Bioactive Secondary Metabolites from Marine Streptomyces griseorubens f8: Isolation, Identification and Biological Activity Assay
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of Marine Microorganisms
2.2. Genomic DNA Extraction and Taxonomic Identity of Strain f8
2.3. Morphological Observation of S. griseorubens f8
2.4. Fermentation and Compound Extraction and Isolation
2.5. Structural Analysis of the Compounds
2.6. Antibacterial Activity Assay
2.7. Cytotoxicity Assays
2.8. Cell Invasion and Migration Assays
2.9. Method of Statistical Calculations
3. Results
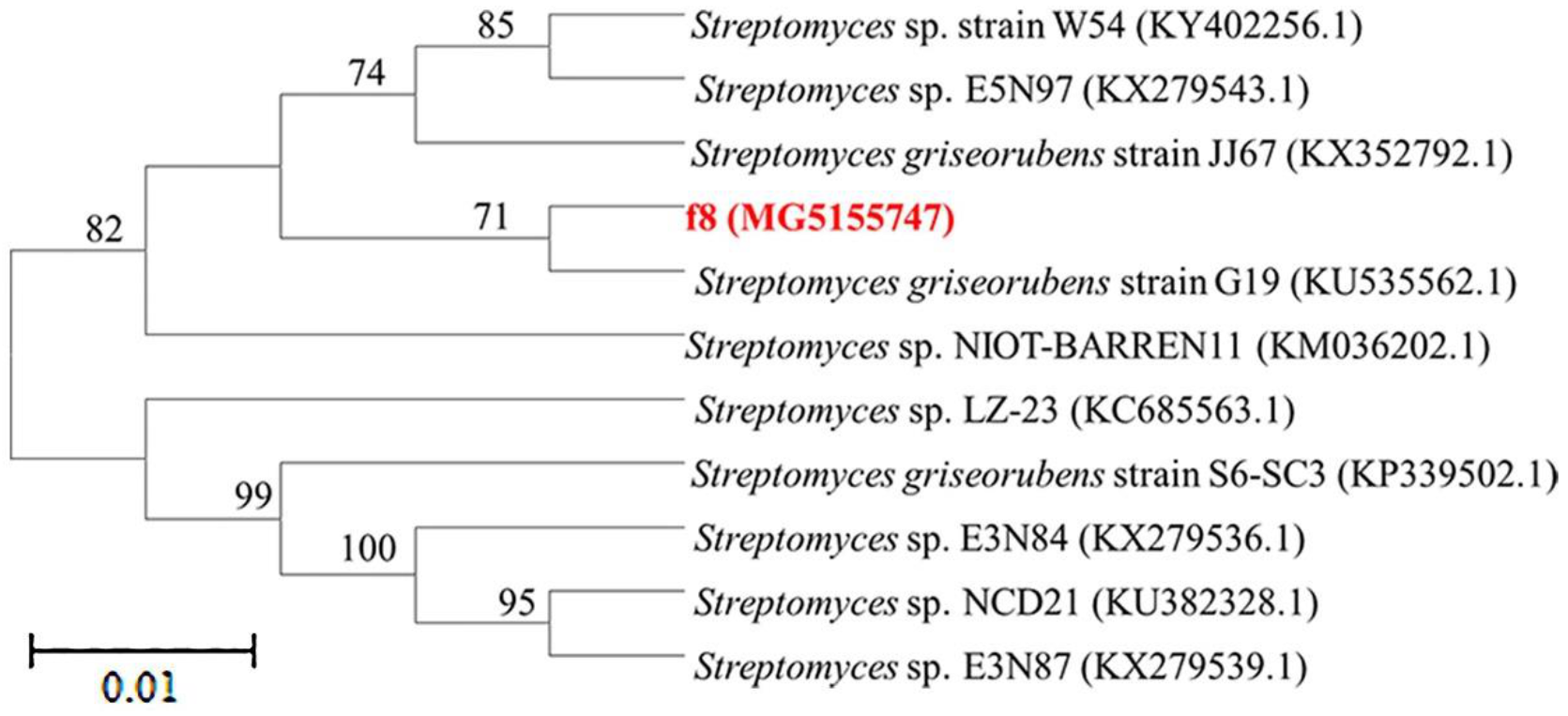
3.1. Isolation and Taxonomy of Strain f8
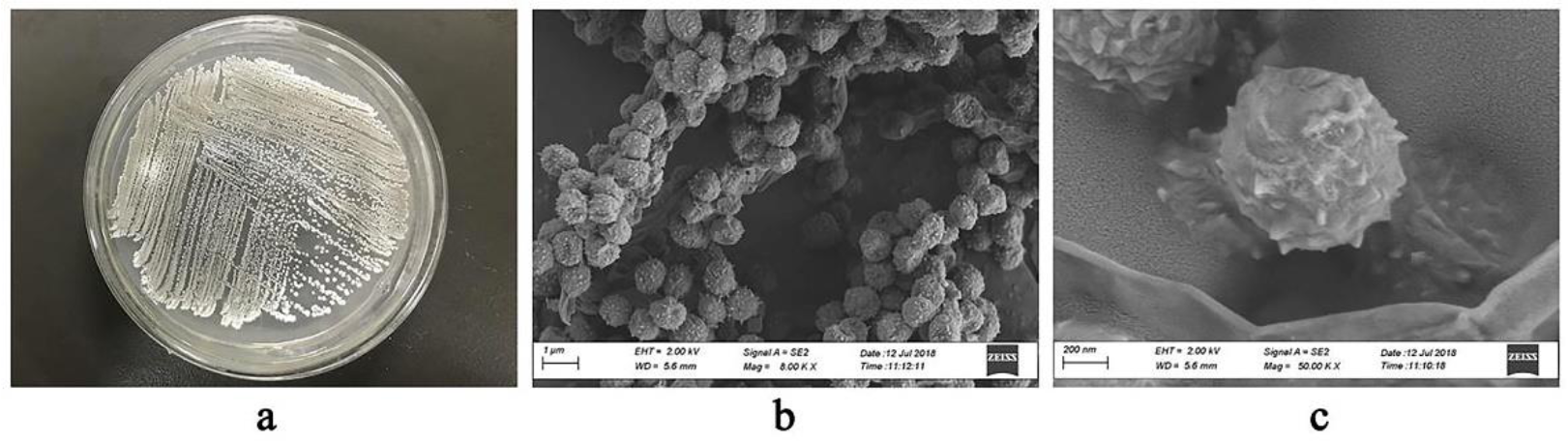
3.2. Morphology of Strain S. griseorubens f8
3.3. Structure Elucidation
3.4. Antimicrobial Activity
3.5. Cytotoxic Activity
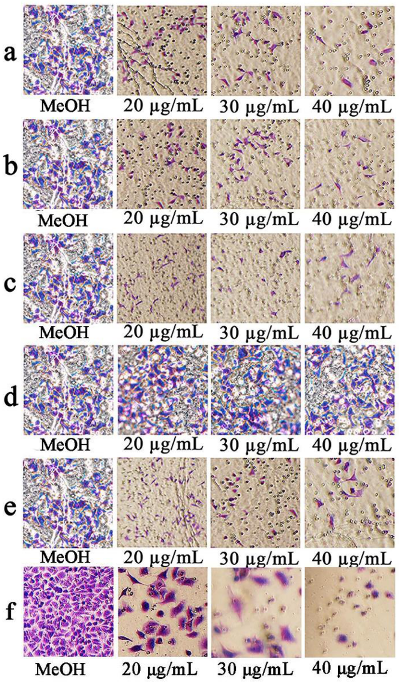
3.6. Effects of the Compounds on Cell Invasion and Migration
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ventura, M.; Canchaya, C.; Tauch, A.; Chandra, G.; Fitzgerald, G.F.; Chater, K.F.; van Sinderen, D. Genomics of actinobacteria: Tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 2007, 71, 495–548. [Google Scholar] [CrossRef]
- Matsuo, H.; Nakanishi, J.; Noguchi, Y.; Kitagawa, K.; Shigemura, K.; Sunazuka, T.; Takahashi, Y.; Omura, S.; Nakashima, T. Nanaomycin K, a new epithelial–mesenchymal transition inhibitor produced by the actinomycete “Streptomyces rosa subsp. notoensis” OS-3966. J. Biosci. Bioeng. 2020, 129, 291–295. [Google Scholar] [CrossRef]
- Jenifer, O.; Khatabi, B.; Nybo, S.E.; Kharel, M.K. Renewed interests in the discovery of bioactive actinomycetes metabolites driven by emerging technologies. J. Appl. Microbiol. 2021. [Google Scholar] [CrossRef]
- Bull, A.T.; Stach, J.E.M. Marine actinobacteria: New opportunities for natural product search and discovery. Trends Microbiol. 2007, 15, 491–499. [Google Scholar] [CrossRef]
- Busarakam, K.; Brown, R.; Bull, A.T.; Tan, G.Y.A.; Zucchi, T.D.; Silva, Z.L.J.; Souza, W.R.; Goodfellow, M. Classification of thermophilic actinobacteria isolated from arid desert soils, including the description of Amycolatopsis deserti sp. nov. Antonie Van Leeuwenhoek 2016, 109, 319–334. [Google Scholar] [CrossRef]
