Folate-Mediated One-Carbon Metabolism in the Crustacean Copepod Calanus finmarchicus: Identification of Transcripts and Relative Expression across Development
Abstract
1. Introduction
2. Materials and Methods
2.1. In Silico Mining for Transcripts Encoding Enzymes Involved in the One-Carbon Metabolic Pathway
2.2. Cladogram of Calanus finmarchicus Transcripts Encoding Enzymes Involved in the Folate and Methionine Cycles, with Other Marine Organisms
2.3. Expression of Calanus finmarchicus Transcripts Encoding Enzymes across Development
3. Results
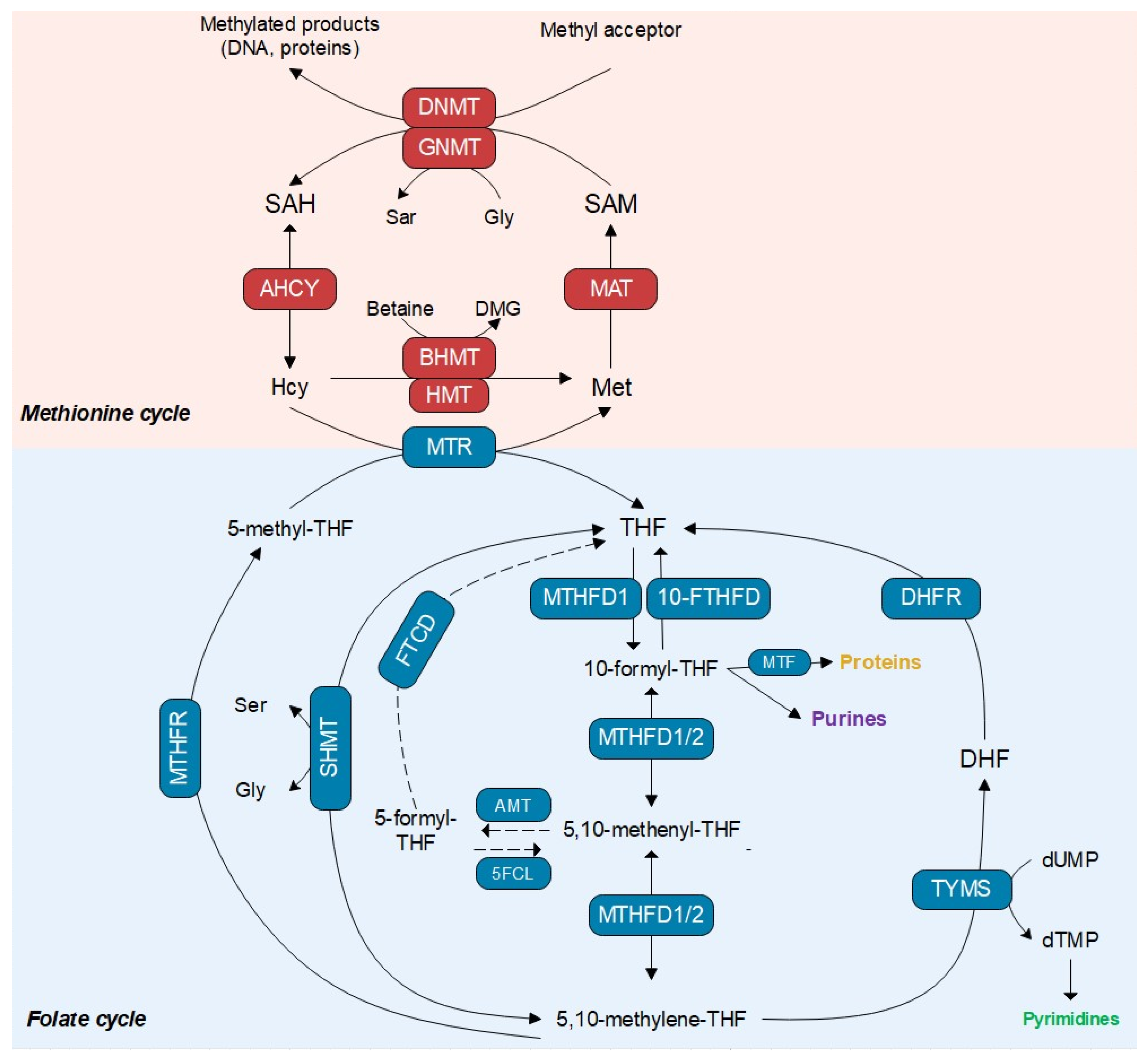
3.1. Identification of Calanus finmarchicus Transcripts Encoding Enzymes in the Folate Cycle
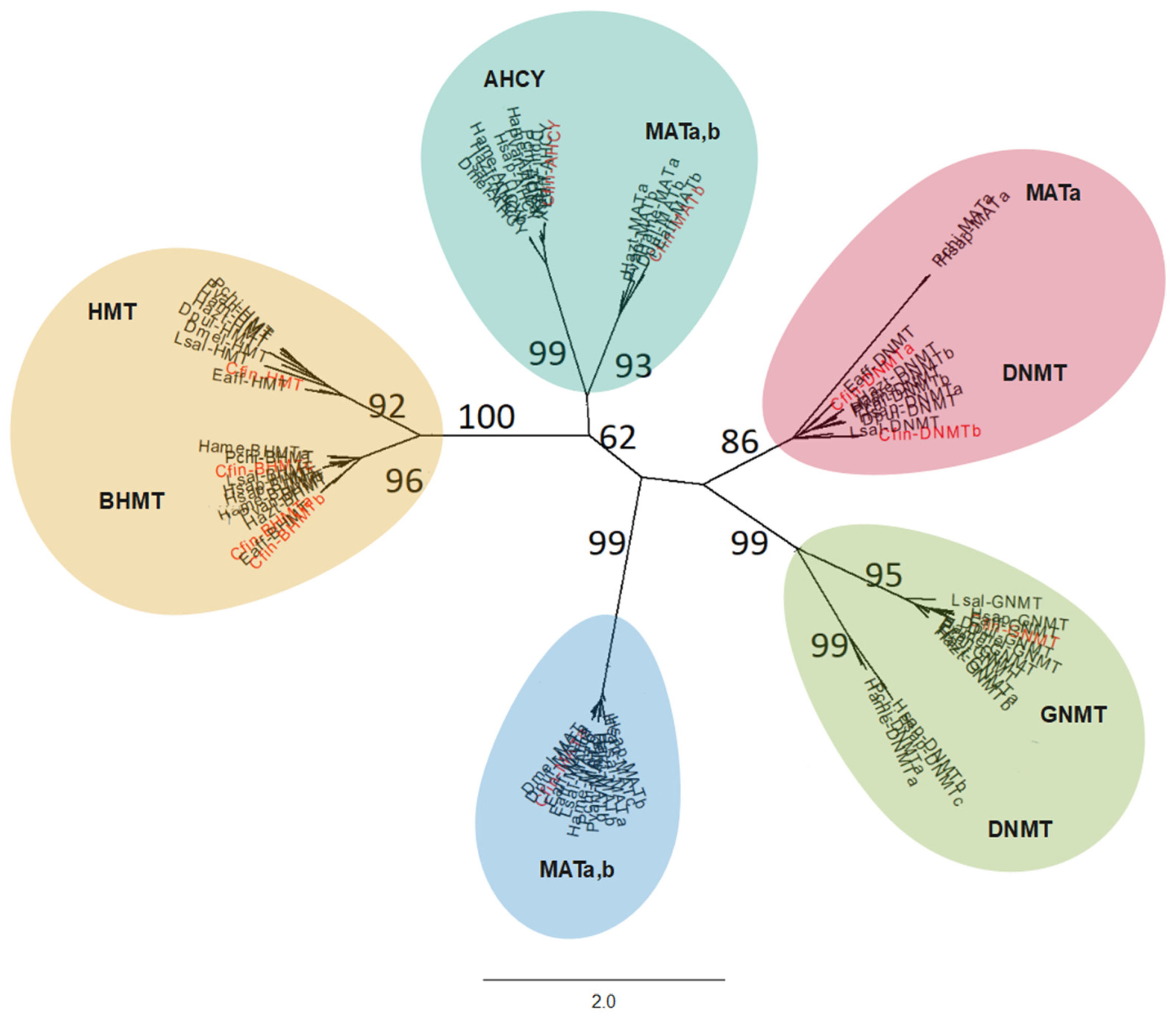
3.2. Comparison of C. finmarchicus Enzymes in the 1C Metabolism with Other Marine Organisms
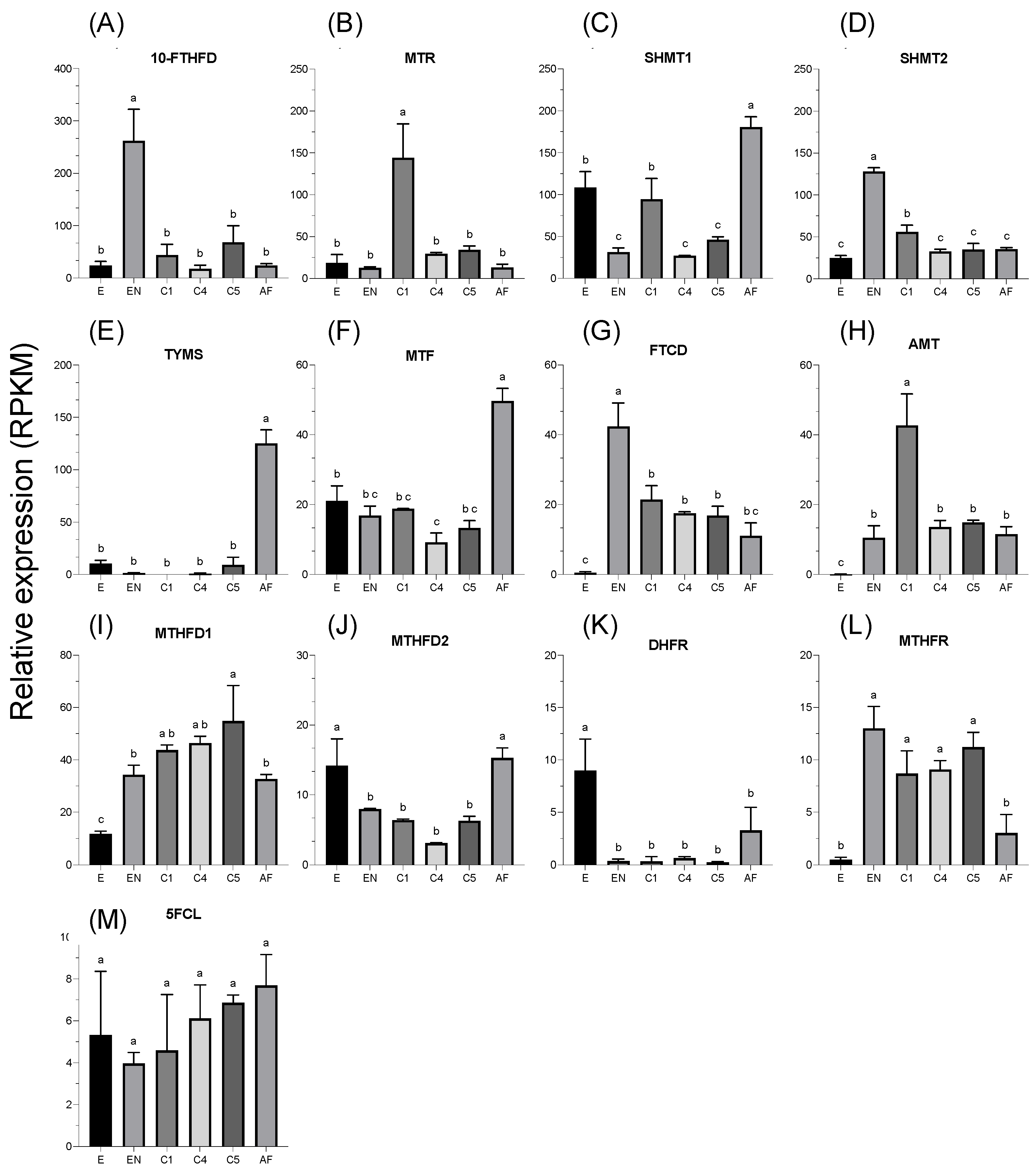
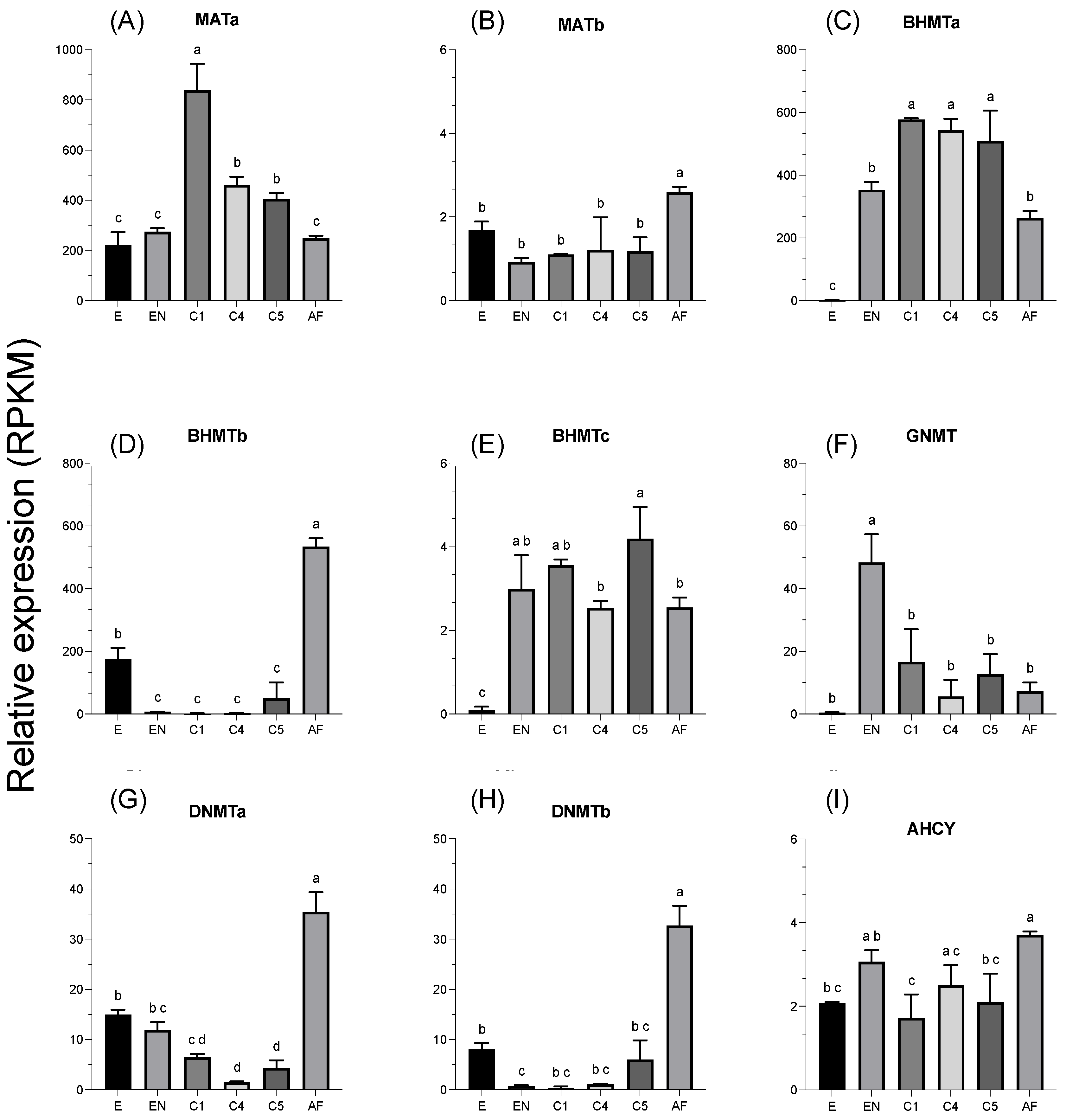
3.3. Relative Expression of Enzymes across Development
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rébeillé, F.; Ravanel, S.; Jabrin, S.; Douce, R.; Storozhenko, S.; Van Der Straeten, D. Folates in plants: Biosynthesis, distribution, and enhancement. Physiol. Plant. 2006, 126, 330–342. [Google Scholar] [CrossRef]
- Ravanel, S.; Douce, R.; Rébeillé, F. Metabolism of folates in plants. In Advances in Botanical Research; Elsevier: Amsterdam, The Netherlands, 2011; Volume 59, pp. 67–106. [Google Scholar]
- Fox, J.T.; Stover, P.J. Folate-mediated one-carbon metabolism. Vitam. Horm. 2008, 79, 1–44. [Google Scholar] [PubMed]
- Zheng, Y.; Cantley, L.C. Toward a better understanding of folate metabolism in health and disease. J. Exp. Med. 2019, 216, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Korsmo, H.W.; Jiang, X. One carbon metabolism and early development: A diet-dependent destiny. Trends Endocrinol. Metab. 2021, 32, 579–593. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.S.; Bonner, J.R.; Bernard, D.J.; Sanchez, E.L.; Sause, E.T.; Prentice, R.R.; Burgess, S.M.; Brody, L.C. Disruption of the folate pathway in zebrafish causes developmental defects. BMC Dev. Biol. 2012, 12, 12. [Google Scholar] [CrossRef] [PubMed]
