Abstract
This work deals with the identification of a predominant thraustochytrid strain, the optimization of culture conditions, the synthesis of nanoparticles, and the evaluation of antioxidant and antimicrobial activities in biomass extracts and nanoparticles. Thraustochytrium kinnei was identified as a predominant strain from decomposing mangrove leaves, and its culture conditions were optimized for maximum biomass production of 13.53 g·L−1, with total lipids of 41.33% and DHA of 39.16% of total fatty acids. Furthermore, the strain was shown to synthesize gold and silver nanoparticles in the size ranges of 10–85 nm and 5–90 nm, respectively. Silver nanoparticles exhibited higher total antioxidant and DPPH activities than gold nanoparticles and methanol extract of the strain. The silver nanoparticles showed higher antimicrobial activity than gold nanoparticles and petroleum ether extract of the strain. Thus, Thraustochytrium kinnei is proven to be promising for synthesis of silver nanoparticles with high antioxidant and antimicrobial activity.
1. Introduction
Thraustochytrids are important fungus-like protists of class Labyrinthulomycetes and family Thraustochytriaceae. They are unicellular and heterotrophic protists, abundantly present in decomposing mangrove litter [1]. The thraustochytrids are known to produce polyunsaturated fatty acids (PUFAs), such as docosahexaenoic acid (DHA) and eicosapentaenoic acid (EPA), which are promising for pharmaceutical and industrial applications, especially in aquaculture [2,3,4]. They contain 10–50% lipids, of which 30–70% is DHA [5]. The DHA plays a vital role as a component of phospholipids in cell membrane structure and function [6]; it is essential for development of neural, retinal and immune systems in fetal life and infants [7] as well for the prevention of asthma, arthritis, and neural, cardiovascular and skin diseases in adulthood [8,9]. In thraustochytrids, up to 5% of lipids occur as phospholipids, while 70–90% of lipids are present as neutral lipids or triacylglycerols. The accumulation of DHA in the form of neutral lipids is essential for the metabolic process of thraustochytrids [10].
The culture conditions of thraustochytrids must be optimized to obtain maximum biomass with a high yield of omega-3 fatty acids. This can be done either through modification of carbon and nitrogen sources in the culture media or manipulation of the physicochemical conditions such as pH, salinity and temperature of the culture media [4,11,12,13].
Nanoparticles are the starting point for preparing many nano-structured materials and devices [14]. Since noble metal nanoparticles such as gold, silver and platinum are widely applied, there is a growing need to develop eco-friendly protocols for nanoparticle synthesis [15]. The applications of nanotechnology are fast growing in the areas of agriculture, aquaculture and pharmaceuticals [16]. The synthesis of nanoparticles using biological entities has generated great interest due to their unusual optical, chemical, photo electrochemical and electronic properties [17,18]. Biological methods for nanoparticle synthesis using microorganisms, enzymes and plant extracts have been suggested as possible eco-friendly alternatives to chemical and physical methods [19,20].
Thraustochytrids are explored for the production of a wide range of bioactive compounds as a supplement for the production of animal feed [21], antimicrobials [2], antioxidants [3], biodiesel [22], enzymes [23], carotenoids [24], squalene [25], exopolysaccharides [26], vaccines [27], and cancer drugs [28]. There are no reports of thrautochytrids producing toxic chemicals [29]. In spite of various potential applications, the utilization of thraustochytrids in synthesis of silver and gold nanoparticles has not been properly attempted [30,31]. Therefore, the present work was undertaken to isolate a predominant thraustochytrid strain from decomposing mangrove leaves of Rhizophora annamalayana, to optimize its culture conditions and to evaluate its efficacy in the synthesis of nanoparticles with antioxidant and antimicrobial activities.
2. Materials and Methods
2.1. Isolation and Maintenance of Thraustochytrids from Decomposing Mangrove Leaves
Thraustochytrids were isolated from decomposing mangrove leaves of Rhizophora annamalayana Kathir., collected from a mangrove forest in Pichavaram on the southeast coast of India, by the pour plate method on GPYAS culture medium containing Glucose (3 g·L−1), Peptone (1.25 g·L−1), Yeast extract (1.25 g·L−1), and Agar (12 g·L−1) in sterile natural seawater with the addition of antibacterial (streptomycin 100 µg·L−1) and antifungal (fluconazole 100 µg·L−1) agents to prevent contaminations, adjusted to a pH of 7.2 and incubated at 28 °C for 2–7 days [2,3]. The pure cultures were found after 3 repeated sub-cultures in 2–7 days, and culture stocks were maintained on agar slants. Then, the predominant thraustochytrid strain TSKK1 was selected for morphological and molecular identifications.
2.2. Morphological Identification of Thraustochytrids Species
Thraustochytrid strain TSKK1 was morphologically identified [32] after acriflavine staining at a concentration of 0.02 µg·mL−1 and by observing under a light microscope, followed by a Scanning Electron Microscope.
2.3. Genomic DNA Extraction and PCR Amplification of Thraustochytrids
Genomic DNA was extracted by the method of [33], with slight modification. Briefly, 3 mL of thraustochytrid strain TSKK1 cell culture (4–7 days) was centrifuged at 10,000 rpm for 5 min at 4 °C. Cells were lysed using lysis buffer (0.25 M Tris-HCl with pH 8.2, 0.1 M EDTA, 2% Sodium Dodecyl Sulfate, 0.1 M NaCl) and then kept in a water bath at 55 °C for 30 min for efficient homogenization. Chloroform-isoamyl alcohol was added (24:1), and the DNA was precipitated with chilled isopropanol. Extracted DNA was dissolved with 30 µL of TE buffer and kept at −20 °C for further use. The small subunit rRNA gene was amplified in a thermal cycler (Tech GeneTM, South San Francisco, California, USA) using 18S rDNA specific primers 18S001 (5′AACCTGGTTGATCCTGCCAGTA3′) and 18S13 (5′CCTTGTTACGACTTCACCTTCCTC T3′) [34]. The amplification cycle consisted of an initial denaturation step of 94 °C for 5 min, followed by 35 cycles of denaturation (94 °C for 60 s), annealing (50 °C for 60 s), elongation (72 °C for 120 s) and a final 10 min elongation step at 72 °C. The molecular weight was determined using the molecular weight marker (100 bp ladder). The amplicons were tested in 1.5% agarose gel.
2.4. Sequencing and Phylogenetic Analyses of 18S rRNA Gene
PCR product purification and DNA sequencing were carried using a high throughput Mega Bace sequencer (YAAZH XENOMICS, Chennai, India). The obtained sequences were edited based on the electropherogram peak clarities in MEGA 6.0 software [35]. Sequences with noisy peaks were excluded from the analysis. The amplified sequences belonging to 18S rRNA gene were confirmed by the percentage of similarity in the NCBI’s BLAST program. The species identity was confirmed based on 95–100% similarity with the reference sequences.
The sequence was submitted to NCBI Genbank (Accession No. KT716334). The evolutionary analysis was conducted in MEGA6 software. The successfully sequenced rDNA region of thraustochytrids was blasted against the complete non-redundant NCBI Genbank database and aligned using the CLUSTAL W Multiple Sequences Alignment Program and analyzed. The phylogenetic tree was constructed, including the reference thrausochytrids gene sequences along with the sequences determined in this study by using the maximum likelihood method. Species relations were evaluated with statistical analysis through the bootstrap protocol. To group the phylogenetic analysis, the labyrinthula genus was used as the out group.
