Abstract
Metagenomics studies, as well as genomics studies of polyploid species such as wheat, deal with the analysis of high variation data. Such data contain sequences from similar, but distinct genetic chains. This fact presents an obstacle to analysis and research. In particular, the detection of instrumentation errors during the digitalization of the sequences may be hindered, as they can be indistinguishable from the real biological variation inside the digital data. This can prevent the determination of the correct sequences, while at the same time make variant studies significantly more difficult. This paper details a collection of ML-based models used to distinguish a real variant from an erroneous one. The focus is on using this model directly, but experiments are also done in combination with other predictors that isolate a pool of error candidates.
1. Introduction
Metagenomics studies and studies of polyploid organisms are two areas of research in genomic analysis that are forced to deal with datasets containing high variation among the individual genetic sequences, which negatively affects the data analysis.
Metagenomics studies communities of micro-organisms, like the ones forming the human microbiome [1] or the microbiome of urban environments [2], and are the focus of studies of viral evolution [3]. Understanding of the human microbiome is essential for the future of medicine [4], is a significant part of nutrition studies [5], affects human space flight [6], and the proper study of disease outbreaks [7]; metagenomics studies are also critical for agricultural development [8].
From a computational perspective, however, the presence of genetic sequences from multiple similar organisms in the same sample—a feature of great importance from a scientific standpoint—is a big obstacle to the data analysis. The inevitable presence of machine errors in the data, which can be masquerading as legitimate inter-species variation, cannot always be accounted for. To make matters worse, not even the number of organisms in the samples is always known, and it is not uncommon for these samples to be comprised of altogether unknown organisms, as the majority of micro-organisms in the world are not known. The errors cause inaccuracies in the final study results [9,10] and are often resolved by discarding reads suspected to contain errors [11] in their entirety.
Polyploid organisms, most notably the genome of hexaploid wheat [12], present a similar challenge, albeit a less pronounced one. This is due to the fact that it contains three distinct, yet very similar, subgenomes with three pairs of chromosomes that wheat inherited from three different ancestor organisms [13]. This is less than the number of genomes present in a metagenomics study, but significantly more than the number of genomes present in the study of any typical diploid organism. As a staple food grown more than any other crop in the world [14], the importance of understanding its genome is tremendous, yet the analysis has been slowed down due to its hexaploid nature.
There is a lack of good computational tools to filter errors in this type of dataset, as most such tools target individual monoploid or diploid organisms. Artificial intelligence and in particular machine learning (ML) offer a suitable tool to approach this problem, especially in light of the obscure characteristics of the datasets. Models based on ML, such as artificial neural networks (ANN) [15] and random forests (RF) [16], can be trained to classify examples based on unknown features of the dataset, expressed either in numerical or non-numerical form. This is possible even when the systematic relationship between the input and the result is not yet known, but there are enough training examples on which the models can be trained to recognize this relationship. Random forests are becoming more and more popular in bioinformatics [17], where there is a rapid influx of poorly-studied raw data.
This paper extends the earlier published [18], focusing on creating an ML-based approach, comprised of several identically-trained ML-based models, to classify bases into groups of sequencing errors and correct bases. In addition, it also discusses an earlier approach to discover errors numerically—or, in the case of this paper, error candidates—based on a more complex estimate of the frequency of appearance of individual nucleotide bases across the different genetic sequences and offers this approach as a tool to select candidates for testing with the ML-based model, while also evaluating the ability to use the ML-based models on their own. The details of the ML-based model are extended to include most of the tests performed for the choice of the model, as well as the details on the input construction.
2. Input Data and Problem Statement
The products of next-generation sequencing (NGS) are sets of character strings, based on a four-letter alphabet, which represents the four nucleotide bases of DNA (deoxyribonucleic acid) in the physical DNA chain. In the raw data produced by NGS, these sequences might be randomly-sampled fragments from different sites and loci in the DNA chain. After data preprocessing—which is a computational process, incorporated in the sequencing machines and the subsequent bioinformatics preliminary analysis—the operational datasets are comprised of aligned DNA sequences containing specific sites of genes or regions.
The metagenomics datasets used in the study were clustered using the CD-HIT [19] software package and underwent multiple sequence alignment using the MAFFT [20] software package. The obtained aligned clusters included strings representing the genetic sequences from the same region of the (meta)genome of a variety of micro-organisms, with the meaningful parts of the sequences aligned into the same string positions (columns) by the use of gap characters (“-”). The raw data included 30,000–50,000 character sequences with lengths of 300–500 characters. The working test clusters were comprised of between 1000 and 6000 such sequences; all sequences started from the same position in the micro-organism’s genome.
For the Triticum aestivum L. (hexaploid wheat) datasets, preliminary genome assembly [21] was executed. The target of this bioinformatics processing was to cluster the genetic fragments into sites. This process provides a potential alignment to a representative or consensus sequence of the respective gene. The sequences have shorter lengths, up to 100 nucleotide bases (characters), and are starting at various positions in the gene with varying overlaps. Up to 30–40 sequences cover the same region of the gene and may belong to up to three distinct hexaploid wheat subgenomes that have a similar, but different genetic code.