- Lahoum, A.; Verheecke-Vaessen, C.; Bouras, N.; Sabaou, N.; Mathieu, F. Taxonomy of mycelial actinobacteria isolated from Saharan soils and their efficiency to reduce aflatoxin B1 content in a solid-based medium. Ann. Microbiol. 2017, 67, 231–237. [Google Scholar] [CrossRef]
- Azman, A.S.; Othman, I.; Fang, C.M.; Chan, K.G.; Goh, B.H.; Lee, L.H. Antibacterial, anticancer and neuroprotective activities of rare actinobacteria from mangrove forest soils. Indian J. Microbiol. 2017, 57, 177–187. [Google Scholar] [CrossRef]
- Kasarla, S.; Sampath, G.; Govindarajan, R.K.; Ameen, F.; Alwakeel, S.; Gwaiz, H.I.A. Antimicrobial and antifungal activity of soil actinomycetes isolated from coal mine sites. Saudi J. Biol. Sci. 2021, 28, 3553–3558. [Google Scholar]
- Sveta, J.; Manemann, E.M.; Rowe, S.E.; Callender, M.C.; Soto, W. Marine actinomycetes, new sources of biotechnological products. Mar. Drugs 2021, 19, 365. [Google Scholar]
- Ranjani, A.; Gopinath, P.M.; Rajesh, K.; Dhanasekaran, D.; Priyadharsin, P. Diversity of silver nanoparticle synthesizing actinobacteria isolated from marine soil, Tamil Nadu, India. Arab. J. Sci. Eng. 2016, 41, 25–32. [Google Scholar] [CrossRef]
- Sarmiento-Vizcaíno, A.; González, V.; Braña, A.F.; Palacios, J.J.; Otero, L.; Fernández, J.; Molina, A.; Kulik, A.; Vázquez, F.; Acuña, J.L.; et al. Pharmacological potential of phylogenetically diverse actinobacteria isolated from deep-sea coral ecosystems of the submarine avilés canyon in the Cantabrian Sea. Microb. Ecol. 2017, 73, 338–352. [Google Scholar] [CrossRef]
- Al-Dhabi, N.A.; Esmailet, G.A.; Ghilan, A.M.; Arasu, M.V.; Duraipandiyan, V.; Ponmurugan, K. Chemical constituents of Streptomyces sp. strain Al-Dhabi-97 isolated from the marine region of Saudi Arabia with antibacterial and anticancer properties. J. Infect. Public. Health 2020, 13, 235–243. [Google Scholar] [CrossRef]
- Becerril-Espinosa, A.; Freel, K.C.; Jensen, P.R.; Soria-Mercado, I.E. Marine actinobacteria from the Gulf of California: Diversity, abundance and secondary metabolite biosynthetic potential. Antonie Van Leeuwenhoek 2013, 103, 809–819. [Google Scholar] [CrossRef]
- Maldonado, L.A.; Fragoso-Yañez, D.; Pérez-García, A.; Rosellón-Druker, J.; Quintana, E.T. Actinobacterial diversity from marine sediments collected in Mexico. Antonie Van Leeuwenhoek 2009, 95, 111–120. [Google Scholar] [CrossRef]