2.5. Mass Production of Thraustochytrid
The Erlenmeyer flasks containing GYPS broth medium were inoculated with thraustochytrid strain TSKK1 and incubated in an orbital shaker and agitated at 150 rpm (Orbit Shaker, Labline Instrument Incorporated, Dubuque, Iowa, USA) at 28 °C for 2–7 days. The experimental condition of the biomass production was adopted from the industrial statistical model of response surface methodology [13]. Biomass production was estimated as milligram of dry weight biomass per one liter of medium. The biomass was filtered through filter paper (Whatman No. 1) and washed with distilled water to remove medium components from the biomass and then freeze-dried. Biomass was weighed and stored in a sealed container at −4 °C for further study.
2.6. Prediction of Important Factors for Biomass Production Using Plackett-Burman Design
Biomass production depends on basal media composition and environmental factors, and hence these factors were studied for biomass production in thraustochytrid strain TSKK1 by using 2 phases of the response surface methodology (RSM) method (Plackett-Burman and Central Composite Design). In the first phase, 16 runs of Plackett-Burman design were maintained. The factors studied were temperature, pH, salinity, carbon sources (glucose, sugarcane molasses, bread crumbs) and nitrogen sources (yeast extract, peptone, monosodium glutamate). The Plackett-Burman design was used to determine the important factors that influenced the biomass production, using statistical modeling and the actual and coded values. After selection of the important factors for the biomass production, further optimization was continued for the second phase.
The Plackett-Burrman experimental design is based on the first order model, as given in Equation (1).
where Y is the response, β0 is the intercept coefficient, βi is the variable estimate, and Xi is the independent variable.
Y = β0 + Σ βi Xi
All experiments were carried out in triplicate and the average was taken as a response.
2.7. Optimization of Media Components for Maximm Biomass Production by Using Central Composite Design (CCD)
The important factors (temperature, pH, salinity, carbon and nitrogen sources) for biomass production were optimized using CCD [11]. For the optimization of the specific level, different concentrations, individuals and interaction effect of the factors were tested using the 30 runs batch experimental technique as derived from RSM. The experimental and predicted responses are in grams of dry weight biomass production. The coded values and actual factor values were calculated using the following equations.
where Y is the predicted response, Xi and Xj are independent variables, β0 is the interception coefficient, βi is the linear coefficient, βii is the quadratic coefficient and βij is the interaction coefficient. In this study, the independent variables were coded as X1, X2 and X3. Thus, the second order polynomial equation could be presented as Equation (3):
Thraustochytrid strain Y = β0 + Σ βi Xi + Σ βii Xi2 + Σ βijXiXj
Y = β0 + β1 X1 + β2 X2 + β3 X3 + β4 X4 + β11 X12 + β22 X22 + β33 X32 + β44 X42 + β12
X1X2 + β13 X1X3 + β14 X1X4 + β23 X2X3 + β24 X2X4 + β34 X3X4
X1X2 + β13 X1X3 + β14 X1X4 + β23 X2X3 + β24 X2X4 + β34 X3X4
The fit of the Central Composite Design (CCD) with the multi-factorial depth model was tested, and it was used for determination of fit of the statistical model. The variables of individual and interaction were significant. This revealed that the model was fit to analyze the interaction and individual effect on biomass production. Therefore, the 3D surface plot and perturbation plot was drawn further for optimization of the factors. The statistical program package Design Expert 8.0.6 was used for regression analysis and to estimate the coefficient of the regression equation. The equations were analyzed using ANOVA.
2.8. Lipid and Fatty Acid Methyl Ester (FAME) Extraction and Analysis
Total lipid content in thraustochytrid strain TSKK1 biomass was analyzed by the method specified in [36]. The freeze-dried thraustochytrid sample was ground finely using a mortar and pestle. Preparation and analysis of fatty acid methyl ester (FAME) in the sample was performed according to the method described earlier [37], with some modifications. In brief, 1 g of powder sample was added to 1 mL of 1.2 M NaOH in 50% aqueous methanol in a screw cap tube and then incubated at 100 °C for 30 min in a water bath. The saponified sample was cooled at 30 °C for 25 min and methylated by adding 2 mL of 54% 6 N HCl with 46% aqueous methanol and incubated at 80 °C for 10 min in a water bath. After rapid cooling, the methylated fatty acids were extracted with 1.25 mL of 50% diethyl ether in hexane. Each sample was mixed for 10 min; the bottom phase was removed using a glass syringe, and the top phase was washed with 3 mL of 0.3 M NaOH for 5 min and collected. The FAMEs were cleaned in anhydrous sodium sulphate, and then the sample was transferred to Gas Chromatography Mass Spectroscopy (GC Clarus 500 Perkin Elmer, Waltham, Massachusetts, USA) for fatty acid analysis. The FAMEs were identified according to their retention time and area of percentage. The FAME peaks were identified and quantified using MS Library and NIST Version-Year 2005.
2.9. Biosynthesis of Silver Nanoparticles
Ten grams of fresh wet biomass was harvested from 96-hour-old culture of thraustochytrid strain TSKK1 and agitated with 200 mL of Milli-Q deionized water for 72 h at 25 °C in an Erlenmeyer flask at 120 rpm. The filtered cells were obtained by passing through a Whatman No. 1 filter paper and washed twice with distilled water to remove medium components from the aqueous methanol biomass. This cell filtrate was further used for silver and gold nanoparticle synthesis. For the synthesis of silver nanoparticles, 50 mL of 1 mM of silver nitrate was mixed with 50 mL of cell filtrate in a 250 mL Erlenmeyer flask and agitated at 25 °C in darkness at 120 rpm. Then, the whole mixture was centrifuged at 5000 rpm for 30 min and 1 mL of the sample was withdrawn for measuring its optical density at different wavelengths of broad range 300 to 800 nm and a narrow range 400 to 500 nm using UV-visible spectrophotometer (Elico, Chennai, India), values were plotted on a graph.
2.10. Biosynthesis of Gold Nanoparticles
An aliquot of 50 mL of cell filtrate of thraustochytrid strain TSKK1 was transferred to 50 mL of 1 mM chloroauric acid solution, and the whole mixture was kept in a shaker at 28 °C at 120 rpm. During the process of biosynthesis, flasks were observed for visual color change from yellow to pinkish purple. Then, the whole mixture was centrifuged at 5000 rpm for 30 min and 1 mL of sample was taken and measured for its optical density at different wavelengths (300–800 nm).
For purification of nanoparticles, the suspended solution was again centrifuged at 5000 rpm for 20 min. The resulting pellet was removed and resuspended in double distilled water. The supernatant was again centrifuged at 5000 rpm for 20 min. Finally, the nanoparticles were washed with distilled water, and the pure particles were separated and dried in vacuum and stored in a dark bottle at 4 °C.