In both phases of the sequencing and subsequent bioinformatics processing, the character strings representing sequences contained errors. The errors may be substitutions, insertions, or deletions; the sequences may contain letters that differ from the real nucleotide base in the given position, or may have extra or missing letters. The main goal of this paper is to suggest an approach to detect these errors by distinguishing them from the real biological variation, caused by the presence of multiple biological species or subgenomes.
This problem could be considered as a classification one, which separates the signal-variation in the real sequences from the noise-variation introduced in the error by the sequencing devices. Nucleotide bases, treated as regular characters, were classified individually, and erroneous insertions and deletions were dealt with by treating the gap characters (“-”) as a fifth letter in the allowed alphabet, which is evaluated like the remaining four.
3. Using a Machine Learning Model
The presence of the complex unknown structure inside the genetic chains of the studies species suggested the use of machine learning-based models. Since ML-based models like artificial neural networks are a very powerful tool to detect unknown and unstudied relations between the input variables; they make a perfect candidate to attempt classification of the data, especially after the analytical ways to approach the problem did not seem to yield sufficiently good results.
To select the most appropriate ML-based model, during the study, the same input was provided to different models: an artificial neural network, a random forest [16] model, as well as various other decision tree and classification models.
3.1. Machine Learning Input
The learning input was composed based on two presumptions:
- The frequency of a base in a given column is the most significant factor in whether it might be erroneous, as errors would be among the rarest bases in their position/column.
- The frequency of the base among locally-similar sequences is more important than the frequency among locally-dissimilar sequences, making the factor of similarity a good criterion to split the sequences into multiple bins to produce the input values.
For each nucleotide base in each sequence, represented by a single character in the sequence’s string, a learning input example will be constructed. For each such base, the nucleotide sequences from the dataset will be placed in multiple bins depending on how similar they are to the evaluated sequence. A triplet of such bins will be created for a pair of equidistant bases neighboring the evaluated one. These auxiliary pairs of bases will serve as the criteria of local similarity between the sequence with the evaluated base and the remaining sequences. For each such pair, the set of sequences can be split into three groups: the sequences that concur with both bases in both positions, the sequences that differ from the bases in both positions, and the sequences that concur with only one of them.
Around each evaluated character in position i, a window of radius w will be constructed inside the evaluated sequence r, containing the neighboring characters of . Then, for each offset inside the window, , the pair of characters will be selected, and three bins will be constructed: the subset of character sequences p for which , the subset of character sequences p for which , and all the remaining sequences, which would differ in either position or , but not both.
The frequency of base at position i will be recorded for each of these three subsets. This frequency is the count of characters for which , among the reads p in the bin, where p is not r, divided by the total number of the sequences in the bin. Thus, we would define . Here, S is the set of sequences found in the bin.
The computed three frequencies will be added to the input for the ML-based models, which will be comprised of a series of such triplets, creating an input vector of length .
Figure 1 shows an example of a learning input to evaluate the middle base T, using a window w of three bases. For the closest pair of neighboring bases T and G, the entire dataset has been split into three. Among the sequences that contain T and G in both of those positions, confirms the middle base T; among the sequences that contain only one of them in their position, confirms the middle base T; and among the sequences that contain neither, only confirms it. These three values are used as the first three values inside the input vector that will be classified by the ML-based models. This is repeated for the next pair of characters A and T and then for the most distant one: C and A.
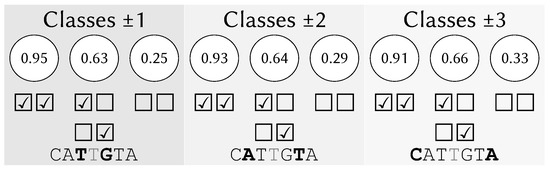
Figure 1.
Learning example input vector for the ML-based models.
Let us assume that there are n sequences of average length m, and we need to compute the input vectors for approximately all m characters inside all n sequences. Without complex, potentially inexact, optimizations, this will have a time complexity of and a space complexity of . The time complexity is reasonable, since would be the time to loop over the learning inputs once.
Because speed was not the main aim of this paper, during the experiments, a slower algorithm with a time complexity of and a space complexity of was used, as it was more straightforward to verify that it constructs the examples exactly as defined here.
3.2. Training with Virtual Errors
The data from metagenomics and bread (6n) wheat sequencing bring two challenges that prevent the use of real errors to construct training examples. On the one hand, the errors subject to evaluation were unknown, and on the other hand, the error frequency was quite low, so the real errors would not provide enough positive training examples [22] for errors. For that reason, instead of training on examples of real errors, the models were trained to recognize an artificially-introduced error at each position.
In other words, the dataset was unchanged, and no errors were simulated at large. Instead, only the individual base used to construct the error example was substituted with an erroneous one and then reverted back for the construction of the remaining examples. This was referred to as a “virtual” error by the research team, as no permanent errors were added to the dataset for the training.
This technique is easier to implement than the simulation of errors. It allows the crafting of a very high number of error examples and ensures that the trained models are not overfitted to the patterns of simulated errors, although they may still be overfitted to other features of the particular genomic dataset.
This training technique produces artificial examples of errors at arbitrary positions of the real genomic data. This should produce a good enough training set for the model to be able to recognize an error at any arbitrary position. However, it may result in a bias in the model, particularly when it comes to the false negatives. The examples of non-errors were constructed out of real data, so the model was trained on adequate examples of non-errors; however, the real errors would not have a uniform distribution inside the sequences like the artificial error training examples would.