- Blunt, J.W.; Copp, B.R.; Munro, M.H.; Northcote, P.T.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2006, 23, 26–78. [Google Scholar] [CrossRef]
- Williams, P.G. Panning for chemical gold: Marine bacteria as a source of new therapeutics. Trends Biotechnol. 2009, 27, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.W.; Copp, B.R.; Hu, W.P.; Munro, M.H.; Northcote, P.T.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2009, 26, 170–244. [Google Scholar] [CrossRef] [PubMed]
- Fenical, W.; Sethn, K.M.; Lloyd, G.K. Marine microorganisms as a developing resource for drug discovery. Pharm. News 2002, 9, 489–494. [Google Scholar]
- Goodfellow, M.; Fiedler, H.P. A guide to successful bioprospecting: Informed by actinobacterial systematics. Antonie Van Leeuwenhoek 2010, 98, 119–142. [Google Scholar] [CrossRef]
- Wang, C.; Lu, Y.; Cao, S. Antimicrobial compounds from marine actinomycetes. Arch. Pharm. Res. 2020, 43, 677–704. [Google Scholar] [CrossRef]
- Davies-Bolorunduro, O.F.; Adeleye, I.A.; Akinleye, M.O.; Wang, P.G. Anticancer potential of metabolic compounds from marine actinomycetes isolated from Lagos Lagoon sediment. J. Pharm. Anal. 2019, 9, 201–208. [Google Scholar] [CrossRef]
- Subramanian, D.; Kim, M.S.; Kim, D.H.; Heo, M.S. Isolation, characterization, antioxidant, antimicrobial and cytotoxic effect of marine actinomycete, streptomyces carpaticus MK-01, against fish pathogens. Braz. Arch. Biol. Technol. 2017, 60, 1–9. [Google Scholar] [CrossRef]
- Rahman, H.; Austin, B.; Mitchell, W.J.; Morris, P.C.; Jamieson, D.J.; Adams, D.R.; Spragg, A.M.; Schweizer, M. Novel anti-infective compounds from marine bacteria. Mar. Drugs 2010, 8, 498–518. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Z.Q.; Wang, J.F.; Hao, Y.Y.; Wang, Y. Recent advances in the discovery and development of marine microbial natural products. Mar. Drugs 2013, 11, 700–717. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Yu, Y.; Li, R.; Dong, N.; Zhang, X.H. Phylogenetic diversity and biological activity of actinobacteria isolated from the Chukchi shelf marine sediments in the Arctic Ocean. Mar. Drugs 2014, 12, 1281–1297. [Google Scholar] [CrossRef]
