the pre-retrieval reformulated query using BM25.
Post-Retrieval Query Reformulation:
- Take the top k clinical trials retrieved by BM25 using the pre-retrieval reformulated query.
- Select document concepts that match the concepts associated to query terms.
- Select neighbor concepts—restricted to neop, gngm, and aapp semantic types—that present a hierarchical or associative relation within UMLS with matched concepts.
- Obtain from neighbor concepts all the name variants belonging to the knowledge sources contained within UMLS.
- Expand the pre-retrieval reformulated query with the name variants of neighbor concepts.
Second Round of Retrieval:
- Weight expansion terms with a value .
- Perform retrieval with the post-retrieval reformulated query using BM25.
Filtering
- Filter out candidate clinical trials for which the patient is not eligible.
We considered three different combinations of the above procedure to address the objectives of this study. The first combination (base) performs indexing, retrieval, and filtering—that is, pre- and post-retrieval query expansions are not applied. The second combination (QE) adds the pre-retrieval query expansion to the pipeline, whereas the third one (QE/PRF) includes all the previous steps as well as the post-retrieval query expansion. In this way, we were able to evaluate how iterative—and increasingly aggressive—query expansions affect performance and whether the effectiveness of the retrieval approach can be correlated with the relational information stored within UMLS.
4.1.4. Parameters
We used the Whoosh python library (https://whoosh.readthedocs.io/, accessed on 23 September 2021) as a search engine using a BM25 retrieval model with default settings for the parameters and . We kept the default values as they are within the optimal range suggested by Robertson and Zaragoza [41] and they are equal to the default values of most of the main search engines (e.g., Lucene (https://lucene.apache.org, accessed on 23 September 2021), Terrier (http://terrier.org, accessed on 23 September 2021), ElasticSearch (https://www.elastic.co/elasticsearch/, accessed on 23 September 2021)). The search for optimal or ideal parameters, such as the work of Lipani et al. in [44], in combination with query reformulation approaches is left for future work. Regarding the PRF based query expansion, we set the number of feedback documents .
4.2. Experimental Results
The summary of the results for the TREC PM 2018 Track were provided by TREC in terms of best, median, and worst values for each topic. In Table 1, we present the results of the three considered models—base, QE, and QE/PRF—for P@10, Rprec, and infNDCG averaged across topics, as well as the median values for the Clinical Trials task.

Table 1.
Retrieval performances on the TREC PM 2018 Clinical Trials task. Retrieval performances of the considered models on the TREC PM 2018 Clinical Trials task. The first combination (base) performs indexing, retrieval, and filtering only. The second combination (QE) adds the pre-retrieval query expansion to the pipeline, whereas the third one (QE/PRF) includes all the previous steps as well as the post-retrieval query expansion. Median refers to the average median values of the Clinical Trials task and it is computed considering all the runs submitted to the task. Precision at rank 10 (P@10), inferred nDCG (infNDCG), and R-precision (Rprec) are shown. Bold values refer to the highest scores between models and median.
The results from Table 1 show that BM25 performs best when none of the considered query expansions are used. On average, the base model outperforms median values by a large margin—with an average gap grater than or equal to for all measures. Conversely, the use of both pre- and post-retrieval query expansions significantly worsens performances. QE and QE/PRF models achieve scores lower than the median values for all measures. In particular, QE/PRF shows the lowest performances among the three models considered. This suggests that the developed knowledge-based techniques are sensitive to topic drift—which often occurs when the query is expanded with terms that are not pertinent to the information need [45].
We performed a per-topic analysis to better understand the performances of the proposed models. The analysis compares, for each measure, the three models with the task median values. Figure 1, Figure 2 and Figure 3 display, topic by topic, the difference in performance between each model and the median values. For a positive difference (model better than median), a green bar plot is shown, whereas for a negative difference (model worse than median), a red bar plot is shown.
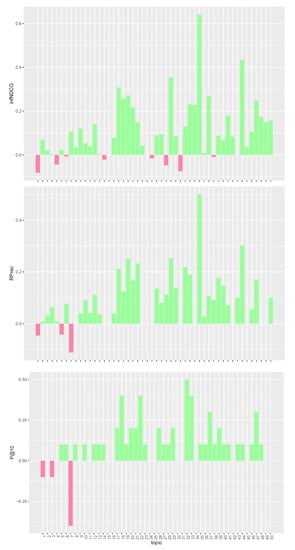
Figure 1.
Per-topic difference between base model and clinical trials median values. A topic is a synthetic case built by oncologists. Inferred nDCG (infNDCG), R-precision (Rprec), and precision at rank 10 (P@10) are shown. For a positive difference (model better than median), a green bar plot is shown, whereas for a negative difference (model worse than median), a red bar plot is shown.
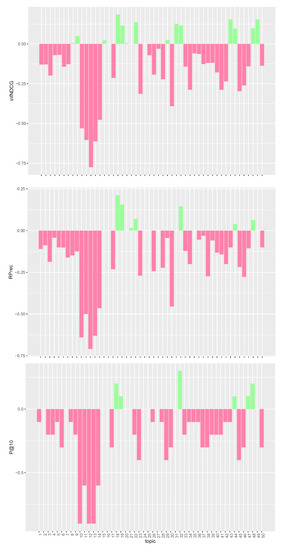
Figure 2.
Per-topic difference between QE model and clinical trials median values. A topic is a synthetic case built by precision oncologists. Inferred nDCG (infNDCG), R-precision (Rprec), and precision at rank 10 (P@10) are shown. For a positive difference (model better than median), a green bar plot is shown, whereas for a negative difference (model worse than median), a red bar plot is shown.
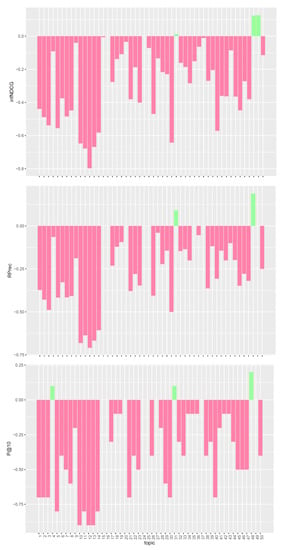
Figure 3.
Per-topic difference between QE/PRF model and clinical trials median values. A topic is a synthetic case built by precision oncologists. Inferred nDCG (infNDCG), R-precision (Rprec), and precision at rank 10 (P@10) are shown. For a positive difference (model better than median), a green bar plot is shown, whereas for a negative difference (model worse than median), a red bar plot is shown.
