Bonamia exitiosa in European Flat Oyster (Ostrea edulis) on the Croatian Adriatic Coast from 2016 to 2020
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Tissue Processing, Cytological and Histological Examination
2.3. Molecular Detection and Characterization
2.3.1. DNA Extraction
2.3.2. PCR and Sequencing
2.3.3. Real-Time PCR
2.3.4. Sequencing and Phylogeny
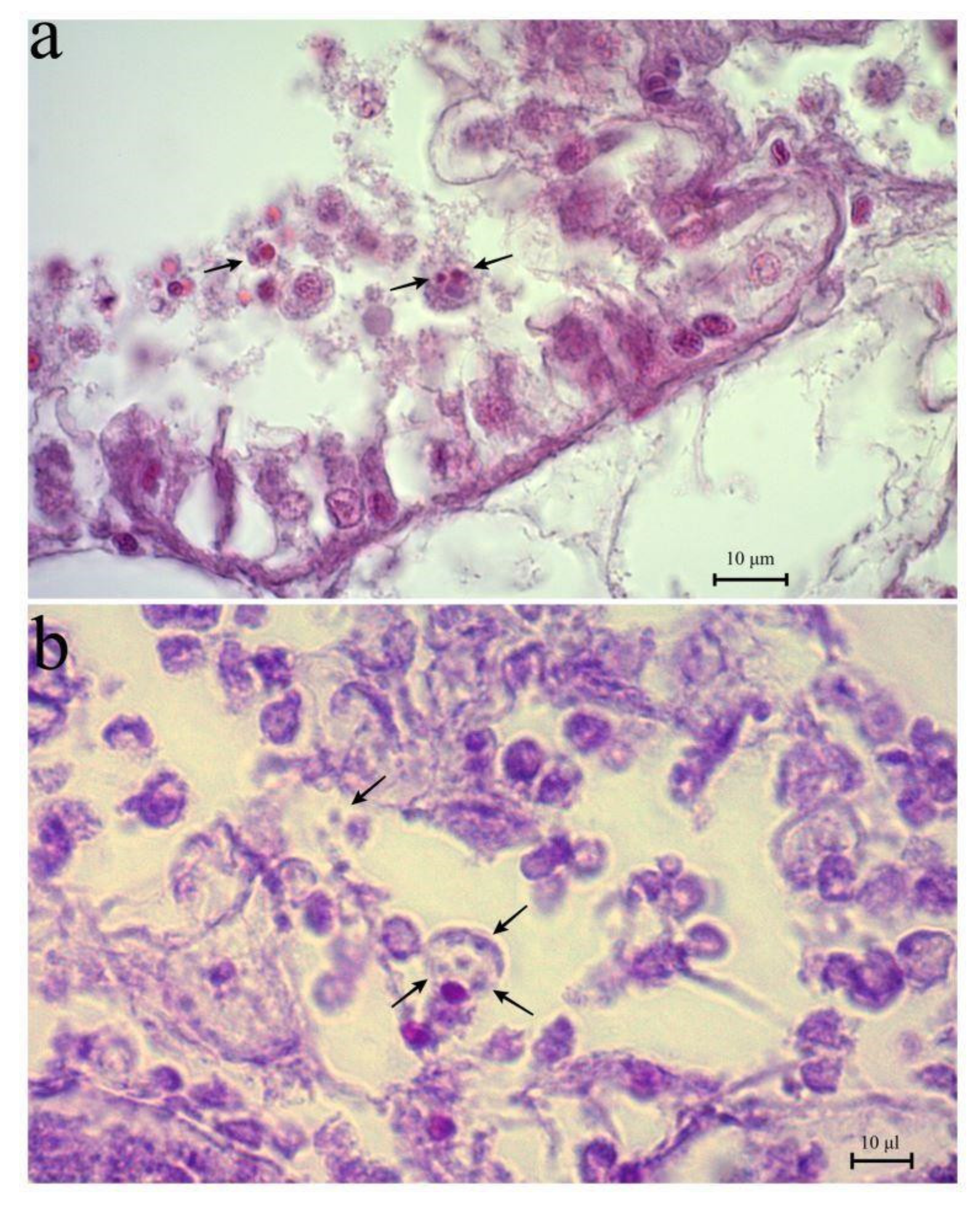
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- MP-Uprava Ribarstva. 2019. Available online: https://ribarstvo.mps.hr/default.aspx?id=14 (accessed on 17 August 2021).