- Padmanabhan, N.; Jia, D.; Geary-Joo, C.; Wu, X.; Ferguson-Smith, A.C.; Fung, E.; Bieda, M.C.; Snyder, F.F.; Gravel, R.A.; Cross, J.C. Mutation in folate metabolism causes epigenetic instability and transgenerational effects on development. Cell 2013, 155, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Greene, N.D.; Stanier, P.; Copp, A.J. Genetics of human neural tube defects. Hum. Mol. Genet. 2009, 18, R113–R129. [Google Scholar] [CrossRef] [PubMed]
- Affleck, J.G.; Neumann, K.; Wong, L.; Walker, V.K. The effects of methotrexate on Drosophila development, female fecundity, and gene expression. Toxicol. Sci. 2006, 89, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Affleck, J.G.; Walker, V.K. Transgenic rescue of methotrexate-induced teratogenicity in Drosophila melanogaster. Toxicol. Sci. 2007, 99, 522–531. [Google Scholar] [CrossRef]
- Hernandorena, A. The patterning function of purines in the brine shrimp Artemia. Comptes Rendus L’académie Sci. Ser. III-Sci. Vie 1999, 322, 289–301. [Google Scholar] [CrossRef]
- Asai, S.; Sanges, R.; Lauritano, C.; Lindeque, P.K.; Esposito, F.; Ianora, A.; Carotenuto, Y. De novo transcriptome assembly and gene expression profiling of the copepod Calanus helgolandicus feeding on the PUA-producing diatom Skeletonema marinoi. Mar. Drugs 2020, 18, 392. [Google Scholar] [CrossRef] [PubMed]
- Hansen, B.H.; Tarrant, A.M.; Lenz, P.H.; Roncalli, V.; Almeda, R.; Broch, O.J.; Altin, D.; Tollefsen, K.E. Effects of petrogenic pollutants on North Atlantic and Arctic Calanus copepods: From molecular mechanisms to population impacts. Aquat. Toxicol. 2023, 267, 106825. [Google Scholar] [CrossRef] [PubMed]