2.11. Characterization of Nanoparticles: SEM and Dynamic Light Scattering (DLS) Analysis
SEM analysis of silver and gold nanoparticles synthesized by thraustochytrid strain TSKK1 was carried out. SEM observation was performed under a JEOL JEM IT-200SX Scanning Electron Microscope with the accelerating voltage at 15 Kv. The dynamic light-scattering (DLS) measurement was performed 24 h after preparation of the nanoparticle suspension for analyzing size group using a nano ZS apparatus at 25 °C.
2.12. In Vitro Antioxidant Assays of Nanoparticles
The silver and gold nanoparticles synthesized from thraustochytrid strain TSKK1 were freeze-dried and extracted in aqueous solution, following centrifugation at 8000 rpm for 5 min. The extraction procedure was repeated three times, and all supernatants were combined. Subsequently, the lyophilized thraustochytrid strain TSKK1 was extracted with 80% methanol. Then, these methanolic and aqueous extracts, prepared separately in different concentrations of 50, 100, 150, 200, and 250 μg·mL−1, were tested using standard antioxidant assay methods. Each experiment was done in triplicate.
2.12.1. Total Antioxidant Capacity (TAC)
Total antioxidant capacity (TAC) of the methanolic extract of thraustochytrid strain TSKK1 and the silver and gold nanoparticle synthesized extract of thraustochytrid strain TSKK1 was estimated by the method specified [38], with modification. An aliquot of 0.3 mL was taken separately for different concentrations of the extract. To this, 3 mL of reagent solution, containing 0.6 M sulfuric acid, 28 mM sodium phosphate and 4 mM ammonium molybdate, was added. The reaction mixture was incubated at 95 °C for 90 min in a water bath and then cooled to room temperature. Absorbance of the sample mixtures was measured at 695 nm. Total antioxidant activity is expressed in percent, as butylated hydroxyl toluene (BHT) equivalent in mg/g of extract.
2.12.2. DPPH Radical Scavenging Activity
Free radical scavenging activity of methanolic extract of thraustochytrid strain TSKK1 and silver and gold nanoparticle synthesized thraustochytrid strain TSKK1 extracts was analyzed using the DPPH (1,2-diphenyl-1-picrylhydrazyl) free radical method, modified from Yu et al. [38]. An amount of 0.1 mL of the extract at different concentrations was mixed separately in 0.9 mL methanolic solution of DPPH solution (120 µM). DPPH is a stable, nitrogen-centered free radical that produces violet color in methanol solution. When a substrate that can donate a hydrogen atom is added to DPPH solution, it is reduced to a yellow colored product, diphenylpicryl hydrazine. The reaction mixture was incubated in darkness at 37 °C for 30 min, and its absorbance was measured at 517 nm. DPPH radical scavenging activity is expressed in percent, as butylated hydroxyl toluene (BHT) equivalent in mg/g of extracts.
2.13. In Vitro Antibacterial Activity of Nanoparticles
The lyophilized silver and gold nanoparticles synthesized from thraustochytrid strain TSKK1 were extracted in aqueous solution. The aqueous extract was filtered in Whatman No. 1 filter paper and centrifuged at 5000 rpm for 10 min. Supernatant of the extract was collected and tested for antibacterial activity by the agar disc diffusion method [2] against 10 clinical bacterial strains: Proteus mirabilis (P. mirabilis), Staphylococcus aureus (S. aureus), Streptococcus pyogens (S. pyogens), Vibrio cholerae (V. cholerae), Vibrio parahaemolyticus (V. parahaemolyticus), Escherichia coli (E. coli), Klebsiella pneumonia (K. pneumonia), Klebsiella oxytoca (K. oxytoca), Salmonella typhi (S. typhi), and Salmonella paratyphi (S. paratyphi), obtained from Raja Muthiah Medical College, Annamalai University, India. The 24-hour-old bacterial culture cell suspension (100 μL) of each of the strains was poured onto the nutrient agar plates and spread using a glass spreader aseptically. A sterile disc (6 mm) was soaked with 100 µL of silver and gold nanoparticle extract of thraustochytrid strain TSKK1 separately. Lyophilized thraustochytrid strain was extracted with petroleum ether and tested for antibacterial activity [2] separately for the comparison of synthesized nanoparticles and normal solvent extract. Ampicillin antibiotic was used as a positive control and distilled water as a negative control. The discs were then placed on inoculated agar plates and incubated at 37 °C for 24 h. Antimicrobial activity was measured as the diameter of the zone of inhibition in mm, excluding the paper disc diameter. All the experiments were performed in triplicate.
3. Results
3.1. Morphological and Molecular Identification
The thraustochytrid strain TSKK1 was identified as Thraustochytrium sp., as shown in Figure 1, according to its size, shape and zoospore formation, when visualized under a light microscope under 4×, 10×, 40× and 100× illumination (Figure 1a,b,d–f) and also under a Scanning Electron Microscope (Figure 1c), referring to morphological characters as proposed by [4,39]. The cytoplasmic content of the cell acted as sporangia to release zoospores and intracellular organelles such as vacuoles, nucleus, Golgi bodies and lipids (Figure 1d–f).
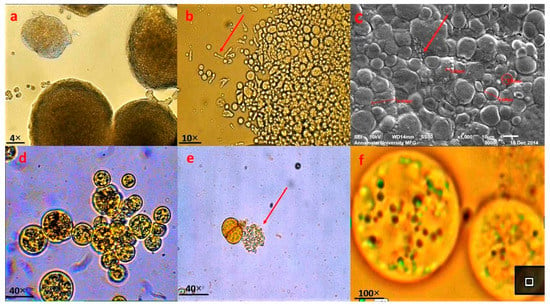
Figure 1.
Light and scanning electron microscopic (SEM) analysis of thraustochytrid strain TSKK1 (Thraustochytrium kinnei). (a) Clusters of vegetative cells dividing through binary cell division. (b) Clusters of smaller mature vegetative cells and many active zoospores and larger amoeboid cells. (c) Mature vegetative cells, active zoospores and larger amoeboid cells. (d) Mature zoosporangium with active zoospores as it is about to explode. (e) Vegetative cells and sporangiophores (just exploded in zoosporangium). (f) Many intercellular organelles, such as Golgi bodies, vacuoles, lipids, and nucleus.
To evaluate the phylogenetic relationship, a search of homologous sequences (Megablast algorithm) was performed. The 18S region of rRNA from thraustochytrid strain TSKK1 was sequenced as a result of the final alignment of 1796 bp, which was used for further comparison and conformation in NCBI analysis. Based on the NCBI BLAST analysis, the thraustochytrid strain TSKK1 (Genbank No. KT716334) was 100% similar to Thraustochytrium kinnei (Genbank No. DQ367053), confirming a close taxonomic classification to a member of the thraustochytrids family (Figure 2).
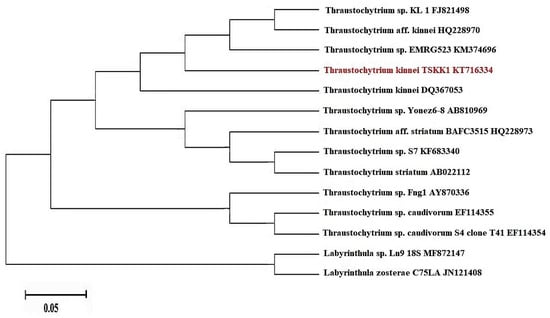
Figure 2.