We considered this non-critical for three reasons. First, a major part of the non-uniformness is a result of the way the sequencing machine works, making the error positions in the genetic sequence less significant. Second, while we expected it to affect the identification of some of the real errors, it should be limited to the inability of the model to factor the genetic location of those errors to further improve their classification. Third, a test with errors simulated at a low frequency with the same distribution as the real errors was performed to confirm that their identification was unaffected by the bias.
3.3. Imputation
Another problem was faced during the input set construction. There was not always data to construct all values inside the input vectors. There were cases where the bins would be empty, because no other sequence but the evaluated one would contain a given base that was used to construct the bin. In such cases where there would be no other sequences in the bin, the computation of would result in a division by zero.
One way to solve this problem would be to provide the nominator and denominator of independently to the ML-based model, but in this work, the choice was made to instead use imputation and put the average value for other bases inside all the bins S having the same offset and number of matches at the offset. Because the computation of that average for all the sequences is resource-intensive, it was computed only for a random subset of a hundred sequences.
The rationale of the imputation of averages chosen over two separate inputs was that the three groups of bins represent subsets that are very different in nature, and it was considered wrong to default to the same value, two inputs of zero, for two bins that have the opposite meaning: one containing matching sequences and the other containing mismatching sequences. Especially when in at least one of those two bins, a default value would be significantly, yet accidentally different from the values commonly seen by the ML model, it was thus considered safest to provide these common average values in places where the value was a total unknown.
Since the models are empirical in nature and judged by their final performance, shortcomings in this approach would be observed in that performance. On the one hand, this means that there is not a need for a strict verification that the particular choice of imputation is appropriate. On the other hand, it provides one potential place where a significant improvement to the model might be possible by significant adjustment. The potential alternatives include two nominator and denominator inputs; inclusion of the evaluated sequence in the counts so that the denominator is never zero; and a different choice of imputation.
Evaluation of these options was not attempted in the work preceding this paper, but they provide one potential avenue to increase accuracy to the point where an ML-based model can be used on its own.
It should be noted that one of the sources for missing values was the exceptionally high variation found in metagenomics, which provided both highly unusual values and large gaps caused by these mismatches where there would be effectively no sequence data from the remaining sequences at all. This was rarely true for the hexaploid wheat genome samples; hence, imputation was used much less there, which may be one of the reasons for the significantly better results observed for wheat.
A note should be made about the reason for the use of only a subset of the reads to compute the averages for imputation. Should all individual bases be turned into examples to be passed through the learning model for evaluation, there would be no performance penalty in computing all averages, as all bases would be fully evaluated anyway. However, training only uses a random subset of sequences, and the additional filtering with a weighted frequency to select good error candidates presented in Section 4.3 only works with a small number of bases, requiring the full computation of the input values for bases that would never be evaluated by the ML-based model, which can take a significantly long time.
4. Filtering Using Weighted Frequencies
The results in Section 5.6 would suggest that the ML-based approach described thus far provides high enough accuracy for error detection in hexaploid wheat, especially for the purpose of variant detection. Similarly, the results in Section 5.4 would suggest that the use of a random forest model may be good enough even for metagenomics.
However, it was overall observed that the accuracy of an ANN and other models came short when detecting errors in metagenomics data: almost all of the errors were correctly detected, but the low error rate, together with the natural variation, made the models detect errors in multiples of the error rate, which is very unsatisfactory. Should those predictions be used for correction, they would only decrease the overall accuracy of the data. Thus, in metagenomics, the ML-based models (with the possible exception of RF) are only useful as general selection of possible errors, due to low specificity.
To address this issue, we employed an earlier threshold-based error detection described in detail in [23] as a filter to select error candidates to be fed to the ML-based models. The approach is based on computing similarity-weighted frequencies for the bases. This constitutes a measure of how common a base is in a given position, but only among the locally-similar sequences; using the same rationale as for the ML input from Section 3.1.
4.1. Weighted Frequency
Let us say that a frequency of a base in position k of read r, among all bases in column k among the set of the remaining reads , is computed as:
Instead of computing this in a subset with certain characteristics for similarity like in Section 3.1, a measure of the local similarity between reads r and p around position k is introduced and added as a weight in (1):
Once weighted by the similarity, the result would reflect how common the base is among reads similar to r, as opposed to all reads p, which can come from different organisms in metagenomics, or from different subgenomes in hexaploid wheat.
Unlike the bins used to generate the ML-based model input where the sequences were separated into distinct bins, here, the similarity acts as non-binary separation. It is a form of fuzzy clustering, and the frequency of is computed in the fuzzy set of reads locally similar to r.
4.2. Similarity Functions
The localization of the similarity by computing it only around the evaluated base in position k helps with the short partially-overlapping reads in the hexaploid wheat and also makes it easier to distinguish the hypervariable from highly-conserved RNA regions in the metagenomics samples. It is only computed inside a window of radius w around evaluated base , and the effect of the more distant bases is further reduced using a weight factor q.
Therefore, the following similarity function based on the formula of the Hamming distance—a relatively simple and commonly-used measure of distance between DNA sequences—is used. It is modified by the addition of a decay factor q ().