The analysis of Figure 1, Figure 2 and Figure 3 confirms the trend found for average performances. The base model performs consistently better than task median values for most queries. Conversely, both pre- and post-retrieval expansions significantly worsen the performances for most queries. We attribute this performance drop to two main reasons: (i) we did not apply a weighting scheme on the query terms, and (ii) we employed all the knowledge resources contained within UMLS. Applying a weighting scheme on query terms can reduce the impact of noisy terms when performing retrieval, while selecting a specific subset of knowledge resources can improve the quality of the extracted concepts and terms. For this reason, in Section 5, we performed an in-depth analysis of pre-retrieval query reformulation techniques to understand if the use of weighting schemes and tailored knowledge resources can be beneficial for retrieval effectiveness. Besides, improving the effectiveness of pre-retrieval techniques has a positive effect also on post-retrieval ones, like PRF, as the number of relevant documents retrieved in the first round grows larger.
Comparison with TREC PM 2018 Top Systems
When we look at the detailed analysis in the TREC PM 2018 overview [21], we see that the base model is one of the top 10 performing systems for all the evaluation measures in the Clinical Trials task. Specifically, it is the second best system for P@10 and Rprec, and the third one for infNDCG. Besides, the performance variations of our model across topics are among the smallest ones of all top-performing systems (see “IMS_TERM” run in Figure 3 of TREC PM 2018 overview [21]). This is a promising result, as it highlights the robustness of our baseline model to the set of topics considered. Thus, in the next section, we kept this model as a core component to investigate the effectiveness of pre-retrieval query reformulations relying on weighting schemes and specific knowledge resources.
5. In-Depth Analysis of Query Reformulations
Given the outcomes of the preliminary study presented in Section 4, we performed an in-depth analysis of pre-retrieval query reformulations. Compared to the previous study, we performed the analysis on both tasks of TREC Precision Medicine—that is, Scientific Literature and Clinical Trials—and we considered the test collections from 2017 and 2018 tracks. In this way, we leveraged the dual nature of TREC PM collections, and we evaluated several pre-retrieval query expansion and reduction techniques to investigate whether a particular combination can be helpful in both scientific literature and clinical trials retrieval.
5.1. Experimental Setup
5.1.1. Test Collections and Knowledge Resources
We considered TREC PM 2017 (PM17) [20] and 2018 (PM18) [21] Tracks. We performed experiments on both Scientific Literature and Clinical Trials collections using the 30 and 50 topics provided, respectively, in 2017 and 2018. For query expansion, we adopted the following knowledge resources: NCI thesaurus [35], MeSH thesaurus [34], SNOMED CT [33], and UMLS metathesaurus [36]. For each resource, we considered the version contained within the 2018AA release of UMLS. For query reduction, we relied on the Cancer Biomarkers database [37]. In this case, we adopted the database version of 17 January 2018.
5.1.2. Evaluation Measures
Compared to the official measures presented in Section 4.1, we could not compute the infNDCG for the 2017 Clinical Trials task because the sampled relevance judgments are not available, and we do not report P@5 and P@15 since they were used only for the 2017 Clinical Trials task (subsequently replaced by infNDCG and Rprec in 2018 and 2019).
5.1.3. Experimental Procedure
The procedure representing (part of) the methodology presented in Section 3 is as follows.
Indexing:
- Index clinical trials with the following fields: <docid>, <text>, <max_age>, <min_age>, and <gender>.
- Index scientific literature with the following fields: <docid> and <text>.
Pre-Retrieval Query Reformulation:
- Extract from queries the UMLS concepts belonging to neop for <disease>, gngm/comd for <gene>, and all for <other> using MetaMap.
- Get the name variants of extracted concepts from NCI, MeSH, SNOMED CT, and UMLS metathesaurus knowledge resources.
- Expand topics not mentioning blood-related cancers with the term “solid”.
- Reduce topics by removing, whenever present, gene mutations from the <gene> field.
- Remove the <other> query field whenever present.
Retrieval:
- Adopt any combination of the previous reformulation strategies.
- Weight expansion terms with a value .
- Perform a search using reformulated queries with BM25.
Filtering:
- Filter out candidate clinical trials for which the patient is not eligible.
Regarding pre-retrieval query reformulations, query expansion and reduction techniques can be used alone or in combination with each other.
5.1.4. Parameters
We used the same parameters setting presented in Section 4.1.4.
5.2. Experimental Results
In Table 2, we report the results of our experiments (upper part) and compare them with the top-performing systems at TREC PM 2017 and 2018 (lower part). For each year and task, we present the five top performing query reformulations in terms of P@10. Each line depicts a particular combination (yes or no values) of semantic types (neop, comd, gngm), usage and expansion of <other> field (oth, oth_exp), query reduction (orig), and expansion using weighted “solid” (tumor) keyword. We report the results for both Scientific Literature (sl) and Clinical Trials (ct) tasks. We highlighted in bold the top 3 scores for each measure, and we used the symbols † and ‡ to indicate two combinations that perform well in both years. Regarding TREC PM systems, we selected systems from those participants who submitted runs in both years and reached top 10 performances for at least two measures [20,21]. The results reported in the lower part of Table 2 indicate the best score obtained by a particular system for a specific measure; in general, the best results of a participant’s system are often related to different runs. The symbol ‘−’ means that the measure is not available, while ‘<’ indicates that none of the runs submitted by the participant achieved top 10 performances. For comparison, we added for each measure the lowest score required to enter the top 10 TREC results list, and the score obtained by our best combination. The combination is indicated by the line number, which refers to its position in the upper part of Table 2.

Table 2.
The results of the experiments (upper part) and a comparison with the top-performing systems at TREC PM 2017 and 2018 (lower part). In the upper part, each line depicts a particular combination (yes or no values) of semantic types (neop, comd, gngm), usage and expansion of <other> field (oth, oth_exp), query reduction (orig), and expansion using weighted “solid” (tumor) keyword. We report the results for both Scientific Literature (sl) and Clinical Trials (ct) tasks. We highlighted in bold the top 3 scores for each measure, and we used the symbols † and ‡ to indicate two combinations that perform well in both years. The results reported in the lower part indicate the best score obtained by a particular system for a specific measure. The symbol ‘−’ means that the value for that measure is not available, while ‘<’ indicates that none of the runs submitted by the participant achieved top 10 performances. As a reference point, we added for each measure the lowest score required to enter the top 10 TREC results list, and the score obtained by our best combination.
5.2.1. Analysis of Query Reformulations
The results from Table 2 (upper part) highlight different trends in 2018 and 2017. In 2018, there is a clear distinction in terms of performances among the combinations that achieve the best results for Scientific Literature and Clinical Trials tasks. For Scientific Literature, considering the semantic type neop expansion without using the umbrella term “solid” provides the best performances for all the measures considered. On the other hand, two of the best three runs for Clinical Trials (lines 5 and 9) use no semantic type expansion, but rely on the “solid” (tumor) expansion with weight .