Neighbor joining phylogenetic tree of thraustochytrid strain TSKK1 (Thraustochytrium kinnei) using 18S rRNA gene sequence data.
3.2. Selection of the Important Factors and Their Optimization for Biomass Production
The Plackett-Burman design was used to determine the important factors that influenced the biomass production using statistical modeling, and the actual and coded values are presented in Table 1. The factors that influenced biomass production were detected based on the coefficient value resulting from the two-level factorial design. The important factors of Thraustochytrium kinnei were found to be pH, days of incubation, bread crumbs and peptone (Table 2).

Table 1.
Experimental range and levels of independent variables.

Table 2.
Selection of the important biomass production influencing factors for the strains of Thraustochytrium kinnei (SE Coefficient = 1).
The application of RSM was based on the estimates of the parameters for biomass production in the strain as indicated as an experimental relationship between the response and input variables, expressed by the following quadratic model equation.
where Y is response of biomass production, A is pH, B is days of incubation, C is bread crumbs and D is peptone.
Y— Thraustochytrium kinnei biomass (g · dry wt · L−1) = 13.53 − 0.311A + 0.476B
− 0.208C + 0.485D − 0.381AB − 0.086AC + 0.058AD + 0.173BC − 0.041BD −
0.088CD − 3.113A2 − 2.566B2 − 1.868C2 − 1.965D2
− 0.208C + 0.485D − 0.381AB − 0.086AC + 0.058AD + 0.173BC − 0.041BD −
0.088CD − 3.113A2 − 2.566B2 − 1.868C2 − 1.965D2
The above selected factors were further optimized for maximum biomass production using 30 runs of Central Composite Design. The data are presented as experimental values along with predicted values of biomass production (Table 3) and tested by a quadratic model (Figure 3A–D).

Table 3.
Central composite design for the optimization of factors for biomass production by Thraustochytrium kinnei.
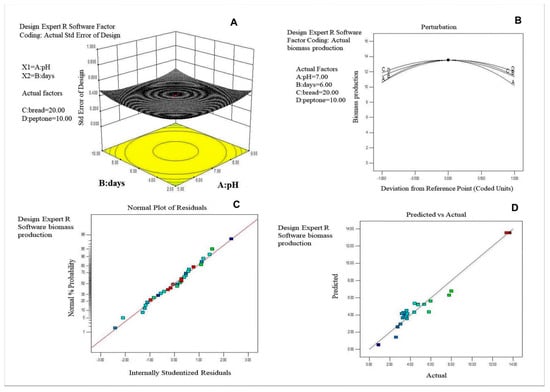
Figure 3.
(A) Three−dimensional standard error plot for biomass production of Thraustochytrid strain TSKK1 (Thraustochytrium kinnei). (B) Perturbation plot for biomass production. (C) Normal plot for the residuals and normal percentage of probability for the response of predicted and experimental values. (D) Predicted and actual experimental response for the biomass production.
Analysis of variance (ANOVA) of the regression model and interaction and combined effects of the factors on the biomass production is given in Table 4. The graphic representation of the interaction effects of the factors on biomass production is shown in Figure 4A–F. The maximum biomass production could be attained at the following optimal conditions: 7.16 pH, 6.22 days of incubation, 20.54 g·L−1 of bread crumbs and 10.14 g·L−1 of peptone. Under these optimized conditions, a maximum biomass of 13.53 g·L−1 was produced. The biomass was harvested through centrifugation, freeze-dried and stored at −4 °C until fatty acid analysis.

Table 4.
Analysis of variance for response of biomass production by Thraustochytrium kinnei.
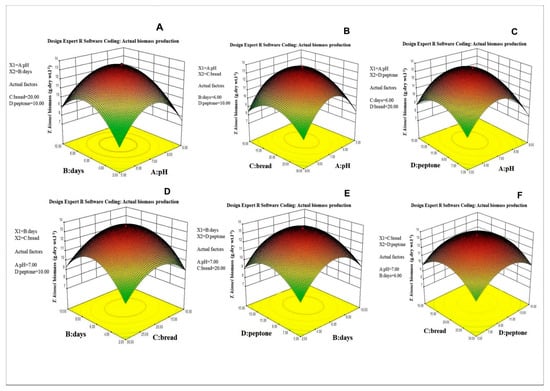
Figure 4.
Three−dimensional response surface plot for the (A) effect of incubation period (days) and pH, (B) effect of bread crumbs and pH, (C) effect of peptone and pH, (D) effect of incubation period (days) and bread crumbs, (E) effect of peptone and incubation period (days), and (F) effect of bread crumbs and peptone on biomass production of Thraustochytrid strain TSKK1 (Thraustochytrium kinnei).
3.3. Lipid and Fatty Acid Profiles of Thraustochytrid
The freeze-dried biomass of Thraustochytrium kinnei was analyzed for fatty acid profile using gas chromatography and mass spectrum (GC-MS). The total lipid (TL) of the strain was 41.33%. The major fatty acid component was palmitic acid (16:0), a saturated fatty acid (SFA) making up 31.81% of total fatty acids (TFA). The omega-3 poly unsaturated fatty acids (PUFAs) were eicosapendaenioc acid comprising up to 7.76%, docosapentaenoic acid comprising up to 2.09%, and docosahexaenoic acid comprising up to 39.16% (Table 5).

Table 5.
Fatty acid composition (expressed as % of the total fatty acids) of Thraustochytrium kinnei.
3.4. Synthesis and Characterization of Silver and Gold Nanoparticles
The color change of silver and gold nanoparticles was noted by visual observation in Thraustochytrium kinnei culture broth when incubated with solutions of silver nitrate and chloroauric acid in darkness. The solution showed a change in color to yellowish brown for silver and pinkish for gold, with intensity increasing during the period of incubation up to 16 days for silver nanoparticles (AgNPs) and 12 days for gold nanoparticles (AuNPs) (Figure 5a,b). The absorbance peak for the nanoparticles was observed at 430 nm for AgNPs and around 600 nm for AuNPs (Figure 6a,b). The shape and size of nanoparticles were confirmed by SEM. The micrograph showed nanoparticles that were cubical in nature. The particle size of AgNPs ranged from 5 to 90 nm, and AuNPs from 10 to 85 nm (Figure 7a,b). Dynamic light scattering (DLS) also revealed the size of nanoparticles in the range of 5–85 nm for AgNPs and 10–78 nm for AuNPs (Figure 8a,b).

Figure 5.
Nanoparticles synthesized by Thraustochytrium kinnei: (a) silver nanoparticles; (b) gold nanoparticles.
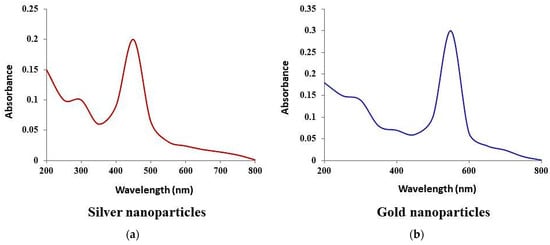
Figure 6.
UV−Vis spectrum of plasmon resonance of nanoparticles synthesized by Thraustochytrium kinnei: (a) silver; (b) gold.