- Melle, W.; Runge, J.; Head, E.; Plourde, S.; Castellani, C.; Licandro, P.; Pierson, J.; Jonasdottir, S.; Johnson, C.; Broms, C. The North Atlantic Ocean as habitat for Calanus finmarchicus: Environmental factors and life history traits. Prog. Oceanogr. 2014, 129, 244–284. [Google Scholar] [CrossRef]
- Christie, A.E.; Fontanilla, T.M.; Nesbit, K.T.; Lenz, P.H. Prediction of the protein components of a putative Calanus finmarchicus (Crustacea, Copepoda) circadian signaling system using a de novo assembled transcriptome. Comp. Biochem. Physiol. Part D Genom. Proteom. 2013, 8, 165–193. [Google Scholar] [CrossRef] [PubMed]
- Lenz, P.H.; Roncalli, V.; Hassett, R.P.; Wu, L.-S.; Cieslak, M.C.; Hartline, D.K.; Christie, A.E. De novo assembly of a transcriptome for Calanus finmarchicus (Crustacea, Copepoda)–the dominant zooplankter of the North Atlantic Ocean. PLoS ONE 2014, 9, e88589. [Google Scholar] [CrossRef] [PubMed]
- Roncalli, V.; Cieslak, M.C.; Passamaneck, Y.; Christie, A.E.; Lenz, P.H. Glutathione S-transferase (GST) gene diversity in the crustacean Calanus finmarchicus–contributors to cellular detoxification. PLoS ONE 2015, 10, e0123322. [Google Scholar]
- Tarrant, A.M.; Baumgartner, M.F.; Hansen, B.H.; Altin, D.; Nordtug, T.; Olsen, A.J. Transcriptional profiling of reproductive development, lipid storage and molting throughout the last juvenile stage of the marine copepod Calanus finmarchicus. Front. Zool. 2014, 11, 91. [Google Scholar] [CrossRef] [PubMed]
- Tarrant, A.M.; Baumgartner, M.F.; Lysiak, N.S.; Altin, D.; Størseth, T.R.; Hansen, B.H. Transcriptional profiling of metabolic transitions during development and diapause preparation in the copepod Calanus finmarchicus. Integr. Comp. Biol. 2016, 56, 1157–1169. [Google Scholar] [CrossRef]