- Edwards, U.; Rogall, T.; Ende, M.; Bottger, E.C. Isolation and direct sequencing of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989, 17, 7843–7853. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Cockerill, F.R.; Wikler, M.A.; Alder, J.; Dudley, M.N.; Eliopoulos, G.M.; Ferraro, M.J. M07–A9: Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically; Approved Standard, 9th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012. [Google Scholar]
- Oo, T.Z.; Cole, N.; Garthwaite, L.; Willcox, M.D.; Zhu, H. Evaluation of synergistic activity of bovine lactoferricin with antibiotics in corneal infection. J. Antimicrob. Chemother. 2010, 65, 1243–1251. [Google Scholar] [CrossRef]
- Mehnaz, S.; Saleem, R.S.Z.; Yamee, B.; Pianet, I.; Schnakenburg, G.; Pietraszkiewicz, H.; Valeriote, F.; Josten, M.; Sahl, H.-G.; Franzblau, S.G.; et al. Lahorenoic acids A−C, ortho-dialkyl-substituted aromatic acids from the biocontrol strain. Pseudomonas aurantiaca PB-St2. J. Nat. Prod. 2013, 76, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Dong, J.; Zhou, X.; Yang, X.; Lee, J.K.; Wang, L.; Zhang, S.; Liu, Y. Proline-containing dipeptides from a marine sponge of a Callyspongia species. Helv. Chim. Acta 2010, 92, 1112–1117. [Google Scholar] [CrossRef]
- Zhao, P.J.; Wang, H.X.; Li, G.H.; Li, H.D.; Liu, J.; Shen, Y.M. Secondary metabolites from endophytic Streptomyces sp. Lz531. Chem. Biodivers. 2010, 4, 899–904. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Fu, S.; Sun, X.; Li, P.; Wu, J.; Dong, T.; He, F.; Deng, Y. Identification of cyclic dipeptides from Escherichia coli as new antimicrobial agents against Ralstonia Solanacearum. Molecules 2018, 23, 214. [Google Scholar] [CrossRef]
- Dharmaraj, S. Marine Streptomyces as a novel source of bioactive substances. World J. Microbiol. Biotechnol. 2010, 26, 2123–2139. [Google Scholar] [CrossRef]
- Miller, E.D.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Piperazimycins cytotoxic hexadepsipeptides from a marine derived bacterium of the genus Streptomyces. J. Org. Chem. 2007, 72, 323–330. [Google Scholar] [CrossRef] [PubMed]
- Moore, B.S.; Trischman, J.A.; Seng, D.; Kho, D.; Jensen, P.R.; Fenical, W. Salinamides, anti-inflammatory depsipeptides from a marine Streptomyces. J. Org. Chem. 1999, 64, 1145–1150. [Google Scholar] [CrossRef]