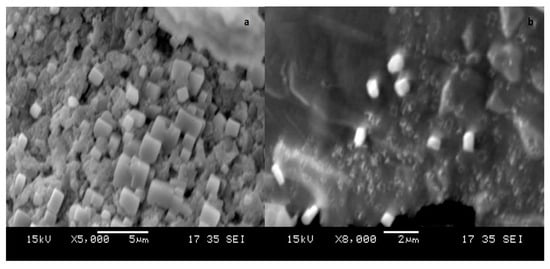
Figure 7.
SEM micrograph of (a) silver and (b) gold nanoparticles synthesized by Thraustochytrium kinnei.
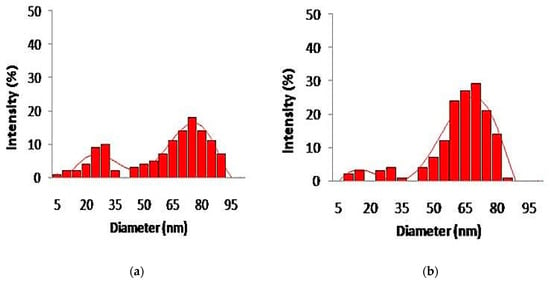
Figure 8.
The size distribution of nanoparticles synthesized by Thraustochytrium kinnei through Dynamic Light Scattering (DLS): (a) silver; (b) gold.
3.5. Antioxidant Activity
The nanoparticles synthesized from Thraustochytrium kinnei exhibited a high total antioxidant capacity (TAC) of 86.37 ± 1.52% for silver and 84.09 ± 2.39 for gold at higher concentrations of the extract (Figure 9a). The decrease in absorbance of the radical was due to hydrogen donation. It was visible, with color change from purple to yellow. DPPH-free radical scavenging activity was higher (87.27 ± 2.08%) for silver than that (85.54 ± 1.73) for gold at 500 µL/mL (Figure 9b). The methanolic extract of Thraustochytrium kinnei exhibited maximum total antioxidant capacity (TAC) (81.45 ± 2.03%), and DPPH-free radical scavenging activity (82.29 ± 1.73) (Figure 9a). All antioxidant activities were significant between the concentrations (p < 0.05) as compared with butylated hydroxyl toluene (BHT) as a control. Thus, the antioxidant activity was recorded as the highest for silver nanoparticles, followed by gold nanoparticles and petroleum ether extract.
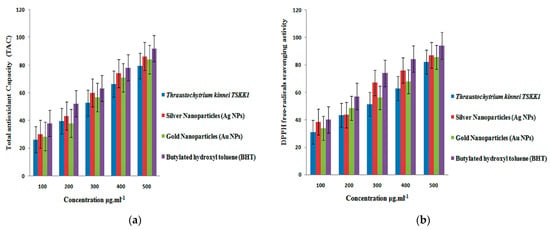
Figure 9.
In vitro antioxidant activity of silver and gold nanoparticles synthesized from mangrove-derived strain Thraustochytrium kinnei TSKK1 at different concentrations: (a) total antioxidant capacity; (b) DPPH free-radical scavenging activity.
3.6. Antibacterial Activity
The nanoparticles synthesized from Thraustochytrium kinnei showed antibacterial activity against 10 clinical pathogens. Among the nanoparticle extracts, the antibacterial activity was found to be high with silver nanoparticles compared to gold nanoparticles. The maximum (22.16 ± 1.52 mm) and the minimum activities (9.28 ± 1.36) were recorded against Klebsiella pneumonia (Figure 10) and Escherichia coli, respectively.
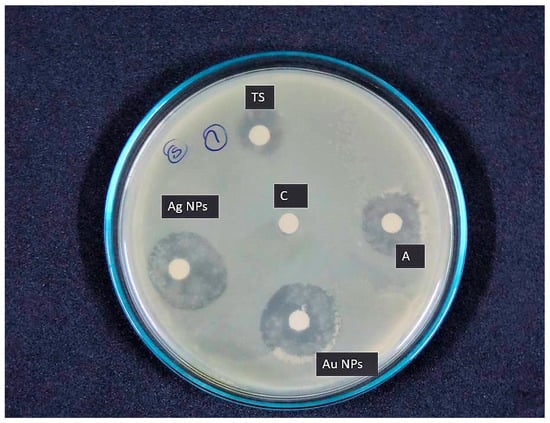
Figure 10.
In vitro antibacterial activity of silver and gold nanoparticles synthesized from mangrove-derived strain Thraustochytrium kinnei TSKK1 against Klebsiella pneumonia.
The gold nanoparticle extract showed the maximum activity (21.84 ± 2.00 mm) against Klebsiella pneumonia (Figure 10) and the minimum activity (8.06 ± 1.00 mm) against Escherichia coli. Interestingly, both the nanoparticles showed maximum and minimum activity for the same bacterial species. The antibacterial activity of Thraustochytrium kinnei extracted with petroleum ether showed maximum activity (13.10 ± 1.00 mm) against Staphylococcus aureus and minimum (6.69 ± 0.57 mm) against Escherichia coli (Figure 10). Ampicillin was used as a positive control, and it exhibited the maximum activity against Salmonella typhi (21 ± 1.52 mm) and the minimum activity against Streptococcus pyogens (14 ± 1.52 mm) (Table 6). Thus, the antibacterial activity was recorded as the highest for silver nanoparticle extract, followed by gold nanoparticle extract and petroleum ether extract.

Table 6.
Antibacterial activity (as indicated by inhibition zones) of nanoparticles and extracts of Thraustochytrium kinnei against clinical pathogens.
4. Discussion
Thraustochytrid taxonomy is generally based on morphology and lifecycle [39]. The identification of thraustochytrids is very difficult at the species level, due to the morphological similarities among them. Many morphological characters that are used for their classification overlap with each other [4]. In this regard, 18S rDNA sequences are important in understanding the taxonomy of thraustochytrids [32]. Moreover, the non-availability of much reference gene sequence data in Genbank is also a major drawback. Hence, the present study used both morphological and molecular techniques and identified the marine thraustochytrid strain as Thraustochytrium kinnei (NCBI Accession No. KT716334). The blast results showed high 18S sequence similarity with sequences of other thraustochytrids available in the Genbank database. The phylogenetic analyses confirmed that thraustochytrid strain TSKK1 was closely related to Thraustochytrium kinnei (Genbank No. DQ367053), Thraustochytrium aff. kinnei (Genbank: HQ228970), Thraustochytrium sp. KL1 (Genbank: FJ821498) and Thraustochytrium sp. EMRG523 (Genbank: KM374696).
Biomass productions of thraustochytrids are dependent on media composition and environmental conditions. Morphological characteristics of thraustochytrids are also changed by growth conditions, including nutrients, temperature, pH and salinity [40]. The statistically optimized conditions for maximum biomass production were pH of 7.16, 6.22 days of incubation and the media composition of bread crumbs (20.54 g·L−1) and peptone (10.14 g·L−1) (Table 3). The present study is in accordance with earlier reports that the biomass production of thraustochytrid species is highly dependent on carbon and nitrogen sources in the culture medium as well pH and days of incubation [12]. The maximum biomass production of 13.53 g·L−1 recorded in Thraustochytrium kinnei falls within the range of 0.8–48 g·L−1 under the temperature of 25–28 °C and incubation time of 4–12 days, as earlier reported [10,41].