- Grizel, H.; Mialhe, E.; Chagot, D.; Boulo, V.; Bachère, E. Bonamiosis: A model study of diseases in marine molluscs. Am. Fish. Soc. Spec. Publ. 1988, 18, 1–4. [Google Scholar]
- Robert, R.; Borel, M.; Pichot, Y.; Gilles, T. Growth and mortality of the European oyster Ostrea edulis in the Bay of Arcachón (France). Aquat. Living Resour. 1991, 4, 265–274. [Google Scholar] [CrossRef]
- Hudson, E.B.; Hill, B.J. Impact and spread of bonamiosis in the UK. Aquaculture 1991, 93, 279–285. [Google Scholar] [CrossRef]
- Carnegie, R.B.; Cochennec-Laureau, N. Microcell parasites of oysters: Recent insights and future trends. Aquat. Living Resour. 2004, 17, 519–528. [Google Scholar] [CrossRef]
- O’Neill, G.; Culloty, S.C.; Mulcahy, M.F. The effectiveness of two routine diagnostic techniques for the detection of the protozoan parasite Bonamia ostreae. Bull. Eur. Ass. Fish. Pathol. 1998, 18, 117–120. [Google Scholar]
- Culloty, S.; Cronin, M.; Mulcahy, M. Possible limitations of diagnostic methods recommended for the detection of the protistan, Bonamia ostreae in the European flat oyster, Ostrea edulis. Bull. Eur. Assoc. Fish. Pathol. 2003, 199, 67–71. [Google Scholar]
- Da Silva, P.M.; Villalba, A. Comparison of light microscopic techniques for the diagnosis of the infection of the European flat oyster Ostrea edulis by the protozoan Bonamia ostreae. J. Invertebr. Pathol. 2004, 85, 97–104. [Google Scholar] [CrossRef]
- Marty, G.D.; Bower, S.M.; Clarke, K.R.; Meyer, G.; Lowe, G.; Osborn, A.L.; Chow, E.P.; Hannah, H.; Byrne, S.; Sojonky, K.; et al. Histopathology and a real-time PCR assay for detection of Bonamia ostreae in Ostrea edulis cultured in western Canada. Aquaculture 2006, 261, 33–42. [Google Scholar] [CrossRef]
- Engelsma, M.Y.; Culloty, S.C.; Lynch, S.A.; Arzul, I.; Carnegie, R.B. Bonamia parasites: A rapidly changing perspective on a genus of important mollusc pathogens. Dis. Aquat. Org. 2014, 110, 5–23. [Google Scholar] [CrossRef] [Green Version]
- Cochennec, N.; LeRoux, F.; Berthe, F.; Gerard, A. Detection of Bonamia ostreae based on small subunit ribosomal probe. J. Invert. Pathol. 2000, 76, 26–32. [Google Scholar] [CrossRef]
- Cochennec-Laureau, N.; Reece, K.S.; Berthe, F.C.J.; Hine, P.M. Mykrocytos roughleyi taxonomic affiliation leads to the genus Bonamia (Haplosporidia). Dis. Aquat. Org. 2003, 54, 209–217. [Google Scholar] [CrossRef] [Green Version]
- Arzul, I. Chapter 2.4.3. Infection with Bonamia ostreae. In Manual of Diagnostic Tests for Aquatic Animals; World Organisation for Animal Health (OIE): Paris, France, 2012. [Google Scholar]
- Grizel, H.; Comps, M.; Raguenes, D.; Leborgne, Y.; Tigè, G.; Martin, A.G. Bilan des essais d’acclimatation d’Ostrea chilensis sur les côtes de Bretagne. Rev. Trav. Inst. Pêches Marit. 1983, 46, 209–225. [Google Scholar]