However, the Hamming distance has one disadvantage when trying to give the rationale for its use: the similarity decreases linearly with each additional difference, and even less once the decay factor disregards the more distant bases. On the other hand, the probability that the two sequences are from the same species or subgenome, and that the difference is accidentally caused by an error, decreases exponentially with each each such difference. For that reason, a second, “sharp” similarity measure was also evaluated:
Here, q is some arbitrarily-chosen parameter. A decay is still used as power of q. Without it, the sharp similarity would result in the desired exponential decrease in the similarity. However, to make the measure unbiased in instances of varying overlaps, the decay needs to be kept.
4.3. Application as a Filter for the ML-Based Model
When a threshold cutoff was used on the proposed weighted frequency to predict which bases are errors in [23], it was revealed to be a very poor predictor of errors. The specificity was always very low; and if a lower threshold was used to increase it, the sensitivity suffered, and no threshold was able to give good error predictions, even though a variable threshold computed from the predicted local error rate around each base was used.
However, even with poor specificity, the use of a high threshold against the weighted frequency measure created a significantly reduced pool of potential errors. This justified its use to select a limited pool of bases to test with the ML-based models explicitly, instead of running the ML-based model against all bases present in the data. This improves the specificity of the ML-based results for metagenomics significantly.
5. Results
To verify the chosen design of the ML input examples, as well as the non-standard way to craft training examples, models constructed using them were tested on metagenomics data and, then, through a later experiment, on hexaploid wheat data.
5.1. Artificial Neural Networks on Metagenomics Data
Two artificial neural networks (ANN) were trained using two sets of metagenomics input data using the proposed “virtual” errors. The only preprocessing activities performed on the input data were clustering and multiple sequence alignment. To measure the performance of the neural network, three different approaches were taken: splitting of the training set into a training and a testing set with a 2:1 ratio; constructing a testing set from a subset of the same sequences used to build the training set, but corrected using a rudimentary analytic error detection [23]; and two testing sets constructed on a completely different dataset of sequences: one on uncorrected and one on sequences corrected by the aforementioned rudimentary approach. The results of these experiments are shown in Table 1.

Table 1.
Accuracy of error classification in metagenomics using artificial neural networks.
It is obvious that the applied ANN models were trained in a manner that provided a satisfactory sensitivity, as it correctly classified the examples corresponding to the artificial errors with a very high accuracy. This is even true when the testing set was constructed using a completely different set of biological data (referred to as C1 and C3 in the table). Unfortunately, the specificity—while still high—was not satisfactory. Even with high error rates, the errors were still found in disproportionately fewer numbers than the correct nucleotide bases, starting with an already high signal-to-noise ratio.
For the ANN model to be usable for correction purposes, the specificity needs to be at least higher than the frequency of correct bases. This prompted the earlier work focused on the pre-filtering of classification examples using analytical approaches [24], but also leaves the ANN model applicable for data digitalized using sequencing technologies that have much lower signal-to-noise ratios, such as Oxford Nanopores [25]. In addition, when the same models were trained on wheat datasets, a much higher specificity was observed, justifying the direct use of this model, as is shown in Section 5.6.
These results also show a significant decrease in the specificity once the model is applied to a biological dataset different from the one on which it was trained. Given the proposed method of training, it should be possible to re-train the model for each dataset and overlook that difference. The virtual errors introduced in the dataset were random and statistically independent from the actual errors. The models were trained to recognize those virtual errors in a small sample of bases in the dataset, making the possibility of using such dataset-targeted training every time. This may itself be a potentially valid approach to process future samples, allowing us to take advantage of a specificity that is almost twice as good.
It should be also noted that neither the training nor the testing data were perfect, as they already contained errors before the virtual errors had been introduced. This would lead to some minimal bias during training, but it would also lead to underestimation of the specificity of the models: since there are already errors in the data and since the models would detect those errors, we would incorrectly count them as falsely identified. This would be most significant for the random forest results in Section 5.4 and the wheat results in Section 5.6.
5.2. Varying the ANN Model Parameters
The results given in Table 1 were achieved after an initial selection of the ANN model parameters: 30 input neurons (corresponding to 10 pairs of nucleotide bases neighboring the evaluated one) and 16 hidden neurons. This seems to be a reasonable choice: a radius of 10 bases gives enough information about the local variation to the ANN model, which will allow the neural network to determine which part of the datasets that corroborates the evaluated nucleotide is made of relevant sequences. A number of hidden neurons that is approximately half the number of input neurons is a common initial choice, as well. However, ANN models are first and foremost a heuristic and empirical tool, where the results are dependent on experiments, not on some hard set rules, so some experiments supporting the choice of parameters or exploring other potential values have to be conducted.
In our experiment, we first varied the radius of the window around the evaluated base to determine if 10 pairs of nucleotide bases give enough local context to the ANN model, thus varying the number of input neurons away from the initial 30. Table 2 shows the results when radii of 1–6 (3–18 input neurons) were used. In addition, tests were made with only the outer 2, 4, and 6 pairs of bases inside a 10-base window to test if the closest bases provided the most relevant context.

Table 2.
Change of accuracy with different ANN input neuron counts.
It is evident that increasing the number of input neurons improved the detection accuracy, particularly the sensitivity. The specificity was also affected. With a radius of one base, or only three input neurons corresponding to the two adjacent bases, the accuracy was substantially weakened; the sensitivity did not become reasonable until at least two base-radii, where six input neurons summarized information about the four neighboring nucleotide bases. Beyond six bases, there was saturation of accuracy, as adding more bases did not lead to a dramatic improvement, demonstrating that inputting 18 neurons is enough.