In 2017, the situation is completely different. Lines 12 and 13 show two combinations that achieve top 3 performances for both Scientific Literature and Clinical Trials tasks. These two combinations use query reduction and a weighted 0.1 “solid” (tumor) expansion. The use of a weighted “solid” expansion, as well as a reduced query (orig = n), seems to improve performances consistently for all measures in 2017. The semantic type gngm seems more effective than neop, while comd does not seem to have a positive effect at all.
Thus, the analysis of query reformulations shows that no clear pattern emerges for both tasks. Overall, a query expansion approach using a selected set of semantic types helps the retrieval of scientific literature. On the other hand, a query reduction approach and a “solid” (tumor) expansion improve performances on clinical trials retrieval. Nevertheless, most of the proposed query reformulations perform well for both tasks. Besides, we found that a particular combination (marked as ‡ in Table 2) could have been one of the top 10 performing runs for many evaluation measures in both TREC PM 2017 and 2018.
5.2.2. Comparison with TREC PM Systems
The results in Table 2 (lower part) mark a clear distinction between the performances of participants’ systems for Scientific Literature and Clinical Trials tasks. For Scientific Literature, many of the participants’ runs do not reach the top 10 threshold for the considered measures, especially in 2017. The only exceptions are the systems of the research group from the University of Delaware (see udel_fang in Table 2), whose best runs always achieve top 10 performances for this task. Conversely, most of the participants’ runs achieve top 10 performances for Clinical Trials. In particular, all participants’ runs surpass the top 10 threshold in 2018.
When we consider the top 3 query reformulations from Table 2 (upper part), we see that they achieve performances higher than the top 10 threshold for most measures. The only exception is P@10 in the 2018 Scientific Literature task, where none of the three best query reformulations reaches the top 10 threshold. Compared to the participants’ systems, most of our query reformulations achieve higher performances for most measures in both the 2017 and 2018 Clinical Trials tasks. The only notable exception is P@10 in 2017, where the system of the research group from the University of Texas at Dallas (see UTDHLTRI in Table 2) outperforms our top 5 query reformulations. A different situation occurs for Scientific Literature tasks, where our query reformulations do not achieve best performances for any measure in both 2017 and 2018. Nevertheless, the top 3 query reformulations achieve better results than most of the considered participants’ systems.
Thus, the in-depth analysis, which stemmed from our preliminary study on the TREC PM 2018 Track, shows the effectiveness of applying a weighting scheme on expansion terms and selecting tailored knowledge resources for query expansion and reduction techniques. In particular, the results highlight the robustness of our approach across different collections and tasks. Therefore, in the next section, we conducted a validation study on the TREC PM 2019 Track to investigate whether the findings of this analysis remain valid. In other words, we evaluated how the proposed query reformulations generalize to TREC PM 2019 collections, how they affect retrieval performances, and if the trends found are confirmed.
6. Validation Study: TREC Precision Medicine 2019
Given the outcomes of the in-depth analysis of query reformulations, we conducted a validation study on the TREC PM 2019 Track. We performed experiments on both tasks, with a particular focus on the Clinical Trials task. The objective of the study is twofold: First, we wanted to validate the effectiveness of the top query reformulations found in the previous analysis for the 2019 track. Second, we wanted to verify if combining the rankings obtained using such query reformulations proves effective.
6.1. Experimental Setup
6.1.1. Test Collection and Knowledge Resources
We considered the TREC PM 2019 (PM19) Track [22]. We performed experiments on both Scientific Literature and Clinical Trials collections using the 40 topics provided. For query expansion and reduction, we considered the same knowledge resources used in Section 5.1.1.
6.1.2. Evaluation Measures
We adopted the official measures used in the TREC PM 2019 Track, which are infNDCG, Rprec, and P@10.
6.1.3. Experimental Procedure
We adopted the same procedure presented in Section 5.1.3, plus the rank fusion step that we summarize below.
Rank Fusion:
- Perform rank fusion using CombSUM and min-max normalization over the three most effective query reformulations for Clinical Trials.
- Perform rank fusion using CombSUM and min-max normalization over the three most effective query reformulations for Scientific Literature.
For each task, we considered five different combinations of the above procedure to address the objectives of this study.
Clinical Trials:
- base: refers to the baseline model, that is BM25 plus filtering.
- neop/reduced: refers to neop expansion over reduced queries.
- solid/original: refers to “solid” expansion over original queries.
- solid/reduced: refers to “solid” expansion over reduced queries.
- qrefs/combined: refers to the combination of the above query reformulations using CombSUM.
Scientific Literature:
- base: refers to the baseline model, that is BM25.
- neop/original: refers to neop expansion over original queries.
- neop+comd/original: refers to neop and comd expansions over original queries.
- neop+gngm/original: refers to the neop and gngm expansions over original queries.
- qrefs/combined: refers to the combination of the above query reformulations using CombSUM.
6.1.4. Parameters
We used two different search engine libraries to index, retrieve, and filter the given collections: Whoosh for Clinical Trials and ElasticSearch for Scientific Literature. We moved from Whoosh to ElasticSearch for Scientific Literature because Whoosh could not efficiently handle the increased collection size—which presents over two million documents more than the 2017 and 2018 collections. As for BM25, we kept the same parameters used in Section 4 and Section 5.
6.2. Experimental Results
The organizers of the TREC PM 2019 Track provided the summary of the results in terms of best, median, and worst value for each topic for P@10, infNDCG, and Rprec. In Table 3a,b, we report the results of the five considered models, as well as the median values, for the Clinical Trials and Scientific Literature tasks, respectively.

Table 3.
Retrieval performances on the TREC PM 2019 Clinical Trials task a and Scientific Literature task b. Retrieval performances of the considered models on the TREC PM 2019 Scientific Literature task. Base refers to the baseline model (BM25 plus filtering); neop/reduced refers to neop expansion over reduced queries; solid/original refers to “solid” expansion over original queries; solid/reduced refers to “solid” expansion over reduced queries; qrefs/combined refers to the combination of the above query reformulations using CombSUM; neop/original refers to neop expansion over original queries; neop+comd/original: refers to neop and comd expansions over original queries; neop+gngm/original: refers to the neop and gngm expansions over original queries. Median refers to the average median values of the Scientific Literature task and it is computed considering all the runs submitted to the task. Bold values represent the highest scores among models and median.