- Christie, A.E.; Fontanilla, T.M.; Roncalli, V.; Cieslak, M.C.; Lenz, P.H. Identification and developmental expression of the enzymes responsible for dopamine, histamine, octopamine and serotonin biosynthesis in the copepod crustacean Calanus finmarchicus. Gen. Comp. Endocrinol. 2014, 195, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Christie, A.E.; Fontanilla, T.M.; Roncalli, V.; Cieslak, M.C.; Lenz, P.H. Diffusible gas transmitter signaling in the copepod crustacean Calanus finmarchicus: Identification of the biosynthetic enzymes of nitric oxide (NO), carbon monoxide (CO) and hydrogen sulfide (H2S) using a de novo assembled transcriptome. Gen. Comp. Endocrinol. 2014, 202, 76–86. [Google Scholar] [CrossRef]
- Porter, M.L.; Steck, M.; Roncalli, V.; Lenz, P.H. Molecular characterization of copepod photoreception. Biol. Bull. 2017, 233, 96–110. [Google Scholar] [CrossRef] [PubMed]
- Roncalli, V.; Uttieri, M.; Capua, I.D.; Lauritano, C.; Carotenuto, Y. Chemosensory-Related Genes in Marine Copepods. Mar. Drugs 2022, 20, 681. [Google Scholar] [CrossRef]
- Roncalli, V.; Lauritano, C.; Carotenuto, Y. First report of OvoA gene in marine arthropods: A new candidate stress biomarker in copepods. Mar. Drugs 2021, 19, 647. [Google Scholar] [CrossRef] [PubMed]
- Roncalli, V.; Uttieri, M.; Carotenuto, Y. The distribution of ferritins in marine copepods. J. Mar. Sci. Eng. 2023, 11, 1187. [Google Scholar] [CrossRef]
- Roncalli, V.; Christie, A.E.; Sommer, S.A.; Cieslak, M.C.; Hartline, D.K.; Lenz, P.H. A deep transcriptomic resource for the copepod crustacean Labidocera madurae: A potential indicator species for assessing near shore ecosystem health. PLoS ONE 2017, 12, e0186794. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Cieslak, M.C.; Castelfranco, A.M.; Roncalli, V.; Lenz, P.H.; Hartline, D.K. t-Distributed Stochastic Neighbor Embedding (t-SNE): A tool for eco-physiological transcriptomic analysis. Mar. Genom. 2020, 51, 100723. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Hien, D.; Doolgindachbaporn, S. Effect of niacin and folic acid in feed rations on growth and live weights of green catfish (Mystus nemurus Valenciennes 1840). Pak. J. Biol. Sci. 2011, 14, 64–68. [Google Scholar] [CrossRef]