This is only valid when the dataset-targeted would be used, as the result on a different biological dataset was highly inconsistent. One can presume the possibility of overfitting the model towards the biological features in the training set, a problem that seems less pronounced when a dataset-targeted training is to be used.
It should also be noted that using the data for only distant bases led to a significant and consistent decrease in the accuracy, both on a split of the training set and on a different biological dataset. This confirms the expectations that the nearby bases are the most important and also suggests that the nearest four bases may be enough, but are also necessary to make an accurate determination using this ANN model.
Table 3 shows the effect of varying the number of neurons on the hidden layer. For this experiment, the input neurons were fixed to 30, and the hidden neurons were increased and decreased from 16 to see if that would affect the result positively or negatively.

Table 3.
Change of accuracy with different ANN hidden neuron counts.
The variation of the number of neurons shows that 16 can be regarded as an optimal number of hidden neurons. The varying of their number lead to a decrease in accuracy, and in seven out of eight cases, 16 hidden neurons were better than 15 or 17 hidden neurons. We would not consider these empirically-obtained numbers as a definite mark for successful analysis, but in this particular case, it supported the assumption that 16 hidden neurons was enough to postpone modifications of the hidden layer as a means to improve the results.
5.3. Evaluation of Other Potential Changes
In Section 3.3, it was mentioned that imputation presents one area of potential improvement, as the ML-based error evaluation in the presence of missing values no longer stands on input that properly represents the base being evaluated. Another problem that is discussed throughout the paper is the bias resulting from the disproportionate number of correct and incorrect bases. In addition to that, for the evaluated metagenomics samples, the quality varied by position, dropping significantly at the tail of the sequences, affecting both the quality of training data itself and the error rate, together with the aforementioned disproportionality.
Alternatives to the imputation have not been sought in this work, and the disproportional number of errors was only addressed by the introduction of an additional filter to select rare bases for testing through the analytical model discussed in Section 4.
However, an experiment was performed to evaluate how the ANN model would react if it was additionally provided the number of the current position as an extra input. Unfortunately, this could not possibly address the varying disproportionality, which was the biggest effect the current position had on the evaluation, but it was tested in the hopes that it might allow the ANN model to distinguish the highly-noisy examples at the end of the sequences from the pristine examples at the start of the sequences. In particular, examples in a higher position would be subject to imputation, as well as randomly-shifting values caused by adjacent errors. It seemed reasonable that the model should treat those differently.
In addition, an experiment was performed to estimate the effect of shuffling the training examples. While sequences for training were sampled randomly from the genomic dataset, in the original training, the learning examples generated for each bases were passed to the neural network in the order they appeared in the sequence. To make sure this was not affecting the results, the training example generator was modified to provide them in a completely random order, and the change in accuracy was then evaluated.
The result from both experiments—the introduction of a new position input neuron to the ANN and shuffling the bases—is shown in Table 4.

Table 4.
Change of accuracy of error classification with other modifications of the ANN training.
It was oddly observed that while the use of shuffling improved the accuracy on a split of the training dataset, the accuracy surprisingly declined on a different dataset. Both the improvement or decline were too small to be considered significant, but regardless of significance, the use of shuffling did not add any noticeable accuracy, as was originally expected over training in the base order.
Similarly, the introduction of a position input neuron only resulted in insignificant decline in the accuracy, except in a small number of cases where it provided an insignificant improvement. It was thus concluded that adding providing the position to the ML-based models was unnecessary, and the expectation that it might help separate noisy and pristine examples was not supported by experiment.
5.4. Application of Other ML-Based Models
During the development of a threshold-based analytical approach that was presented in [23], it was noted that thresholds may be a good way to separate the signal (correct bases) from the noise (errors). Since decision tree models over numerical data rely on the use of empirically-determined thresholds, this inspired us to test a decision tree algorithm and then subsequently test 20 different ML-based models, focusing on the algorithms that use trees. The models were selected among those available in the Weka [26] AI system using default parameters, and all the tests were also conducted using it.
Table 5 shows the results with all the tree-based models that were selected in the Weka software, as well as other classification models like RIPPER. The table is sorted by specificity on a different biological dataset, in an effort to select the model that was best suited to be used directly to detect the errors, without the need to preselect error candidates, by finding the highest possible specificity.

Table 5.
Accuracy of error classification in metagenomics using various ML-based models.
The observed results confirmed the expectations that decision trees are also a good tool to approach this problem. Half of the decision tree algorithms produced specificity higher than the ANN when used on a different set of biological data.
The random forest model achieved the highest accuracy overall, with a specificity of over 99.9%. This means less than one falsely-selected error for every 1000 bases. This suggests RF is the best model to be used directly to detect errors in the biological data, without the use of further tools to improve the accuracy. On the contrary, from the results in Section 5.5, we would learn that the RF model became an ill-suited choice when it was combined with error candidate pre-selection.
To allow further comparison, when trained on a separate test set, the 60-tree RF model’s F score was and the balanced Matthews correlation coefficient (MCC) , and the ROC Area Under Curve (AUC) reported by Weka exceeded . For a data split of the training set, the F score increased to and the MCC to . Likewise, for the ANN model, the F score on the test set was , the MCC , and the computed AUC . Using the data split of the training set, the F score increased to , and the MCC increased to .