The results from Table 3a show that the top query reformulations, identified in the in-depth analysis from Section 5, remain effective for precision-oriented measures in clinical trials retrieval. Indeed, all the knowledge-enhanced models outperform the baseline by a margin greater than 2% for P@10. On the other hand, the improvements are less marked for infNDCG and Rprec—where only the model employing “solid” expansion (solid/original) and the combined model (qrefs/combined) outperform the baseline. We attribute the drop in performance of the other knowledge-enhanced models to the use of reduction techniques. In fact, reducing queries helps to focus more on relevant terms—thus increasing precision—but can hamper recall, as fewer terms are used to perform retrieval. Thus, the results suggest that the developed query reformulations are precision-oriented rather than recall-oriented.
To better understand the performances of the considered query reformulations on the Clinical Trials task, we performed a per-topic analysis that compares, for each measure, the five models with the task median values. Figure 4, Figure 5, Figure 6, Figure 7 and Figure 8 display, topic by topic, the difference in performance between each model and the median values. For a positive difference (model better than median), a green bar plot is shown, whereas for a negative difference (model worse than median), a red bar plot is shown.
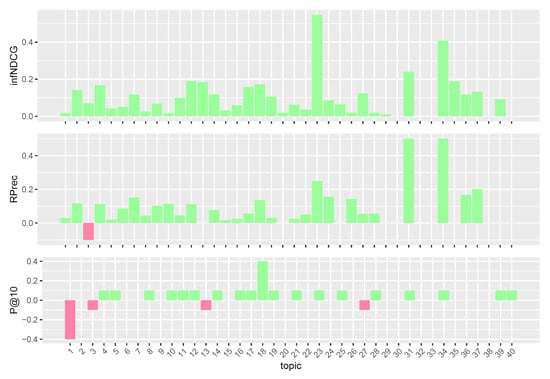
Figure 4.
Per-topic difference between base model and clinical trials median values. A topic is a synthetic case built by precision oncologists. Inferred nDCG (infNDCG), R-precision (Rprec), and precision at rank 10 (P@10) are shown. For a positive difference (model better than median), a green bar plot is shown, whereas for a negative difference (model worse than median), a red bar plot is shown.
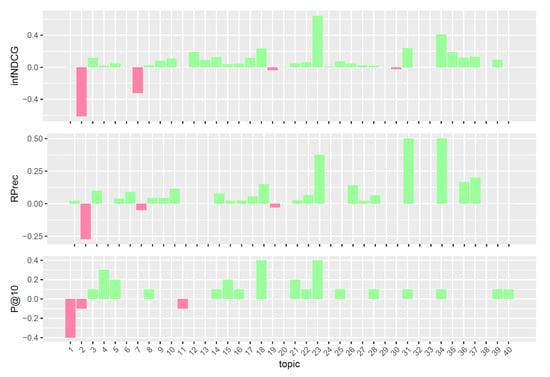
Figure 5.
Per-topic difference between neop/reduced model and clinical trials median values. A topic is a synthetic case built by precision oncologists. Inferred nDCG (infNDCG), R-precision (Rprec), and precision at rank 10 (P@10) are shown. For a positive difference (model better than median), a green bar plot is shown, whereas for a negative difference (model worse than median), a red bar plot is shown.
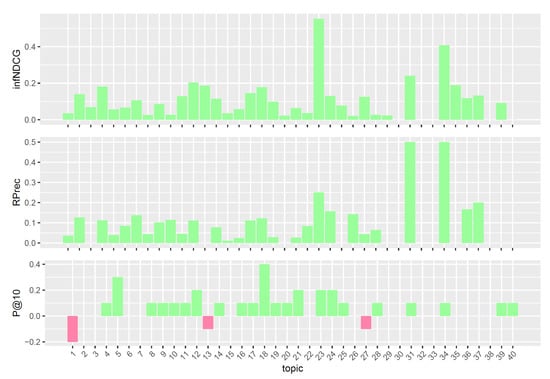
Figure 6.
Per-topic difference between solid/original model and clinical trials median values. A topic is a synthetic case built by precision oncologists. Inferred nDCG (infNDCG), R-precision (Rprec), and precision at rank 10 (P@10) are shown. For a positive difference (model better than median), a green bar plot is shown, whereas for a negative difference (model worse than median), a red bar plot is shown.
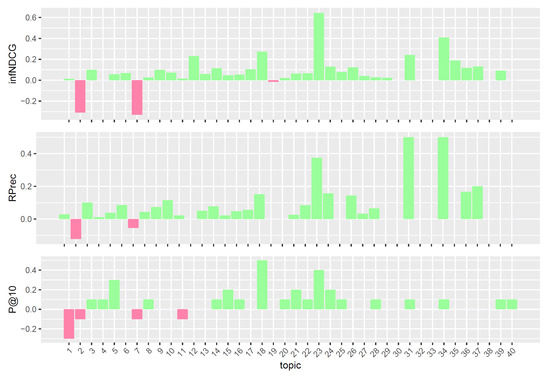
Figure 7.
Per-topic difference between solid/reduced model and clinical trials median values. A topic is a synthetic case built by precision oncologists. Inferred nDCG (infNDCG), R-precision (Rprec), and precision at rank 10 (P@10) are shown. For a positive difference (model better than median), a green bar plot is shown, whereas for a negative difference (model worse than median), a red bar plot is shown.
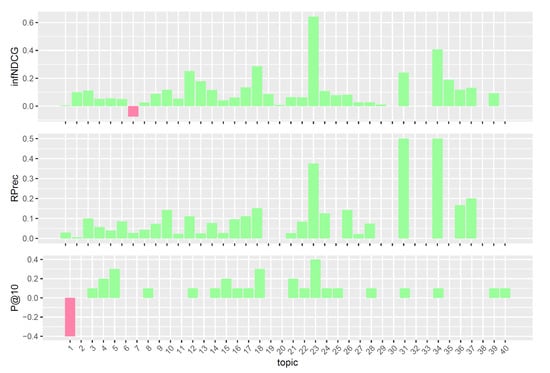
Figure 8.
Per-topic difference between qrefs/combined model and clinical trials median values. A topic is a synthetic case built by precision oncologists. Inferred nDCG (infNDCG), R-precision (Rprec), and precision at rank 10 (P@10) are shown. For a positive difference (model better than median), a green bar plot is shown, whereas for a negative difference (model worse than median), a red bar plot is shown.
The analysis of Figure 4, Figure 5, Figure 6, Figure 7 and Figure 8 highlights an interesting scenario. First, all the considered models achieve performances higher than or equal to median values for most topics. Second, different knowledge-enhanced models achieve top performances on different topics. In other words, there does not exist a query reformulation that provides consistently better results than all the others. This is an interesting outcome, as it shows that the use of different knowledge-based query reformulations improves performances from different angles. However, the results obtained using CombSUM (i.e., qrefs/combined) suggest that more advanced techniques are required to effectively combine the different signals provided by the considered query reformulations. Although effective, CombSUM does not outperform all the individual models.