- Carnegie, R.B.; Burreson, E.M.; Hine, P.M.; Stokes, N.A.; Audemard, C.; Bishop, M.J.; Peterson, C.H. Bonamia perspora n. sp. (Haplosporidia), a parasite of the oyster Ostreola equestris, is the first Bonamia species known to produce spores. J. Eukaryot. Microbiol. 2006, 53, 232–245. [Google Scholar] [CrossRef] [PubMed]
- Carnegie, R.B.; Hill, K.M.; Stokes, N.A.; Burreson, E.M. The haplosporidian Bonamia exitiosa is present in Australia, but the identity of the parasite described as Bonamia (formerly Mikrocytos) roughleyi is uncertain. J. Invertebr. Pathol. 2014, 115, 33–40. [Google Scholar] [CrossRef]
- Culloty, S.C.; Mulcahy, M.F. Bonamia ostreae in the Native Oyster Ostrea edulis: A review; Marine Environment and Health Series No. 29; Marine Institute: Rinville, Ireland, 2007; 40p. [Google Scholar]
- Culloty, S.C.; Cronin, M.A.; Mulcahy, M.F. Potential resistance of a number of populations of the oyster Ostrea edulis to the parasite Bonamia ostreae. Aquaculture 2004, 237, 41–58. [Google Scholar] [CrossRef]
- Hine, P.M.; Cochennec-Laureau, N.; Berthe, F.C.J. Bonamia exitiosus n. sp. (Haplosporidia) infecting flat oysters Ostrea chilensis in New Zealand. Dis. Aquat. Org. 2001, 47, 63–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burreson, E.M.; Stokes, N.A.; Carnegie, R.B.; Bishop, M.J. Bonamia sp. (Haplosporidia) found in non-native oysters Crassostrea ariakensis in Bogue Sound, North Carolina. J. Aquat Anim. Health 2004, 16, 1–9. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Ifremer, EURL-Mollusc, Edition n° 1, Bonamia ostrae and Bonamia exitiosa detection by Taqman® Real-Time Polymerase Chain Reaction. Available online: https://www.eurl-mollusc.eu/content/download/137231/file/B.ostreae%26B.exitiosa%20_TaqmanRealTimePCR_editionN%C2%B01.pdf (accessed on 17 August 2021).
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2013, 41, D36–D42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (ITOL) v4: Recent Updates and New Developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carnegie, R.B. Bonamiosis of Oysters Caused by Bonamia exitiosa; Feist, S., Ed.; ICES Identification Leaflets for Diseases and Parasites of Fish and Shellfish; International Council for the Exploration of Sea: Copenhagen, Denmark, 2017; Volume 66, p. 6. [Google Scholar] [CrossRef]
- Commission Implementing Decision (EU) 2015/1554 of 11 September 2015 Laying down Rules for the Application of Directive 2006/88/EC as Regards Requirements for Surveillance and Diagnostic Methods. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32015D1554&rid=1 (accessed on 17 August 2021).