- Lin, Y.-H.; Lin, H.-Y.; Shiau, S.-Y. Dietary folic acid requirement of grouper, Epinephelus malabaricus, and its effects on non-specific immune responses. Aquaculture 2011, 317, 133–137. [Google Scholar] [CrossRef]
- Badran, M.F.; Ali, M.A. Effects of folic acid on growth performance and blood parameters of flathead grey mullet, Mugil cephalus. Aquaculture 2021, 536, 736459. [Google Scholar] [CrossRef]
- Shiau, S.-Y.; Huang, S.-Y. Dietary folic acid requirement determined for grass shrimp, Penaeus monodon. Aquaculture 2001, 200, 339–347. [Google Scholar] [CrossRef]
- National Research Council, Division on Earth, Life Studies, Committee on the Nutrient Requirements of Fish, & Shrimp. Nutrient Requirements of Fish and Shrimp; National Academies Press: Washington, DC, USA, 2011. [Google Scholar]
- Cripps, G.; Lindeque, P.; Flynn, K.J. Have we been underestimating the effects of ocean acidification in zooplankton? Glob. Chang. Biol. 2014, 20, 3377–3385. [Google Scholar] [CrossRef]
- Turner, J.T. The importance of small planktonic copepods and their roles in pelagic marine food webs. Zool. Stud 2004, 43, 255–266. [Google Scholar]
- Choi, B.-S.; Kim, D.-H.; Kim, M.-S.; Park, J.C.; Lee, Y.H.; Kim, H.-J.; Jeong, C.-B.; Hagiwara, A.; Souissi, S.; Lee, J.-S. The genome of the European estuarine calanoid copepod Eurytemora affinis: Potential use in molecular ecotoxicology. Mar. Pollut. Bull. 2021, 166, 112190. [Google Scholar] [CrossRef]
- Miller, C.B.; Cowles, T.J.; Wiebe, P.H.; Copley, N.J.; Grigg, H. Phenology in Calanus finmarchicus; hypotheses about control mechanisms. Mar. Ecol. Prog. Ser. 1991, 72, 79–91. [Google Scholar] [CrossRef]
- Mauchline, J. The Biology of Calanoid Copepods; Advances in Marine Biology Book Series Volume 33; Academic Press: Cambridge, MA, USA, 1998. [Google Scholar]