The RF model consistently showed higher F, MCC, and ROC area under curve (AUC) than all the other models in either the test set or a split of the training set, except for RIPPER rule learner. On a separate test set, RIPPER performed with an F score of and MCC of , the highest of all the models. Conversely, the ANN model was outperformed by the majority of the tree models.
The F score, the MCC, and the ROC area under curve all represent measures that take into account both false positives and false negatives, giving an equal importance to both types of erroneous classification. However, because of the rarity of errors, the error specificity plays a much more important role in this particular task, as the cost of a false positive is significantly higher. This is why Table 5 is ordered by the specificity instead.
When choosing the most suitable models for the task of error detection, as well as further evaluation, a couple of factors were considered. Several of the tree models, as well as the RIPPER rule learner showed the highest overall accuracy, suggesting them as the most suitable pick for the task altogether. Between RIPPER and RF, RIPPER had the absolute highest accuracy, but RF with 60 trees had the highest sensitivity, which—to avoid as many as possible costly false positives—was selected with higher priority for further testing. It should additionally be noted that if these two models are to be used directly, without a filter like the one proposed in Section 4.3, they seem to be the most suitable choice.
On the other hand, if the filter from Section 4.3 is applied to further improve the specificity, the combination may alter the accuracy ranking of the models, because of interactions between the filter and the models. Additionally, the accuracies were measured on error examples at arbitrary positions, not on a test set constructed from real errors, nor errors simulated with a realistic statistical profile matching the one of expected real errors. This may alter the ranking of the models in regards to their sensitivity, as real errors may, on average, be more or less likely to be detected than arbitrarily-placed ones.
With that in mind, the ANN model was also selected to be tested with a weighted frequency filter as described in Section 4 and was additionally tested using a probabilistic simulation of errors that matched their expected occurrence in Section 5.5. Artificial neural networks have two attractive qualities that support that choice: they are significantly different from the tree models, and unlike trees or rules, they provide a model with a complex continuous non-linear relationship between the inputs and the prediction. Not only would this allow us to test if the ranking is preserved when a filter is added, but the complex nature of the ANN model may decrease the likelihood of its selection overlapping with the rather trivial selection of examples proposed in Section 4.1, allowing the conjunction of the two classifiers to have higher overall accuracy.
5.5. ML Performance on Examples Selected with Weighted Frequencies
Because the validation procedure necessitated by the ML-based model training and testing thus far, relying on virtual errors presented in Section 3.2, differs significantly from the use of the simulation of realistic errors inside the dataset used in our earlier work in Section [23,24], it was not possible to compare the two directly. It was hence not directly possible to estimate how much the specificity had improved when the filter from Section 4 was used to select the error candidates; and devising methods to indirectly measure them, based on their small overlap, is yet to be performed.
The work so far had focused only on using the ML-based model exclusively with the analytical filter, which was published in [24]. In these experiments, the errors were simulated after measuring their patterns and frequency precisely by sequencing datasets of already known data, and the analytical filter and the ML-based models were used to look for them. With the analytical filter, the use of similarity-based weights in the frequencies led to 18% less error predictions compared to using an approach intended for datasets with no variants, and this, as mentioned, still left us with very low specificity.
However when the ANN model was applied on those predictions, it discarded an additional 51.5% of them as non-errors, on average. At the same time, it was not discarding almost any simulated errors. This demonstrated both that half of the weighted frequency predicted errors were actually correct bases, that the ML-based models were a significantly better filter, and that any bias in the virtual error training of the ML-based models did not affect their ability to identify the vast majority of the errors correctly.
In Table 6, more detailed results from some of the performed experiments are shown, all showing how the ML-based model reduced the pool of prediction to a narrow subset that was much closer to the set of actual errors. It can also be observed that while random forests perform better on their own, when combined in an extra weighted frequency (WF) filter, their accuracy decreased, which has been attributed to a higher overlap between their predictions and the WF predictions.

Table 6.
Accuracy of error classification using frequency filter, weighted frequency (WF) filter, and their combination with artificial neural networks (ANN) and random forests (RF).
The work in [24] also tested additional ML-based models, particularly Hoeffding trees [27,28] and the RIPPER propositional rule learner [29]. Their performance was comparable to random forests: better than ANN when used on their own, but worse then combined with WF.
5.6. Application of the ML-Based Models on Wheat
The wheat genome, consisting of three close, but different subgenomes, also poses a challenge similar to that of metagenomics, but because there are only three subgenomes, the problem is less pronounced as the number of variants is much more limited. Given that the specificity of the proposed model seemed to come short on metagenomics data, except for when random forests were used, we decided to attempt to apply the same model on wheat to test if the limited number of variants leads to substantially better results.
It should be noted that while the training and testing scheme was identical, the experiment ended up being conducted with a different set of ML-based models. The experiment was repeated with artificial neural networks, random forests, and naive Bayes, but three SVM model were added.
Table 7 shows the performance of the selected models on wheat. The models were trained using five pairs of neighboring bases, or 15 input values for the models. The testing was performed using cross-validation within the same gene. A more detailed description of both the study and the results was published in [30].

Table 7.
Accuracy of error classification in wheat using various ML-based models.