- Dinamani, P.; Hine, P.M.; Jones, J.B. Occurrence and characteristics of the haemocyte parasite Bonamia sp. in the New Zealand dredge oyster Tiostrea lutaria. Dis. aquat. Org. 1987, 3, 37–44. [Google Scholar] [CrossRef]
- Engelsma, M.Y.; Kerkhoff, S.; Roozenburg, I.; Haenen, O.L.M.; Van Gool, A.; Sistermans, W.; Wijnhoven, S.; Hummel, H. Epidemiology of Bonamia ostreae infecting European flat oyster Ostrea edulis from Lake Grevelingen, The Netherlands. Mar. Ecol. Prog. Ser. 2010, 409, 131–142. [Google Scholar] [CrossRef] [Green Version]
- Abollo, E.; Ramilo, A.; Casas, M.S.; Comesaña, P.; Cao, A.; Carballal, M.J.; Villalba, A. First detection of the protozoan parasite Bonamia exitiosa (Haplosporidia) infecting flat oyter Ostrea edulis grown in European waters. Aquaculture 2008, 274, 201–207. [Google Scholar] [CrossRef]
- Narcisi, V.; Arzul, I.; Cargini, D.; Mosca, F.; Calzetta, A.; Traversa, D.; Robert, M.; Joly, J.P.; Chollet, B.; Renault., T.; et al. Detection of Bonamia ostreae and B. exitiosa (Haplosporidia) in Ostrea edulis from the Adriatic Sea (Italy). Dis. Aquat. Org. 2010, 89, 79–85. [Google Scholar] [CrossRef]
- Carrasco, N.; Villalba, A.; Andree, K.B.; Engelsma, M.Y.; Lacuesta, B.; Ramilo, A.; Gairín, I. Bonamia exitiosa (Haplosporidia) observed infecting the European flat oyster Ostrea edulis cultured on the Spanish Mediterranean coast. J. Invertebr. Pathol. 2012, 110, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Buss, J.J.; Harris, J.O.; Tanner, J.E.; Wiltshire, K.H.; Deveney, M.R. Rapid transmission of Bonamia exitiosa by cohabitation causes mortality in Ostrea angasi. J. Fish. Dis. 2020, 43, 227–237. [Google Scholar] [CrossRef]
- Lynch, S.A.; Mulcahy, M.F.; Culloty, S.C. Efficiency of diagnostic techniques for the parasite, Bonamia ostreae, in the flat oyster, Ostrea edulis. Aquaculture 2008, 281, 17–21. [Google Scholar] [CrossRef]
- Ford, S.E.; Allam, B.; Xu, Z. Using bivalves as particle collectors with PCR detection to investigate the environmental distribution of Haplosporidium nelsoni. Dis. Aquat. Org. 2009, 83, 159–168. [Google Scholar] [CrossRef]