- Marshall, S.M.; Orr, A.P. The Biology of a Marine Copepod: Calanus finmarchicus (Gunnerus); Springer Science & Business Media: Berlin/Heidelberg, Germany, 2013; ISBN 3662131382/9783662131381. [Google Scholar]
- Chang, W.-N.; Lee, G.-H.; Kao, T.-T.; Lin, C.-Y.; Hsiao, T.-H.; Tsai, J.-N.; Chen, B.-H.; Chen, Y.-H.; Wu, H.-R.; Tsai, H.-J. Knocking down 10-Formyltetrahydrofolate dehydrogenase increased oxidati ve stress and impeded zebrafish embryogenesis by obstructing morphogenetic movement. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2014, 1840, 2340–2350. [Google Scholar] [CrossRef]
- Tani, H.; Mito, T.; Velagapudi, V.; Ishikawa, K.; Umehara, M.; Nakada, K.; Suomalainen, A.; Hayashi, J.-I. Disruption of the mouse Shmt2 gene confers embryonic anaemia via foetal liver-specific metabolomic disorders. Sci. Rep. 2019, 9, 16054. [Google Scholar] [CrossRef]
- Roncalli, V.; Sommer, S.A.; Cieslak, M.C.; Clarke, C.; Hopcroft, R.R.; Lenz, P.H. Physiological characterization of the emergence from diapause: A transcriptomics approach. Sci. Rep. 2018, 8, 12577. [Google Scholar] [CrossRef]
- Roy, M.; Leclerc, D.; Wu, Q.; Gupta, S.; Kruger, W.D.; Rozen, R. Valproic acid increases expression of methylenetetrahydrofolate reductase (MTHFR) and induces lower teratogenicity in MTHFR deficiency. J. Cell. Biochem. 2008, 105, 467–476. [Google Scholar] [CrossRef]
- Blatch, S.; Meyer, K.W.; Harrison, J.F. Effects of dietary folic acid level and symbiotic folate production on fitness and development in the fruit fly Drosophila melanogaster. Fly 2010, 4, 312–319. [Google Scholar] [CrossRef]
- Snyder, A.K.; Rio, R.V. “Wigglesworthia morsitans” folate (vitamin B9) biosynthesis contributes to tsetse host fitness. Appl. Environ. Microbiol. 2015, 81, 5375–5386. [Google Scholar] [CrossRef]
- Carotenuto, Y.; Esposito, F.; Pisano, F.; Lauritano, C.; Perna, M.; Miralto, A.; Ianora, A. Multi-generation cultivation of the copepod Calanus helgolandicus in a re-circulating system. J. Exp. Mar. Biol. Ecol. 2012, 418, 46–58. [Google Scholar] [CrossRef]