The total lipid content in Thraustochytrium kinnei was found to be 41.33%, which is comparable with other reports on lipid content of thraustochytrids, ranging from 16 to 50% based on the dry biomass [42,43]. The present study recorded 43.68% saturated fatty acid, 4.90% monounsaturated fatty acid, and 51.42% polyunsaturated fatty acids in Thraustochytrium kinnei, in addition to better fatty acid profile in terms of essential fatty acids such as alpha-linolenic acid (ALA) (0.33%), eicosapentaenoic acid (EPA) (7.76%), docosahexaenoic acid (DHA) (39.16%), arachidonic acid (1.67%) and docosapentaenoic acid (DPA) (2.09%) (Table 5). Thus, thraustochytrids are promising for the production of essential fatty acids, in accordance with earlier reports [1,44].
While microorganisms such as bacteria, actinomycetes, and fungi continue to be investigated in metal nanoparticle synthesis, the use of their marine counterparts is limited [19,45]. The mangrove-derived bacteria, fungi and thraustochytrids were capable of reducing the silver ions at a faster rate with various antimicrobial applications [30]. The present study proved the potential of thraustochytrids for the synthesis of silver and gold nanoparticles. Similarly, Thraustochytrium sp. isolated from mangrove habitat is reported to be efficient in the biosynthesis of silver nanoparticles with the size of 50–100 nm [46]. The shape and size of silver (AgNPs) and gold (AuNPs) nanoparticles produced by Thraustochytrium kinnei were mostly cubical in nature with the size range of 5 to 90 nm. This range of size is in agreement with earlier work on soil fungi, reporting the silver nanoparticle size of 5–50 nm for Trichoderma reesei and gold nanoparticle size of 5–10 nm for Trichoderma koningii [47].
The antioxidant activity of silver and gold nanoparticles synthesized from Thraustochytrium kinnei was found to increase in a dose-dependent manner, which is congruent to the earlier findings with other species of thraustochytrids [3,26,48]. The present study observed DPPH radical scavenging activity of the silver (87.27 ± 2.08%) and gold (85.54 ± 1.73%) nanoparticles and also the total antioxidant capacity (TAC) for silver (86.37 ± 1.52%) and gold (84.09 ± 2.39%) nanoparticles. A similar observation was previously made with mangrove-derived marine thraustochytrids, with the highest activity being 78.95% [3]. Similarly, Schizochytrium sp., and Thraustochytrium striatum, rich in antioxidative components, are proven to be promising for application as health products or cosmetics [26,38]. The antioxidant activity is attributed to polyunsaturated fatty acid derivatives, which help to eliminate toxic free radicals [48]. To the best of our knowledge, this is the first study to evaluate the antioxidant capacity of two nanoparticle synthesized from thraustochytrid strain TSKK1 (Thraustochytrium kinnei) extracts. This result gives some insight into and guidance on the comprehensive and high-value-added utilization of these two nanoparticle synthesized Thraustochytrium kinnei extracts. Further studies need to be conducted to identify which potent antioxidant compounds present in Thraustochytrium kinnei can be further used for appropriate nutraceutical applications.
The silver nanoparticles in the present study showed antibacterial activity with the highest inhibition zone (22.16 ± 1.52 mm) against Klebsiella pneumonia (Table 6); this finding is in agreement with earlier work [49]. The gold nanoparticles also exhibited the maximum activity (21.84 ± 2.00 mm) against Klebsiella pneumonia (Table 6). This inhibitory activity is due to the destruction of the bacterial cell wall, as a result of hydrolytic enzymes produced by thraustochytrids [2,50]. The antibacterial activity is also attributed to fatty acids such as linoleic acid, which are proven to have antibacterial, antiviral, antitumor, anti-inflammatory, anti-proliferative and antioxidant activities [51]. Thraustochytrium striatum is reported to have antibacterial activity against Pseudomonas aeruginosa [26]. The activity varies with the crystallographic nature of nanoparticles [52]. However, the antimicrobial activities of silver and gold nanoparticles synthesized from Thraustochytrium kinnei were compared with distilled water as a negative control, which had no bacterial inhibition (data not shown). Our results recommend that nanoparticle synthesized from thraustochytrium kinnei extract exhibited the greatest inhibition activity when compared with petroleum ether extract. The nanoparticles synthesized from thraustochytrid extract could inhibit the growth of pathogenic bacteria. Further studies will be needed for the bioprospecting potential of thraustochytrids. As a final point we concluded that thraustochytrids from the decomposing mangrove leaves are future sources of DHA as well as antioxidant and antimicrobial compounds and could be considered for nutraceuticals.
5. Conclusions
The present study highlights the importance of mangrove-derived Thraustochytrid strain TSKK1 (Thraustochytrium kinnei) as a potential source for DHA production and nanoparticle synthesis. This work standardized the optimal culture conditions for high biomass production with a high yield of omega-3 fatty acids. Silver and gold nanoparticles synthesized from the strain showed good antioxidant and antibacterial activities. Further studies are required on possible use of Thraustochytrium kinnei in medical applications for omega-3 fatty acid production and as antioxidant and antimicrobial agents.