In all of the cases, all models except for the naive Bayes classifier achieved sensitivity close to 100% and error specificity over 99.5%. The neural network and the random forests were the only two models that got specificity close to 99.9%, with the RF model still outperforming the ANN model.
One thing that was noted during this experiment was the near-100% sensitivity of several of the models. This suggested that the decreased number of variants had positively impacted the general accuracy of the models. Since in this particular experiment, the number of actual errors was unknown, it more strongly suggested the possibility that a high number of the misclassified non-errors were actually accounted for by unknown errors in the data, as hypothesized in Section 5.1. It was already known that in all of these tests, the specificity was underestimated, but with the near-100% sensitivity and the increased specificity of the neural network, the underestimate may be significant. Should this reduction be measured to be high enough, the direct application of this model on wheat datasets without any additional filtering would be justified.
5.7. Comparison to Other Approaches
The same analytical similarity-based isolation was compared to SHREC [31], which was the only viable error detection tool that seemed to account for potential variation at the time of the testing. It seemed to make 6.5% more incorrect error predictions, without detecting more errors. Once the analytical isolation was improved, SHREC’s incorrect predictions became 13.7% more on average, and with the addition of an ML-based model, the difference increased to 27.1%, with the ML-based model detecting 12.7% more of the simulated errors (since SHREC has no parameters to tune, our models were tuned to be close to—or above—SHREC’s detection rates to compare, so each experiment used different tunings, and the figures are not directly comparable).
6. Conclusions
A group of ML-based models was detailed using the same learning input for detecting errors inside datasets with high variation such as found in metagenomics and polyploid genomes. The models were extensively tested, evaluating their parameters, as well as potential improvements. This includes the combination of the models with an analytical filter of error candidates relying on weighted frequencies.
It was shown that the sensitivity of the models was consistently high, as all models were able to detect all or almost all of the errors artificially introduced in the data. This was true for all the tested models and was independent of the parameter tuning of the models. The main shortcoming of the presented models was the insufficient specificity: although very high in absolute value, it was not high enough for the very low error rate in the data.
Several ways to address the specificity were offered, including the application of the models on datasets with lower overall variations, such as hexaploid wheat, where it was demonstrated to be sufficient; using a preliminary filter to select rare bases as error candidates using weighted frequencies; and using the models with the highest specificity—random forests—and further improving any shortcomings in the example generation, such as the imputation of missing data.
In addition, the presented models would be useful for datasets with very high error rates, such as Oxford Nanopores, where the error rates far exceed the ones in the datasets evaluated in this paper.
Author Contributions
Conceptualization, D.V. and M.N.; methodology, M.K. and D.V.; software, M.K.; validation, M.K., D.V. and M.N.; formal analysis, M.K.; investigation, M.K. and D.V.; resources, D.V. and M.N.; data curation, D.V. and M.K.; writing–original draft preparation, M.K.; writing–review and editing, M.N., D.V. and M.K.; visualization, M.K.; supervision, M.N. and D.V.; project administration, M.N. and D.V.; funding acquisition, M.N. and D.V.
Funding
M.K. and D.V. acknowledge financial support by the Bulgarian NSF within the “GloBIG: A Model of Integration of Cloud Framework for Hybrid Massive Parallelism and its Application for Analysis and Automated Semantic Enhancement of Big Heterogeneous Data Collections” Project, Contract DN02/9/2016. M.N. acknowledges financial support by the European Regional Development Fund and the Operational Program “Science and Education for Smart Growth” under Contract No. BG05M2OP001-1.001-0004-C01 (Project “Universities for Science, Informatics and Technologies in the e-Society (UNITe)”).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Nelson, K.; White, B. Metagenomics and Its Applications to the Study of the Human Microbiome. In Metagenomics: Theory, Methods and Applications; Horizon Scientific Press: Poole, UK, 2010; pp. 171–182. [Google Scholar]
- The MetaSUB International Consortium. The Metagenomics and Metadesign of the Subways and Urban Biomes (MetaSUB) International Consortium inaugural meeting report. Microbiome 2016, 4, 24. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, D.; Mushegian, A.; Dolja, V.; Koonin, E. New dimensions of the virus world discovered through metagenomics. Trends Microbiol. 2010, 18, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Allen-Vercoe, E.; Petrof, E.O. The microbiome: What it means for medicine. Br. J. Gen. Pract. 2014, 64, 118–119. [Google Scholar] [CrossRef] [PubMed]