- Arzul, I.; Gagnaire, B.; Bond, C.; Chollet, B.; Morga, B.; Ferrnad, S.; Robert, M.; Renault, T. Effects of temperature and salinity on the survival of Bonamia ostreae a parasite infecting flat oysters O. edulis. Dis. Aquat. Org. 2009, 85, 67–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Audemard, C.; Carnegie, R.B.; Stokes, N.A.; Bishop, M.J.; Peterson, C.H.; Burreson, E.M. Effects of salinity on Bonamia sp. survival in Asian oyster Crassostrea ariakensis. J. Shell. Res. 2008, 27, 535–540. [Google Scholar] [CrossRef]
- Hill, K.M.; Carnegie, R.B.; Aloui-Bejaoui, N.; El Gharsalli, R.; White, D.M.; Stokes, N.A.; Burreson, E.M. Observation of a Bonamia sp. infecting the oyster Ostrea stentina in Tunisia, and a consideration of its phylogenetic affinities. J. Invert. Pathol. 2010, 103, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Lynch, S.A.; Armitage, D.V.; Mulcahy, M.F.; Culloty, S.C. Investigating the possible role of benthic macroinvertebrates and zooplankton in the life cycle of the haplosporidian Bonamia ostreae. Exp. Parasitol. 2007, 115, 359–368. [Google Scholar] [CrossRef]
- Lynch, S.A.; Abolle, E.; Ramilo, A.; Cao, A.; Culloty, S.C.; Villalba, A. Observations raise the question if the Pacific Oyster, Crassostrea gigas, can act as either a carrier or reservoir for Bonamia ostreae or Bonamia exitiosa. Parasitology 2010, 137, 1515–1526. [Google Scholar] [CrossRef]
- Hrs-Brenko, M. Ostrea edulis (Linnaeus) and Crassostrea gigas (Thunberg) larvae in the plankton of Limski kanal in the northern Adriatic Sea. Acta Adriat. 1982, 23, 399–407. [Google Scholar]
- Šegvić-Bubić, T.; Grubišić, L.; Zrnčić, S.; Jozić, S.; Žužul, I.; Talijančić, I.; Oraić, D.; Relić, M.; Katavić, I. Range expansion of the non-native oyster. Acta Adriat. 2016, 57, 321–330. [Google Scholar]
- Hrs-Brenko, M.; Legac, M. Inter-and intra-species relationships of sessile bivalves on the eastern coast of the Adriatic Sea. Nat. Croat. 2006, 15, 203–230. [Google Scholar]
- Vondrak, T. Upotreba Molekularnih Biljega u Identifikaciji Različitih vrsta Kamenica u Jadranu, Diplomski rad, Sveučilište u Zagrebu, Prirodoslovno-Matematički fakultet, Biološki Odsjek. 2017. Available online: https://repozitorij.pmf.unizg.hr/islandora/object/pmf%3A4522/datastream/PDF/view (accessed on 17 August 2021).