Author Contributions
K.K. (Kaliyamoorthy Kalidasan) designed the study, performed the laboratory experiments, prepared the original draft, performed the data analysis, and completed the writing, review, and editing of the manuscript; N.A. and V.G. helped to design the study, wrote the methodology and carried out the laboratory work. K.K. (Kandasamy Kathiresan) and L.D. supervised the work and reviewed, edited and approved the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research work was supported and funded by the Science and Engineering Research Board (SERB), New Delhi, under the National Postdoctoral Fellowship, PDF/2017/002579, dated 4 July 2018.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors are thankful to for DST-SERB fellowship, New Delhi (File Number: PDF/2017/002579) (K.K. (Kaliyamoorthy Kalidasan)) and UGC, New Delhi, for BSR Faculty Fellowship (K.K. (Kandasamy Kathiresan), N.A.). Kaliyamoorthy Kalidasan thanks the authorities of ANCOST-NIOT and Annamalai University, India, for providing the necessary facilities to carry out the project work.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Caamano, E.; Loperena, L.; Hinzpeter, I.; Pradel, P.; Gordillo, F.; Corsini, G.; Tello, M.; Lavini, P.; Gonzaleza, A.R. Isolation and molecular characterization of Thraustochytrium strain isolated from Antarctic Peninsula and its biotechnological potential in the production of fatty acids. Brazil. J. Microbiol. 2017, 48, 671–679. [Google Scholar] [CrossRef]
- Kalidasan, K.; Sunil, K.S.; Kayalvizhi, K.; Kathiresan, K. Polyunsaturated fatty acid-producing marine thraustochytrids: A potential source for antimicrobials. J. Coast. Life Med. 2015, 3, 848–851. [Google Scholar] [CrossRef]
- Kalidasan, K.; Sunil, K.S.; Narendran, R.; Kathiresan, K. Antioxidant activity of mangrove-derived marine thraustochytrids. Mycosphere 2015, 6, 602–611. [Google Scholar] [CrossRef]
- Marchan, F.L.; Chang, K.J.L.; Nichols, P.D.; Mitchell, W.J.; Polglase, J.L.; Gutierrez, T. Taxonomy, ecology and biotechnolog -ical applications of thraustochytrids: A Review. Biotechnol. Adv. 2017, 36, 26–46. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Ward, O.P. Omega-3/6 fatty acids: Alternative sources of production. Process. Biochem. 2005, 40, 3627–3652. [Google Scholar]
- Colomer, R.; Moreno-Nogueira, J.M.; Garcia-Luna, P.P.; Garcia-Peris, P.; Garcia-de-Lorenzo, A.; Zarazaga, A.; Quecedo, L.; del Llano, J.; Usan, L.; Casimiro, C. N-3 fatty acids, cancer and cachexia: A systematic review of the literature. Br. J. Nutr. 2007, 97, 823–831. [Google Scholar] [CrossRef]
- Swanson, D.; Block, R.; Mousa, S. Omega-3 fatty acids EPA and DHA: Health benefits throughout life. Adv. Nutr. An Int. Rev. J. 2012, 3, 1–7. [Google Scholar] [CrossRef]
- Takahata, K.; Monobe, K.; Tada, M.; Weber, P.C. The benefits and risks of n-3 polyunsaturated fatty acids. Biosci. Biotechnol. Biochem. 1998, 62, 2079–2085. [Google Scholar] [CrossRef]
- Horrocks, L.; Farooqui, A. Docosahexaenoic acid in the diet: Its importance in maintenance and restoration of neural membrane function. Prostaglandins Leukot. Essent. Fat. Acids 2004, 70, 361–372. [Google Scholar] [CrossRef]
- Nakahara, T.; Yakochi, T.; Higashihara, S.; Tanaki, T.; Yaguchi, T.; Honda, D. Production of docosahexaenoic and docosapentaenoic acids by Schizochytrium sp. isolated from Yap Island. J. Am. Oil Chem. Soc. 1996, 73, 1421–1426. [Google Scholar] [CrossRef]
- Manikan, K.V.; Hamid, M.S.; Abdul, A. Response surface optimization of culture medium for enhanced docosahexaenoic acid production by a Malaysian thraustochytrid. Sci. Rep. 2015, 5, 8611. [Google Scholar] [CrossRef]
- Sahin, D.; Tas, E.; Altindag, U.H. Enhancement of docosahexaenoic acid (DHA) production from Schizochytrium sp. S31 using different growth medium conditions. AMB Exp. 2018, 8, 7. [Google Scholar] [CrossRef]
- Nazir, Y.; Shuib, S.; Kalil, M.S.; Song, Y.; Hamid, A.A. Optimization of Culture Conditions for Enhanced Growth, Lipid and Docosahexaenoic Acid (DHA) Production of Aurantiochytrium SW1 by Response Surface Methodology. Sci. Rep. 2018, 8, 8909. [Google Scholar] [CrossRef] [PubMed]
- Murali, S.; Ahmad, A.; Khan, M.I.; Rajiv, K. Biosynthesis of metal nanoparticles using fungi and actinomycete. Curr. Sci. 2003, 85, 162–170. [Google Scholar]
- Rai, M.; Ingle, A.P.; Gaikwad, S.; Gupta, I.; Gade, A.; Silvério da Silva, S. Nanotechnology based anti-infective to fight microbial intrusions. J. Appl. Microbiol. 2016, 120, 527–542. [Google Scholar] [CrossRef]
- Saratale, R.G.; Saratale, G.D.; Shin, H.S.; Jacob, J.M.; Pugazhendhi, A.; Bhaisare, M.; Kumar, G. New insights on the green synthesis of metallic nanoparticles using plant and waste biomaterials: Current knowledge, their agricultural and environmental applications. Environ. Sci. Pollut. Res. 2017, 11, 10164–10183. [Google Scholar] [CrossRef] [PubMed]
- Mohanpuria, P.; Rana, N.K.; Yadav, S.K. Biosynthesis of nanoparticles: Technological concepts and future applications. J. Nanopart. Res. 2008, 10, 507–517. [Google Scholar] [CrossRef]
- Wypij, M.; Czarnecka, J.; Świecimska, M.; Dahm, H.; Rai, M.; Golinska, P. Synthesis, characterization and evaluation of antimicrobial and cytotoxic activities of biogenic silver nanoparticles synthesized from Streptomyces xinghaiensis OF1 strain. World J. Microbiol. Biotechnol. 2018, 34, 23. [Google Scholar] [CrossRef] [PubMed]
- Shankar, P.D.; Shobana, S.; Karuppusamy, I.; Pugazhendhi, A.; Ramkumar, V.S.; Arvindnarayan, S.; Kumar, G. A review on the biosynthesis of metallic nanoparticles (gold and silver) using bio-components of microalgae: Formation mechanism and applications. Enzym. Microb. Technol. 2016, 95, 28–44. [Google Scholar] [CrossRef]
- Ramkumar, V.S.; Pugazhendhi, A.; Gopalakrishnan, K.; Sivagurunathan, P.; Saratale, G.D.; Dung, T.N.B.; Kannapiran, E. Bio fabrication and characterization of silver nanoparticles using aqueous extract of seaweed Enteromorpha compressa and its biomedical properties. Biotechnol. Rep. 2017, 14, 1–7. [Google Scholar] [CrossRef]
- Visudtiphole, V.; Phromson, M.; Tala, S.; Bunphimpapha, P.; Raweeratanapong, T.; Sittikankaew, K.; Arayamethakorn, S.; Preedanon, S.; Jangsutthivorawat, W.; Chaiyapechara, S.; et al. Aurantiochytrium limacinum BCC52274 improves growth, hypo-salinity tolerance and swimming strength of Penaeus vannamei post larvae. Aquaculture 2018, 495, 849–857. [Google Scholar] [CrossRef]
- Chen, C.Y.; Lee, M.H.; Leong, Y.K.; Chang, J.S.; Lee, D.J. Biodiesel production from heterotrophic oleaginous microalga Thraustochytrium sp. BM2 with enhanced lipid accumulation using crude glycerol as alternative carbon source. Bioresour. Technol. 2020, 306, 123113. [Google Scholar] [CrossRef] [PubMed]