| Enzyme Name | E.C. | NCBI Transcript # | Blast-p E-Value | Top Hit Species | Deduced Protein Length |
|---|---|---|---|---|---|
| dihydrofolate reductase (DHFR) | 1.5.1.3 | GAXK01099950 | 1.55 × 10−33 | Eurytemora affinis | F |
| methylenetetrahydrofolate dehydrogenase (MTHFD) | 6.3.4.3 (I)/3.5.4.9 (II)/1.5.1.5 (III) | GAXK01161413 | 0 | Eurytemora affinis | F |
| methylenetetrahydrofolate dehydrogenase (MTHFD2) | 1.5.1.15 | GAXK01151792 | 4.04 × 10−131 | Eurytemora affinis | F |
| thymidylate synthase (TYMS) | 2.1.1.45 | GAXK01188281 | 1.47 × 10−92 | Eurytemora affinis | F |
| methylene-THF reductase (MTHFR) | 1.5.1.53 1.5.1.20 | GAXK01056182 | 0 | Penaeus chinensis | F |
| serine hydroxymethyltransferase (SHMT1) | 2.1.2.1 | GAXK01188425-cit | 0 | Eurytemora affinis | F |
| serine hydroxymethyltransferase (SHMT2) | 2.1.2.1 | GAXK01186194-mit | 0 | Eurytemora affinis | F |
| 5-methyl-THF homocysteine methyltransferase (MTR) | 2.1.1.13 | GAXK01188527 | 0 | Eurytemora affinis | F |
| methionyl-tRNA formyltransferase (MTF) | 2.1.2.9 | GAXK01158471 | 1.06 × 10−68 | Harmonia axyridis | P |
| formimidoyltransferase-cyclodeaminase (FTCD) | 2.1.2.5 4.3.1.4 | GAXK01113306 | 0 | Eurytemora affinis | F |
| Aminomethyltransferase (AMT) | 2.1.2.10 | GAXK01044023 | 0 | Eurytemora affinis | F |
| 5-formyl-THF cycloligase (5FCL) | 6.3.3.2 | GAXK01024325 | 1.98 × 10−61 | Amphibalanus amphitrite | F |
| formyl-THF dehydrogenase (10-FTHFDH) | 1.5.1.6 | GAXK01171303 | 0 | Eurytemora affinis | F |
| Enzyme Name | E.C. | NCBI Transcript # | Blasp E-Value | Top Hit Species | Deduced Protein Length |
|---|---|---|---|---|---|
| betaine–homocysteine S-methyltransferase (BHMT) | 2.1.1.5 | GAXK01169231 (a) GAXK01064100 (b) GAXK01101229 (c) | 0 0 0 | Eurytemora affinis Eurytemora affinis Eurytemora affinis | F F F |
| glycine N-methyltransferase (GNMT) | 2.1.1.20 | GAXK01096711 | 1.44 × 10−166 | Eurytemora affinis | F |