- Hill-Spanik, K.M.; McDowell, J.R.; Stokes, N.A.; Reece, K.S.; Burreson, E.M.; Carnegie, R.B. Phylogeographic perspective on the distribution and dispersal of a marine pathogen, the oyster parasite Bonamia exitiosa. Mar. Ecol. Prog. Ser. 2015, 536, 65–76. [Google Scholar] [CrossRef]
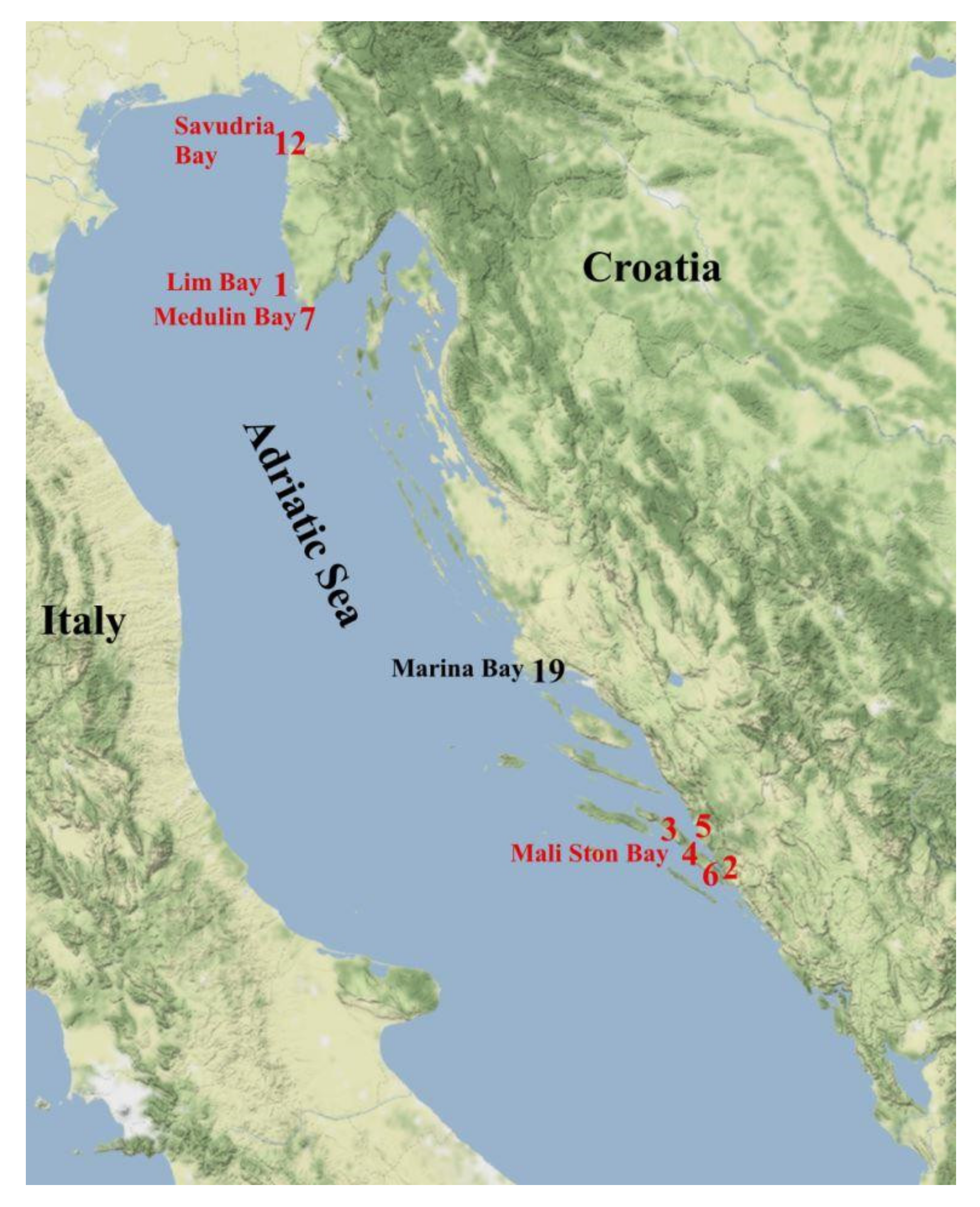


| Species | Flat Oyster (Ostrea edulis) | Pacific Oyster (Magallana gigas) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Production Area | Mali Ston Bay | Medulin Bay | Lim Bay | Savudrija Bay | Marina Bay | Medulin Bay | Lim Bay | |||||||
| Date and number of sample animals | Date M/Y | No | Date M/Y | No | Date M/Y | No | Date M/Y | No | Date M/Y | No | Date M/Y | No | Date M/Y | No |
| 05/2016 | 150 * | 05/2016 | 30 * | 05/2016 | 30 | 05/2016 | 30 | 11/2016 | 30 | |||||
| 11/2016 | 150 | 11/2016 | 30 * | 09/2016 | 30 | 10/2016 | 30 | 06/2017 | 30 | |||||
| 05/2017 | 150 | 07/2017 | 30 * | 05/2017 | 30 | 05/2017 | 30 | 05/2018 | 30 | 07/2017 | 18 | |||
| 11/2017 | 150 | 11/2017 | 30 * | 06/2018 | 30 * | 06/2018 | 30 * | 09/2018 | 30 | |||||
| 11/2018 | 150 | 10/2018 | 30 | 09/2019 | 30 | 10/2019 | 30 | 08/2019 | 30 | 09/2018 | 30 | |||
| 10/2020 | 150 | 10/2020 | 30 * | 09/2020 | 30 | 09/2020 | 30 * | 08/2020 | 30 | |||||
| Totally | 900 | 180 | 180 | 180 | 180 | 18 | 30 | |||||||
| Date of Sampling (M/Y) | Production Area/Name of Sampling Site | Number of Sampled Animals | Sampled Species | Results of Laboratory Analysis | |||||
|---|---|---|---|---|---|---|---|---|---|
| Heart Imprints | PCR | Real Time PCR | |||||||
| MALOSTONSKI BAY AREA | +-ve | % | +-ve | % | +-ve | % | |||
| 05/2016 | 6 * Mali Ston | 30 | Ostrea edulis | 3 | 10 | 5 | 16.7 | n/d | - |
| 05/2016 | 2 * Brijesta | 30 | Ostrea edulis | 3 | 10 | 6 | 2.,0 | n/d | - |
| 05/2016 | 5 * Bistrina | 30 | Ostrea edulis | 2 | 6.7 | 2 | 6.7 | n/d | - |
| 05/2016 | 3 * Bjejevica | 30 | Ostrea edulis | 2 | 6.7 | 3 | 10.0 | n/d | - |
| 05/2016 | 4 * Sutvid | 30 | Ostrea edulis | 1 | 3.3 | 1 | 3.3 | n/d | - |
| MEDULINSKI BAY | |||||||||