Regarding scientific literature retrieval, the results from Table 3b highlight a lower impact of the considered query reformulations on retrieval performances. In particular, none of the considered knowledge-enhanced models outperform the baseline for infNDCG. A possible reason for the marginal impact of query reformulations could lie in the shift from Whoosh to ElasticSearch for the indexing, retrieval, and filtering steps. Indeed, Whoosh and ElasticSearch handle the various steps required by our procedure differently, in particular query term weighting operations. However, further analyses are required to confirm this intuition—which are out of scope given the objectives of this paper.
Comparison with TREC PM 2019 Top Systems
When looking at the detailed analysis in the TREC PM 2019 overview [22], we observe that the best performances obtained by our models surpass the top 10 threshold in both tasks for all the evaluation measures but one. For the Clinical Trials task, the model that relies on “solid” expansion (i.e., solid/original) achieves the second best performance for infNDCG and Rprec, and the third best for P@10 (see “BM25solid01o” in Table 6 Clinical Trials of the TREC PM 2019 overview [22]). As for Scientific Literature, the baseline model and the model employing neop and gngm expansions achieve top 10 performances for infNDCG and Rprec, respectively (see “BM25” and “BM25neopgngm” in Table 6 Literature Articles of the TREC PM 2019 overview [22]). On the other hand, all the proposed models achieve performances lower than median values for P@10, which is consistent with the results found in Section 5 for the 2018 Scientific Literature task.
Thus, the outcomes of this study highlight the effectiveness of the proposed query reformulations for retrieving relevant clinical trials in top positions of the ranking list. This is a promising result for at least two reasons. The first reason is that the proposed query reformulations, along with the weighting scheme applied to expansion terms, prove to be consistent across the years. The second reason regards the robustness of our approach. Indeed, the variation of the performance across topics for solid/original is smaller than any other top 10 system of TREC PM 2019 (see “BM25solid01o” in Figure 3 of the TREC PM 2019 overview [22]). Therefore, the developed query reformulations can be used to build knowledge-enhanced models that are robust to topic variations.
In the next section, given the consistent results achieved by the proposed query reformulations for the Clinical Trials task across the three years of TREC PM, we performed an a posteriori analysis focusing on clinical trials retrieval. The analysis provides an overview of the effectiveness of such techniques over the three years of TREC PM and aims to identify a robust subset of query reformulations specifically tailored to clinical trials retrieval.
7. A Posteriori Analysis of Query Reformulations
Based on the results achieved for the Clinical Trials task in Section 5 and Section 6, we performed an a posteriori analysis on the effectiveness of the proposed query reformulations for clinical trials retrieval over the three years of TREC PM. This systematic analysis compares our approach and those proposed by the research groups that participated in all the three years of TREC PM.
7.1. Experimental Setup
7.1.1. Test Collections and Knowledge Resources
We considered TREC PM 2017 (PM17) [20], 2018 (PM18) [21], and 2019 (PM19) [22] Tracks. We performed experiments on Clinical Trials collections using the 30, 50, and 40 topics provided, respectively, in 2017, 2018, and 2019. For query expansion and reduction, we considered the same knowledge resources used in Section 5.1.1 and Section 6.1.1.
7.1.2. Evaluation Measures
We adopted the official measures used in the TREC PM Tracks, which are infNDCG, Rprec, and P@10.
7.1.3. Experimental Procedure
For the 2017 and 2018 tasks, we relied on the top 5 query reformulations, ordered by P@10, found in the in-depth analysis performed in Section 5. Then, we applied the top 3 reformulations found in the 2017 and 2018 tasks to the 2019 task.
7.1.4. Parameters
We adopted Whoosh to perform indexing, retrieval, and filtering, and we set the rest of the parameters as in Section 5.1.4 and Section 6.1.4.
7.2. Experimental Results
In Table 4, we report the results of our experiments on query reformulation (Part A) and compare them with the results obtained by the research groups that participated at TREC PM 2017, 2018, and 2019 (Part B). For 2017 and 2018 tasks, we presented the five query reformulations with the highest P@10. Then, we reported the effectiveness of the considered reformulations for the 2019 task. Each line shows a particular combination (yes or no values) of semantic types (neop, comd, gngm), usage and expansion of <other> field (oth, oth_exp), query reduction (orig), and expansion using the weighted “solid” (tumor) keyword. We used the symbol ‘·’ to indicate that the features oth, oth_exp are not applicable for the years 2018 and 2019 due to the absence of the <other> field in 2018 and 2019 topics. We highlighted in bold the top 3 scores for each measure, and we used the symbol ‡ to indicate a combination that performs well in all three years. For the TREC PM systems, we selected systems from those participants who submitted runs in all three years and reached top 10 performances in at least one edition for each measure [20,21,22]. The results reported in part B of Table 4 indicate the best score obtained by a particular system for a specific measure—again, note that the best results of a participant’s system are often related to different runs. The symbol ‘−’ means that the measure is not available, while ‘<’ indicates that none of the runs submitted by the participant achieved top 10 performances. For the sake of comparison, we added for each measure the lowest score required to enter the top 10 TREC results list, and the score obtained by our best combination. The combination is indicated by the line number, which refers to its position in Part A of Table 4.

Table 4.
Results of our experiments on query reformulation (Part A) and a comparison with the results obtained by the research groups that participated at TREC PM 2017, 2018, and 2019 (Part B). Each line shows a particular combination (yes or no values) of semantic types (neop, comd, gngm), usage and expansion of <other> field (oth, oth_exp), query reduction (orig), and expansion using the weighted “solid” (tumor) keyword. The symbol ‘·’ indicates that the features oth, oth_exp are not applicable (for 2018 and 2019). We highlighted in bold the top 3 scores for each measure, and we used the symbol ‡ to indicate a combination that performs well in all three years. The results reported in part B indicate the best score obtained by a particular system for a specific measure. The symbol ‘−’ means that the measure is not available, while ‘<’ indicates that none of the runs submitted by the participant achieved top 10 performances. For the sake of comparison, we added for each measure the lowest score required to enter the top 10 TREC results list, and the score obtained by our best combination. The combination is indicated by the line number, which refers to its position in Part A of the table.