| S-adenosylmethionine synthetase (MAT) | 2.5.1.6 | GAXK01168224 (a) GAXK01150749 (b) GAXK01026996 (c) | 0 1.65 × 10−89 0 | Eurytemora affinis Armadillidium nasatum Eurytemora affinis | F F F |
| homocysteine S-methyltransferase 3 (HMT) | 2.1.1.10 | GAXK01160528 | 0 | Eurytemora affinis | F |
| DNA (cytosine-5)-methyltransferase 1 (DNMT) | 2.1.1.37 | GAXK01011690(a) GAXK01002418(b) | 0 | Penaeus chinensis Anneissia japonica | F F |
| Adenosylhomocysteinase (AHCY) | 3.13.2.1 | GAXK01026996 (a) GAXK01168201 (b) | 0 0 | Eurytemora affinis Eurytemora affinis | F F |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ascione, D.; Carotenuto, Y.; Lauritano, C.; Roncalli, V. Folate-Mediated One-Carbon Metabolism in the Crustacean Copepod Calanus finmarchicus: Identification of Transcripts and Relative Expression across Development. J. Mar. Sci. Eng. 2024, 12, 786. https://doi.org/10.3390/jmse12050786
Ascione D, Carotenuto Y, Lauritano C, Roncalli V. Folate-Mediated One-Carbon Metabolism in the Crustacean Copepod Calanus finmarchicus: Identification of Transcripts and Relative Expression across Development. Journal of Marine Science and Engineering. 2024; 12(5):786. https://doi.org/10.3390/jmse12050786
Chicago/Turabian StyleAscione, Daniela, Ylenia Carotenuto, Chiara Lauritano, and Vittoria Roncalli. 2024. "Folate-Mediated One-Carbon Metabolism in the Crustacean Copepod Calanus finmarchicus: Identification of Transcripts and Relative Expression across Development" Journal of Marine Science and Engineering 12, no. 5: 786. https://doi.org/10.3390/jmse12050786
APA StyleAscione, D., Carotenuto, Y., Lauritano, C., & Roncalli, V. (2024). Folate-Mediated One-Carbon Metabolism in the Crustacean Copepod Calanus finmarchicus: Identification of Transcripts and Relative Expression across Development. Journal of Marine Science and Engineering, 12(5), 786. https://doi.org/10.3390/jmse12050786