| 05/2016 | 7 * Medulinski Bay | 30 | Ostrea edulis | 2 | 6.7 | 3 | 6.7 | n/d | - |
| 11/2016 | 7 * Medulinski Bay | 30 | Ostrea edulis | 1 | 3.3 | 1 | 3.3 | n/d | - |
| 07/2017 | 7 * Medulinski Bay | 30 | Ostrea edulis | 2 | 6.7 | 3 | 10.0 | n/d | - |
| 11/2017 | 7 * Medulinski Bay | 30 | Ostrea edulis | 1 | 3.3 | 1 | 3.3 | n/d | - |
| 11/2017 | 7 * Medulinski Bay | 18 | Magallana gigas | 0 | 0 | 0 | 0 | n/d | - |
| 10/2020 | 7 * Medulinski Bay | 30 | Ostrea edulis | 4 | 13.3 | 5 | 16.7 | 5 | 16.7 |
| LIM BAY | |||||||||
| 07/2018 | 1 * Lim Bay | 30 | Ostrea edulis | 1 | 3.3 | 2 | 6.7 | n/d | - |
| 2018 | 1 * Lim Bay | 30 | Magallana gigas | 0 | 0 | 0 | 0 | n/d | - |
| SAVUDRIJA BAY | |||||||||
| 06/2018 | 12 * Savudrija Bay | 30 | Ostrea edulis | 2 | 6.7 | 3 | 10.0 | n/d | - |
| 09/2020 | 12 * Savudrija Bay | 30 | Ostrea edulis | 1 | 6.7 | 4 | 13.3 | 4 | 13.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oraić, D.; Beck, R.; Pavlinec, Ž.; Zupičić, I.G.; Maltar, L.; Miškić, T.; Acinger-Rogić, Ž.; Zrnčić, S. Bonamia exitiosa in European Flat Oyster (Ostrea edulis) on the Croatian Adriatic Coast from 2016 to 2020. J. Mar. Sci. Eng. 2021, 9, 929. https://doi.org/10.3390/jmse9090929
Oraić D, Beck R, Pavlinec Ž, Zupičić IG, Maltar L, Miškić T, Acinger-Rogić Ž, Zrnčić S. Bonamia exitiosa in European Flat Oyster (Ostrea edulis) on the Croatian Adriatic Coast from 2016 to 2020. Journal of Marine Science and Engineering. 2021; 9(9):929. https://doi.org/10.3390/jmse9090929
Chicago/Turabian StyleOraić, Dražen, Relja Beck, Željko Pavlinec, Ivana Giovanna Zupičić, Ljupka Maltar, Tihana Miškić, Žaklin Acinger-Rogić, and Snježana Zrnčić. 2021. "Bonamia exitiosa in European Flat Oyster (Ostrea edulis) on the Croatian Adriatic Coast from 2016 to 2020" Journal of Marine Science and Engineering 9, no. 9: 929. https://doi.org/10.3390/jmse9090929
APA StyleOraić, D., Beck, R., Pavlinec, Ž., Zupičić, I. G., Maltar, L., Miškić, T., Acinger-Rogić, Ž., & Zrnčić, S. (2021). Bonamia exitiosa in European Flat Oyster (Ostrea edulis) on the Croatian Adriatic Coast from 2016 to 2020. Journal of Marine Science and Engineering, 9(9), 929. https://doi.org/10.3390/jmse9090929