7.2.1. Analysis of Query Reformulations
The results from Table 4 (Part A) highlight that the use of “solid” expansions, as well as query gene reductions (orig = n), seems to consistently improve performances in 2017—two of the three best combinations in terms of P@10 (lines 1 and 2) apply both techniques. Regarding knowledge-based expansions, the semantic type gngm (lines 1 and 5) seems more effective than neop (line 3), whereas comd does not seem to have any positive effect at all. All five combinations do not consider the <other> field (oth = n) nor its expansion (oth_exp = n)—confirming our intuition that it might represent a potential source of noise in retrieving precise information for patients. Similarly to 2017, two of the three best combinations in 2018 do not use knowledge-based expansions and rely on “solid” (tumor) expansion (lines 7 and 9). In particular, the reformulation combining query gene reductions and “solid” expansion (marked as ‡) provide the best performances for all the measures considered, both in 2017 and 2018. This suggests that removing over-specialized information (i.e., the gene mutations) or adding general terms (e.g., solid) benefits the retrieval. A possible reason is related to the different level of information contained within clinical trials and queries, as clinical trials often contain general requirements to allow patients to enroll—such as, the umbrella concept “solid tumor”—rather than specific concepts, e.g., tumor types, as in queries [40]. The results obtained in 2019 with the top 3 query reformulations from both 2017 and 2018 confirm this trend. The reformulation combining query gene reductions and “solid” expansion (line 13‡) achieves top 3 performances in 2019, however two query reformulations from 2017 (line 14) and 2018 (line 11) provide better performances. This result shows how difficult the task is. Indeed, even though we found a particular query reformulation approach (marked as ‡) to be highly effective in all three years—especially in 2017 and 2018—it was not the best approach for 2019.
Therefore, this analysis helps to identify a robust subset of query reformulations for clinical trials retrieval. The selected query reformulations can be used at the early stages of the IR pipeline to retrieve relevant clinical trials in top positions of the ranking list. In this way, the different signals, that (knowledge-based) query reformulations provide, can be used (and combined) by multi-stage IR systems to obtain a richer pool of relevant documents, thus reducing the semantic gap between queries and documents.
7.2.2. Comparison with TREC PM Systems
The results from Table 4 (Part B) mark a clear division between the 2017 and 2018 tasks and the 2019 task. In 2017 and 2018, most of the participants’ runs do not reach the top 10 threshold in any of the considered measures—the only exception is the research group from Poznan University of Technology, whose best runs always belong to the top 10 performing runs for the task. Conversely, in 2019 all the participants’ best runs achieve results higher than the top 10 threshold. The reason behind the improvement of participants’ runs in 2019 mostly relates to the use of supervised re-ranking models, which exploit relevance judgments from previous years for training. Thus, participants’ approaches consist of expensive supervised multi-stage systems that, unlike ours, require relevance labels to work.
When we consider the results obtained using the query reformulations from Table 4 (Part A), we see that all query reformulations obtain results higher than the top 10 threshold for all the considered measures in all three years. Furthermore, query reformulations consistently achieve better results than participants’ systems for each measure in all three years. Besides, unlike participants’ systems, our approach operates only in the early stages of the IR pipeline and does not require any labeled data to work. This is an indication of the robustness of our approach across the different collections and also of the effectiveness of the proposed query reformulations for clinical trials retrieval. In particular, it is worth mentioning that models using the (‡) query reformulation achieve performances that belong to the top 3 of the best-performing systems in each year of the TREC PM Track [20,21,22].
8. Conclusions and Future Work
In this paper, we have investigated how to use external knowledge resources to enhance bag-of-words representations and reduce the effect of the semantic gap between queries and documents, focusing on an important use-case in Clinical Decision Support (CDS): providing relevant information to clinicians treating cancer patients. To this end, we have developed simple but effective knowledge-based query reformulations that can be integrated within IR pipelines to help physicians and medical researchers performing their work in a more efficient—and effective—way.
The main findings of these studies and analyses are listed below.
- Preliminary study: The study, conducted on the TREC PM 2018 Clinical Trials task, showed that the proposed query expansion approach introduces noise and significantly decreases retrieval performances. In particular, we found that the detrimental effect of the query expansions depends on the lack of an appropriate weighting scheme on query terms and the uncontrolled use of all the knowledge resources contained within UMLS. Thus, the study highlighted what features are required to build effective query expansions, and what instead should be avoided.
- In-depth analysis: The analysis, performed to investigate approaches that can be effective in both scientific literature and clinical trials retrieval, showed that no strong trend emerges for either task. However, we found query reformulations that perform well in both tasks and achieve top results in several evaluation measures both in TREC PM 2017 and in 2018.
- Validation study: The study, carried out to investigate whether the proposed query reformulations also hold in TREC PM 2019, confirmed the effectiveness of the query reformulations in the Clinical Trials task—with promising performances in precision-oriented measures.
- A posteriori analysis: The analysis, based on the results achieved over the three years of the Clinical Trials task, helped to identify a robust subset of query reformulations for clinical trials retrieval. The selected query reformulations can be used at the early stages of the IR pipeline to retrieve relevant clinical trials in top positions of the ranking list.
As future work, we plan to explore the use of advanced rank fusion techniques based on the combination of large amounts of automatically generated, knowledge-based query reformulations. The aim is to build robust multi-stage IR systems, less sensitive to the problem of topic drift, that combine the different signals provided by knowledge-based query reformulations to obtain a richer pool of relevant documents, thus further reducing the semantic gap between queries and documents.