- Feng, H.; Sun, Y.; Zhi, Y.; Wei, X.; Luo, Y.; Mao, L.; Zhou, P. Identification and characterization of the nitrate assimilation genes in the isolate of Streptomyces griseorubens JSD-1. Microb. Cell. Fact. 2014, 13, 174. [Google Scholar] [CrossRef]
- Chagas, F.O.; Caraballo-Rodríguez, A.M.; Dorrestein, P.C.; Pupo, M.T. Expanding the chemical repertoire of the endophyte Streptomyces albospinus RLe7 reveals amphotericin B as an inducer of a fungal phenotype. J. Nat. Prod. 2017, 80, 1302–1309. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, M.P.; Abraham, W.R. Antimicrobial and biofilm inhibiting diketopiperazines. Curr. Med. Chem. 2012, 19, 3564–3577. [Google Scholar] [CrossRef]
- Shim, S.H. Diketopiperazines from Cultures of Rhodococcus rhodochrous. Chem. Nat. Compd. 2016, 52, 1157–1159. [Google Scholar] [CrossRef]
- Ding, L.; Yuan, W.; Peng, Q.; Sun, H.; Xu, S. Secondary metabolites isolated from the sponge-associated fungus Nigrospora oryzae. Chem. Nat. Compd. 2016, 52, 1–2. [Google Scholar] [CrossRef]





| Indicator | Compound 3 | Compound 5 | AMP |
|---|---|---|---|
| Staphylococcus aureus | 160 µg/mL | 180 µg/mL | 30 µg/mL |
| Klebsiella aerogenes | 100 µg/mL | 130 µg/mL | 20 µg/mL |
| Proteus vulgaris | 120 µg/mL | 150 µg/mL | 20 µg/mL |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, W.; Liang, G.; Sun, Y.; Gong, Z. Bioactive Secondary Metabolites from Marine Streptomyces griseorubens f8: Isolation, Identification and Biological Activity Assay. J. Mar. Sci. Eng. 2021, 9, 978. https://doi.org/10.3390/jmse9090978
Yang W, Liang G, Sun Y, Gong Z. Bioactive Secondary Metabolites from Marine Streptomyces griseorubens f8: Isolation, Identification and Biological Activity Assay. Journal of Marine Science and Engineering. 2021; 9(9):978. https://doi.org/10.3390/jmse9090978
Chicago/Turabian StyleYang, Wenzhi, Guangjie Liang, Yang Sun, and Zhijin Gong. 2021. "Bioactive Secondary Metabolites from Marine Streptomyces griseorubens f8: Isolation, Identification and Biological Activity Assay" Journal of Marine Science and Engineering 9, no. 9: 978. https://doi.org/10.3390/jmse9090978
APA StyleYang, W., Liang, G., Sun, Y., & Gong, Z. (2021). Bioactive Secondary Metabolites from Marine Streptomyces griseorubens f8: Isolation, Identification and Biological Activity Assay. Journal of Marine Science and Engineering, 9(9), 978. https://doi.org/10.3390/jmse9090978