- Bongiorni, L.; Pusceddu, A.; Danovaro, R. Enzymatic activities of epiphytic and benthic thraustochytrids involved inorganic matter degradation. Aquat. Microb. Ecol. 2005, 41, 299–305. [Google Scholar] [CrossRef]
- Park, H.; Kwak, M.; Seo, J.; Ju, J.; Heo, S.; Park, S.; Hong, W. Enhanced production of carotenoids using a Thraustochytrid microalgal strain containing high levels of docosahexaenoic acid-rich oil. Bioprocess Biosyst. Eng. 2018, 41, 1355–1370. [Google Scholar] [CrossRef]
- Patel, A.; Liefeldt, S.; Rova, U.; Christakopoulos, P.; Matsakas, L. Co-production of DHA and squalene by thraustochytrid from forest biomass. Sci. Rep. 2020, 10, 1992. [Google Scholar] [CrossRef]
- Xiao, R.; Xi, Y.; Mi, L.; Xiang, L.; Yanzhang, W.; Min, C.; Arthur, R.; Mark, T.; Junhuan, D.; Yi, Z. Investigation of Composition, Structure and Bioactivity of Extracellular Polymeric Substances from Original and Stress-induced Strains of Thraustochytrium striatum. Carbohyd. Polymer 2018, 195, 515–524. [Google Scholar] [CrossRef]
- Ramos-Vega, A.; Rosales-Mendoza, S.; Bañuelos-Hernández, B.; Angulo, C. Prospects on the Use of Schizochytrium sp. to Develop Oral Vaccines. Front. Microbiol. 2018, 9, 2506. [Google Scholar] [CrossRef]
- Shakeri, S.; Amoozyan, N.; Fekrat, F.; Maleki, M. Antigastric cancer bioactive Aurantiochytrium oil rich in Docosahexaenoic acid: From media optimization to cancer cells cytotoxicity assessment. J. Food. Sci. 2017, 82, 2706–2718. [Google Scholar] [CrossRef]
- Schmitt, D.; Tran, N.; Peach, J.; Edwards, T.; Greeley, M. Toxicologic evaluations of DHA-rich algal oil in rats: Developmental toxicity study and 3-month dietary toxicity study with an in utero exposure phase. Food Chem. Toxicol. 2012, 50, 4149–4157. [Google Scholar] [CrossRef]
- Asmathunisha, N.; Kathiresan, K. A review on biosynthesis of nanoparticles by marine organisms. Colloids. Surf. B Biointerfaces 2013, 103, 283–287. [Google Scholar] [CrossRef]
- Ravinder Singh, C.; Kathiresan, K.; Anandhan, S. A review on marine based nanoparticles and their potential applications. Afr. J. Biotechnol. 2015, 14, 1525–1532. [Google Scholar] [CrossRef]
- Raghukumar, S. Ecology of the marine protists, the labyrinthulomycetes (thraustochytrids and labyrinthulids). Eur. J. Protistol. 2002, 38, 127–145. [Google Scholar] [CrossRef]
- Mo, J.C.; Rinkevich, D.B. Development of a PCR strategy for thraustochytrid identification based on 18S rDNA sequence. Mar. Biol. 2002, 140, 883–889. [Google Scholar] [CrossRef]
- Honda, D.; Yokochi, T.; Nakahara, T.; Raghukumar, S.; Nakagiri, A.; Schaumann, K.; Higashihara, T. Molecular phylogeny of labyrinthulids and thraustochytrids based on the sequencing of 18S ribosomal RNA gene. J. Eukaryot. Microbiol. 1999, 46, 637–647. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Folch, J.; Lees, M.; Sloane Stanley, G.H. A simple method for the isolation and purification of the total lipids from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef]
- Sasser, M. Identification of Bacteria by Gas Chromatography of Cellular Fatty Acids; MIDI Technical Note #101: Newark, DE, USA, 1990. [Google Scholar]
- Yu, J.H.; Wang, Y.; Sun, J.; Bian, F.; Chen, G.; Zhang, Y.; Bi, Y.P.; Wu, Y.J. Antioxidant activity of alcohol aqueous extracts of Crypthecodinium cohnii and Schizochytrium sp. J. Zhejiang Univ. Sci. B 2017, 18, 797–806. [Google Scholar] [CrossRef]
- Booth, T.; Miller, C.E. Comparative morphologic and taxonomic studies in the genus Thraustochytrium. Mycologia 1968, 60, 480–495. [Google Scholar] [CrossRef]
- Wethered, J.M.; Jennings, D.H. Major solutes contributing to solute potential of Thraustochytrium aureum and T. roseum after growth in media of different salinities. Trans. Br. Mycol. Soc. 1985, 85, 439–446. [Google Scholar] [CrossRef]
- Yaguchi, T.; Tanaka, S.; Yokochi, T.; Nakahara, T.; Higashihara, T. Production of high yields of docosahexaenoic acid by Schizochytrium sp. strain SR21. J. Am. Oil Chem. Soc. 1997, 74, 143–1434. [Google Scholar] [CrossRef]
- Iida, I.; Nakahara, T.; Yokochi, T.; Kamisaka, Y.; Yagi, H.; Yamaoka, M.; Suzuki, O. Improvement of docosahexaenoic acid production in a culture of Thraustochytrium aureum by medium optimization. J. Ferment. Bioeng. 1996, 81, 76–78. [Google Scholar] [CrossRef]
- Weete, J.D.; Kim, H.; Gandhi, S.R.; Wang, Y.; Dute, R. Lipids and ultrastructure of Thraustochytrium sp. Lipids 1997, 32, 839–845. [Google Scholar] [CrossRef]
- Kabilan, C.; Roy, R.K.; Chadha, A. Docosahexaenoic acid production by a novel high yielding strain of Thraustochytrium sp. of Indian origin: Isolation and bioprocess optimization studies. Algal Res. 2018, 32, 93–100. [Google Scholar] [CrossRef]
- Mandal, D.; Bolander, M.E.; Mukhopadhyay, D.; Sarkar, G.; Mukherjee, P. The use of microorganisms for the formation of metal nanoparticles and their application. Appl. Microbiol. Biotechnol. 2006, 69, 485–492. [Google Scholar] [CrossRef] [PubMed]
- Gomathi, V. Studies on Thraustochytrids of Mangrove Sediments for Poly Unsaturated Fatty Acids and Nanoparticles Synthesis. Master’s Thesis, Annamai University, Tamil Nadu, India, 2009; pp. 63–65. [Google Scholar]
- Vahabi, K.; Ali Mansoori, G.; Karimi, S. Biosynthesis of silver nanoparticles by fungus Trichoderma Reesei. Insci. J. 2011, 1, 65–79. [Google Scholar] [CrossRef]
- Plaza, M.; Herrero, M.; Cifuentes, A.; Ibanez, E. Innovative natural functional ingredients from microalgae. J. Agric. Food Chem. 2009, 57, 7159–7170. [Google Scholar] [CrossRef] [PubMed]
- Phanjom, P.; Ahmed, G. Effect of different physicochemical conditions on the synthesis of silver nanoparticles using fungal cell filtrate of Aspergillus oryzae (MTCC No. 1846) and their antibacterial effects. Adv. Nat. Sci. Nanosci. Nanotechnol. 2017, 8. [Google Scholar] [CrossRef]
- Taoka, Y.; Nagano, N.; Kai, H.; Hayashi, M. Degradation of distillery lees (shochukasu) by cellulose-producing thraustochytrids. J. Oleo Sci. 2017, 66, 31–40. [Google Scholar] [CrossRef][Green Version]
- Zhang, H.; Zhang, L.; Peng, L.; Dong, X.; Wu, D.; Wu, V.C.; Feng, F. Quantitative structure activity relationships of antimicrobial fatty acids and derivatives against Staphylococcus aureus. J. Zhejiang Univ. Sci. B 2012, 13, 83–93. [Google Scholar] [CrossRef]
- Manjunath, H.M.; Joshi, C.G. Characterization, antioxidant and antimicrobial activity of silver nanoparticles synthesized using marine endophytic fungus Cladosporium cladosporioides. Process. Biochem. 2019, 82, 199–204. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).