Author Contributions
Conceptualization, S.M., G.M.D.N. and M.A.; methodology, S.M., G.M.D.N. and M.A.; software, S.M.; validation, S.M., G.M.D.N. and M.A.; investigation, S.M., G.M.D.N. and M.A.; resources, S.M.; writing—original draft preparation, S.M.; writing—review and editing, S.M., G.M.D.N. and M.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Goeuriot, L.; Jones, G.J.F.; Kelly, L.; Müller, H.; Zobel, J. Medical Information Retrieval: Introduction to the Special Issue. Inf. Retr. J. 2016, 19, 1–5. [Google Scholar] [CrossRef]
- Hersh, W.R. Information Retrieval: A Health and Biomedical Perspective; Health and Informatics Series; Springer: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Edinger, T.; Cohen, A.M.; Bedrick, S.; Ambert, K.H.; Hersh, W.R. Barriers to Retrieving Patient Information from Electronic Health Record Data: Failure Analysis from the TREC Medical Records Track. In Proceedings of the AMIA 2012, American Medical Informatics Association Annual Symposium, Chicago, IL, USA, 3–7 November 2012; AMIA: Bethesda, MD, USA, 2012. [Google Scholar]
- Koopman, B.; Zuccon, G. Why Assessing Relevance in Medical IR is Demanding. In Proceedings of the Medical Information Retrieval Workshop at SIGIR co-located with the 37th annual international ACM SIGIR conference (ACM SIGIR 2014), Gold Coast, Australia, 11 July 2014; Volume 1276, pp. 16–19. [Google Scholar]
- Koopman, B.; Zuccon, G.; Bruza, P.; Sitbon, L.; Lawley, M. Information retrieval as semantic inference: A Graph Inference model applied to medical search. Inf. Retr. J. 2016, 19, 6–37. [Google Scholar] [CrossRef]
- Furnas, G.W.; Landauer, T.K.; Gomez, L.M.; Dumais, S.T. The Vocabulary Problem in Human-System Communication. Commun. ACM 1987, 30, 964–971. [Google Scholar] [CrossRef]
- Crestani, F. Exploiting the Similarity of Non-Matching Terms at Retrieval Time. Inf. Retr. 2000, 2, 23–43. [Google Scholar] [CrossRef]
- Srinivasan, P. Retrieval Feedback in MEDLINE. J. Am. Med. Inform. Assoc. 1996, 3, 157–167. [Google Scholar] [CrossRef][Green Version]
- Srinivasan, P. Query Expansion and MEDLINE. Inf. Process. Manag. 1996, 32, 431–443. [Google Scholar] [CrossRef]
- Aronson, A.R.; Rindflesch, T.C. Query expansion using the UMLS Metathesaurus. In Proceedings of the American Medical Informatics Association Annual Symposium, AMIA 1997, Nashville, TN, USA, 25–29 October 1997; AMIA: Bethesda, MD, USA, 1997. [Google Scholar]
- Hersh, W.R.; Price, S.; Donohoe, L. Assessing Thesaurus-based Query Expansion Using the UMLS Metathesaurus. In Proceedings of the American Medical Informatics Association Annual Symposium, AMIA 2000, Los Angeles, CA, USA, 4–8 November 2000; AMIA: Bethesda, MD, USA, 2000. [Google Scholar]
- Hersh, W.R.; Bhupatiraju, R.T. TREC GENOMICS Track Overview. In Proceedings of the Twelfth Text REtrieval Conference, TREC 2003, Gaithersburg, MD, USA, 18–21 November 2003; pp. 14–23. [Google Scholar]
- Hersh, W.R.; Bhupatiraju, R.T.; Ross, L.; Cohen, A.M.; Kraemer, D.; Johnson, P. TREC 2004 Genomics Track Overview. In Proceedings of the Thirteenth Text REtrieval Conference, TREC 2004, Gaithersburg, MD, USA, 16–19 November 2004; NIST: Gaithersburg, MD, USA, 2004; Volume 500–261. [Google Scholar]
- Hersh, W.R.; Cohen, A.M.; Yang, J.; Bhupatiraju, R.T.; Roberts, P.M.; Hearst, M.A. TREC 2005 Genomics Track Overview. In Proceedings of the Fourteenth Text REtrieval Conference, TREC 2005, Gaithersburg, MD, USA, 15–18 November 2005; NIST: Gaithersburg, MD, USA, 2005; Volume 500–266. [Google Scholar]
- Hersh, W.R.; Cohen, A.M.; Roberts, P.M.; Rekapalli, H.K. TREC 2006 Genomics Track Overview. In Proceedings of the Fifteenth Text REtrieval Conference, TREC 2006, Gaithersburg, MD, USA, 14–17 November 2006; NIST: Gaithersburg, MD, USA, 2006; Volume 500–272. [Google Scholar]
- Hersh, W.R.; Cohen, A.M.; Ruslen, L.; Roberts, P.M. TREC 2007 Genomics Track Overview. In Proceedings of the Sixteenth Text REtrieval Conference, TREC 2007, Gaithersburg, MD, USA, 5–9 November 2007. [Google Scholar]
- Roberts, K.; Simpson, M.; Demner-Fushman, D.; Voorhees, E.; Hersh, W. State-of-the-art in biomedical literature retrieval for clinical cases: A survey of the TREC 2014 CDS track. Inf. Retr. J. 2016, 19, 113–148. [Google Scholar] [CrossRef]
- Roberts, K.; Simpson, M.S.; Voorhees, E.M.; Hersh, W.R. Overview of the TREC 2015 Clinical Decision Support Track. In Proceedings of the Twenty-Fourth Text REtrieval Conference, TREC 2015, Gaithersburg, MD, USA, 17–20 November 2015; NIST: Gaithersburg, MD, USA, 2015. [Google Scholar]
- Roberts, K.; Demner-Fushman, D.; Voorhees, E.M.; Hersh, W.R. Overview of the TREC 2016 Clinical Decision Support Track. In Proceedings of the Twenty-Fifth Text REtrieval Conference, TREC 2016, Gaithersburg, MD, USA, 15–18 November 2016; NIST: Gaithersburg, MD, USA, 2016. [Google Scholar]
- Roberts, K.; Demner-Fushman, D.; Voorhees, E.M.; Hersh, W.R.; Bedrick, S.; Lazar, A.J.; Pant, S. Overview of the TREC 2017 Precision Medicine Track. In Proceedings of the Twenty-Sixth Text REtrieval Conference, TREC 2017, Gaithersburg, MD, USA, 15–17 November 2017; NIST: Gaithersburg, MD, USA, 2017; Volume 26. [Google Scholar]
- Roberts, K.; Demner-Fushman, D.; Voorhees, E.M.; Hersh, W.R.; Bedrick, S.; Lazar, A.J. Overview of the TREC 2018 Precision Medicine Track. In Proceedings of the Twenty-Seventh Text REtrieval Conference, TREC 2018, Gaithersburg, MD, USA, 14–16 November 2018; NIST: Gaithersburg, MD, USA, 2018; Volume 500–331. [Google Scholar]
- Roberts, K.; Demner-Fushman, D.; Voorhees, E.M.; Hersh, W.R.; Bedrick, S.; Lazar, A.J.; Pant, S.; Meric-Bernstam, F. Overview of the TREC 2019 Precision Medicine Track. In Proceedings of the Twenty-Eighth Text REtrieval Conference, TREC 2019, Gaithersburg, MD, USA, 13–15 November 2019; NIST: Gaithersburg, MD, USA, 2019; Volume 1250. [Google Scholar]