- Kau, A.L.; Ahern, P.P.; Griffin, N.W.; Goodman, A.L.; Gordon, J.I. Human nutrition, the gut microbiome, and immune system: Envisioning the future. Nature 2011, 474, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Saei, A.A.; Barzegari, A. The microbiome: The forgotten organ of the astronaut’s body–probiotics beyond terrestrial limits. Future Microbiol. 2012, 7, 1037–1046. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, O.E.; Hansen, T.; Knutsson, R.; Löfström, C.; Granberg, F.; Berg, M. Metagenomic Detection Methods in Biopreparedness Outbreak Scenarios. Biosecur. Bioterrorism Biodef. Strategy Pract. Sci. 2013, 11, S146–S157. [Google Scholar] [CrossRef] [PubMed]
- Li, R.W. (Ed.) Metagenomics and Its Applications in Agriculture, Biomedicine and Environmental Studies; Nova Science Pub Inc.: Hauppauge, NY, USA, 2010. [Google Scholar]
- Kunin, V.; Engelbrektson, A.; Ochman, H.; Hugenholtz, P. Wrinkles in the rare biosphere: Pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ. Microbiol. 2010, 12, 118–123. [Google Scholar] [CrossRef] [PubMed]
- Valverde, J.; Mellado, R. Analysis of Metagenomic Data Containing High Biodiversity Levels. PLoS ONE 2013, 8, e58118. [Google Scholar] [CrossRef] [PubMed]
- Huse, S.; Huber, J.; Morrison, H.; Sogin, M.; Welch, D. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 2007, 8, R143. [Google Scholar] [CrossRef] [PubMed]
- Brenchley, R.; Spannagl, M.; Pfeifer, M.; Barker, G.L.; D’Amore, R.; Allen, A.M.; McKenzie, N.; Kramer, M.; Kerhornou, A.; Bolser, D.; et al. Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 2012, 491, 705–710. [Google Scholar] [CrossRef] [PubMed]
- Marcussen, T.; Sandve, S.R.; Heier, L.; Spannagl, M.; Pfeifer, M.; The International Wheat Genome Sequencing Consortium; Jakobsen, K.S.; Wulff, B.B.H.; Steuernagel, B.; Mayer, K.F.X.; et al. Ancient hybridizations among the ancestral genomes of bread wheat. Science 2014, 345, 1250092. [Google Scholar] [CrossRef] [PubMed]
- United Nations, Food and Agriculture Organization, S.D.F. Crops /World Total /Wheat /Area Harvested. 2014. Available online: https://web.archive.org/web/20150906230329/http://faostat.fao.org/site/567/DesktopDefault.aspx?PageID=567 (accessed on 6 September 2015).
- Rojas, R. Neural Networks: A Systematic Introduction; Springer: Berlin, Germany, 1996. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Qi, Y. Random Forest for Bioinformatics. In Ensemble Machine Learning; Zhang, C., Ma, Y., Eds.; Springer: Boston, MA, USA, 2012; pp. 307–323. [Google Scholar]
- Krachunov, M.; Nisheva, M.; Vassilev, D. Machine Learning-Driven Noise Separation in High Variation Genomics Sequencing Datasets. In Proceedings of the Artificial Intelligence: Methodology, Systems, and Applications (AIMSA 2018), Varna, Bulgaria, 12–14 September 2018; Agre, G., van Genabith, J., Declerck, T., Eds.; Springer: Cham, Switzerland, 2018; Volume 11089. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Kuma, K.; Toh, H.; Miyata, T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleid Acid Res. 2005, 33, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.R.; Koren, S.; Sutton, G. Assembly Algorithms for Next-Generation Sequencing Data. Genomics 2010, 95, 315–327. [Google Scholar] [CrossRef] [PubMed]
- Gilles, A.; Meglécz, E.; Pech, N.; Ferreira, S.; Malausa, T.; Martin, J.F. Accuracy and quality assessment of 454 GS-FLX Titanium pyrosequencing. BMC Genom. 2011, 12, 245. [Google Scholar] [CrossRef] [PubMed]
- Krachunov, M.; Vassilev, D. An approach to a metagenomic data processing workflow. J. Comput. Sci. 2014, 5, 357–362. [Google Scholar] [CrossRef]
- Krachunov, M.; Nisheva, M.; Vassilev, D. Machine learning models in error and variant detection high-variation high-throughput sequencing datasets. Procedia Comput. Sci. 2017, 108C, 1145–1154. [Google Scholar] [CrossRef]
- Laver, T.; Harrison, J.W.; O’Neill, P.; Moore, K.; Farbos, A.; Paszkiewicz, K.; Studholme, D.J. Assessing the performance of the Oxford Nanopore Technologies MinION. Biomol. Detect. Quantif. 2015, 3, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Witten, I.H.; Frank, E.; Hal, M.A. Data Mining: Practical Machine Learning Tools and Techniques, 3rd ed.; Morgan Kaufmann Publishers: San Francisco, CA, USA, 2011. [Google Scholar]
- Hulten, G.; Spencer, L.; Domingos, P. Mining time-changing data streams. In Proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 26–29 August 2001; ACM Press: New York, NY, USA, 2001; pp. 97–106. [Google Scholar]
- Hoeffding, W. Probability Inequalities for Sums of Bounded Random Variables. J. Am. Stat. Assoc. 1963, 58, 13–30. [Google Scholar] [CrossRef]
- Cohen, W.W. Fast Effective Rule Induction. In Proceedings of the Twelfth International Conference on Machine Learning, Tahoe City, CA, USA, 9–12 July 1995; Morgan Kaufmann: Burlington, MA, USA, 1995; pp. 115–123. [Google Scholar]
- Kirov, K.; Krachunov, M.; Kulev, O.; Nisheva, M.; Vassilev, D. Reducing false negatives for errors in SNP detection using a machine learning approach. Comptes Rendus de l’Académie Bulgare des Sciences 2016, 69, 155–160. [Google Scholar]
- Schröder, J.; Schröder, H.; Puglisi, S.J.; Sinha, R.; Schmidt, B. SHREC: A short-read error correction method. Bioinformatics 2009, 25, 2157–2163. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).