- López-García, P.; Oleynik, M.; Kasác, Z.; Schulz, S. TREC 2017 Precision Medicine - Medical University of Graz. In Proceedings of the Twenty-Sixth Text REtrieval Conference, TREC 2017, Gaithersburg, MD, USA, 15–17 November 2017; NIST: Gaithersburg, MD, USA, 2017; Volume 500–324. [Google Scholar]
- Oleynik, M.; Faessler, E.; Sasso, A.M.; Kappattanavar, A.; Bergner, B.; Cruz, H.F.D.; Sachs, J.P.; Datta, S.; Böttinger, E.P. HPI-DHC at TREC 2018 Precision Medicine Track. In Proceedings of the Twenty-Seventh Text REtrieval Conference, TREC 2018, Gaithersburg, MD, USA, 14–16 November 2018; NIST: Gaithersburg, MD, USA, 2018; Volume 500–331. [Google Scholar]
- Sondhi, P.; Sun, J.; Zhai, C.; Sorrentino, R.; Kohn, M.S. Leveraging Medical Thesauri and Physician Feedback for Improving Medical Literature Retrieval for Case Queries. J. Am. Med. Inform. Assoc. 2012, 19, 851–858. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Wu, S.T.; Carterette, B.; Liu, H. Using Large Clinical Corpora for Query Expansion in Text-Based Cohort Identification. J. Biomed. Inform. 2014, 49, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Diao, L.; Yan, H.; Li, F.; Song, S.; Lei, G.; Wang, F. The Research of Query Expansion Based on Medical Terms Reweighting in Medical Information Retrieval. EURASIP J. Wirel. Comm. Netw. 2018, 2018, 105. [Google Scholar] [CrossRef]
- Agosti, M.; Di Nunzio, G.M.; Marchesin, S. The University of Padua IMS Research Group at TREC 2018 Precision Medicine Track. In Proceedings of the Twenty-Seventh Text REtrieval Conference, TREC 2018, Gaithersburg, MD, USA, 14–16 November 2018; NIST: Gaithersburg, MD, USA, 2018; Volume 500–331. [Google Scholar]
- Agosti, M.; Di Nunzio, G.M.; Marchesin, S. An Analysis of Query Reformulation Techniques for Precision Medicine. In Proceedings of the 42nd International ACM SIGIR Conference on Research and Development in Information Retrieval, SIGIR 2019, Paris, France, 21–25 July 2019; ACM: New York, NY, USA, 2019; pp. 973–976. [Google Scholar]
- Di Nunzio, G.M.; Marchesin, S.; Agosti, M. Exploring how to Combine Query Reformulations for Precision Medicine. In Proceedings of the Twenty-Eighth Text REtrieval Conference, TREC 2019, Gaithersburg, MD, USA, 13–15 November 2019; NIST: Gaithersburg, MD, USA, 2019; Volume 1250. [Google Scholar]
- Agosti, M.; Di Nunzio, G.M.; Marchesin, S. A Post-Analysis of Query Reformulation Methods for Clinical Trials Retrieval. In Proceedings of the 28th Italian Symposium on Advanced Database Systems, Villasimius, Sud Sardegna, Italy (Virtual Due to Covid-19 Pandemic), 21–24 June 2020; Volume 2646, pp. 152–159. [Google Scholar]
- Marchesin, S. Developing Unsupervised Knowledge-Enhanced Models to Reduce the Semantic Gap in Information Retrieval. Ph.D. Thesis, Doctoral School in Information Engineering, Department of Information Engineering, University of Padova, Padova, Italy, 2021. [Google Scholar]
- Donnelly, K. SNOMED-CT: The advanced terminology and coding system for eHealth. Stud. Health Technol. Inform. 2006, 121, 279. [Google Scholar] [PubMed]
- Lipscomb, C.E. Medical Subject Headings (MeSH). Bull. Med. Libr. Assoc. 2000, 88, 265. [Google Scholar] [PubMed]
- Sioutos, N.; de Coronado, S.; Haber, M.W.; Hartel, F.W.; Shaiu, W.L.; Wright, L.W. NCI Thesaurus: A semantic model integrating cancer-related clinical and molecular information. J. Biomed. Inform. 2007, 40, 30–43. [Google Scholar] [CrossRef] [PubMed]
- Bodenreider, O. The Unified Medical Language System (UMLS): Integrating biomedical terminology. Nucleic Acids Res. 2004, 32, D267–D270. [Google Scholar] [CrossRef] [PubMed]
- Tamborero, D.; Rubio-Perez, C.; Deu-Pons, J.; Schroeder, M.P.; Vivancos, A.; Rovira, A.; Tusquets, I.; Albanell, J.; Rodon, J.; Tabernero, J. Cancer Genome Interpreter Annotates the Biological and Clinical Relevance of Tumor Alterations. Genome Med. 2018, 10, 25. [Google Scholar] [CrossRef] [PubMed]
- Dienstmann, R.; Jang, I.S.; Bot, B.; Friend, S.; Guinney, J. Database of Genomic Biomarkers for Cancer Drugs and Clinical Targetability in Solid Tumors. Cancer Discov. 2015, 5, 118–123. [Google Scholar] [CrossRef] [PubMed]
- Aronson, A.R. Effective mapping of biomedical text to the UMLS Metathesaurus: The MetaMap program. In Proceedings of the AMIA Symposium, Whasington, DC, USA, 3–7 November 2001; American Medical Informatics Association: Bethesda, MD, USA, 2001; p. 17. [Google Scholar]
- Goodwin, T.R.; Skinner, M.A.; Harabagiu, S.M. UTD HLTRI at TREC 2017: Precision Medicine Track. In Proceedings of the Twenty-Sixth Text REtrieval Conference, TREC 2017, Gaithersburg, MD, USA, 15–17 November 2017; NIST: Gaithersburg, MD, USA, 2017; Volume 500–324. [Google Scholar]
- Robertson, S.E.; Zaragoza, H. The Probabilistic Relevance Framework: BM25 and Beyond. Found. Trends Inf. Retr. 2009, 3, 333–389. [Google Scholar] [CrossRef]
- Gurulingappa, H.; Toldo, L.; Schepers, C.; Bauer, A.; Megaro, G. Semi-Supervised Information Retrieval System for Clinical Decision Support. In Proceedings of the Twenty-Fifth Text REtrieval Conference, TREC 2016, Gaithersburg, MD, USA, 15–18 November 2016; NIST: Gaithersburg, MD, USA, 2016; Volume 500–321. [Google Scholar]
- Shaw, J.A.; Fox, E.A. Combination of Multiple Searches. In Proceedings of the Third Text REtrieval Conference, TREC 1994, Gaithersburg, MD, USA, 2–4 November 1994; NIST: Gaithersburg, MD, USA, 1994; Volume 500–225, pp. 105–108. [Google Scholar]
- Lipani, A.; Lupu, M.; Hanbury, A.; Aizawa, A. Verboseness Fission for BM25 Document Length Normalization. In Proceedings of the 2015 International Conference on The Theory of Information Retrieval, Northampton, Massachusetts, USA, 27–30 September 2015; ACM: New York, NY, USA, 2015. ICTIR ’15. pp. 385–388. [Google Scholar]
- Vechtomova, O. The Role of Multi-word Units in Interactive Information Retrieval. In Proceedings of the 27th European Conference on IR Research, ECIR 2005, Santiago de Compostela, Spain, 21–23 March 2005; Springer: Berlin/Heidelberg, Germany, 2005; Volume 3408, pp. 403–420. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).