Hydrogen Sulfide Production with a Microbial Consortium Isolated from Marine Sediments Offshore
Abstract
:1. Introduction
2. Materials and Methods
2.1. Inoculum Selection Isolated of Marine Sediments for Hydrogen Sulfide Kinetic Tests
2.2. Kinetic Tests in Batch Systems in Hydrogen Sulfide Production
2.3. Analytical Methods
2.4. Mass and Electron Balances
2.5. Sample Preparation for DNA Extraction
2.6. Amplicon Sequencing Analysis of Total DNA
2.7. Amplicon Analysis and Microbial Diversity
3. Results and Discussion
3.1. Inoculum Selection to Kinetic Tests during Hydrogen Sulfide Production
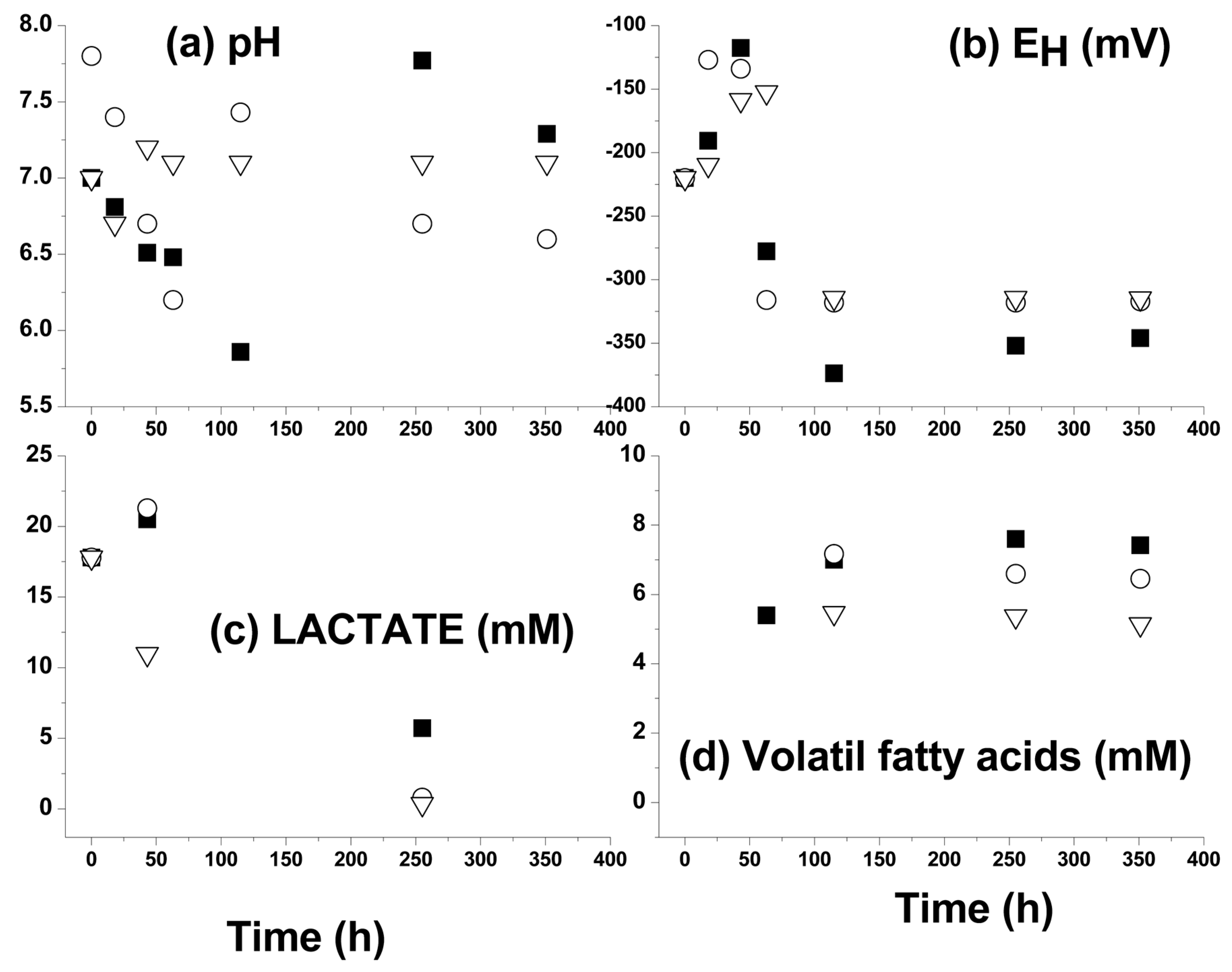
3.2. Kinetic Sulfide Production Tests in Batch Systems
3.3. Diversity of Bacterial 16S rRNA Gene Sequences
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Li, A.; Chu, Y.; Wang, X.; Ren, L.; Yu, J.; Liu, X.; Yan, J.; Zhang, L.; Wu, S.; Li, S. A pyrosequencing-based metagenomic study of methane-producing microbial community in solid-state biogas reactor. Biotechnol. Biofuels 2013, 6, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Preethi, V.; Kanmani, S. Performance of four various shapes of photocatalytic reactors with respect to hydrogen and sulphur recovery from sulphide containing wastestreams. J. Clean. Prod. 2016, 133, 1218–1226. [Google Scholar] [CrossRef]
- Muyzer, G.; Stams, A.J. The ecology and biotechnology of sulphate-reducing bacteria. Nat. Rev. Microbiol. 2008, 6, 441–454. [Google Scholar] [CrossRef] [PubMed]
- Leloup, J.; Fossing, H.; Kohls, K.; Holmkvist, L.; Borowski, C.; Jørgensen, B.B. Sulfate-reducing bacteria in marine sediment (Aarhus Bay, Denmark): Abundance and diversity related to geochemical zonation. Environ. Microbiol. 2009, 11, 1278–1291. [Google Scholar] [CrossRef] [PubMed]
- Sevda, S.; Dominguez-Benetton, X.; Vanbroekhoven, K.; De Wever, H.; Sreekrishnan, T.R.; Pant, D. High strength wastewater treatment accompanied by power generation using air cathode microbial fuel cell. Appl. Energy 2013, 105, 194–206. [Google Scholar] [CrossRef]
- Pandey, P.; Shinde, V.N.; Deopurkar, R.L.; Kale, S.P.; Patil, S.A.; Pant, D. Recent advances in the use of different substrates in microbial fuel cells toward wastewater treatment and simultaneous energy recovery. Appl. Energy 2016, 168, 706–723. [Google Scholar] [CrossRef]
- Barbosa, L.P.; Costa, P.F.; Bertolino, S.M.; Silva, J.C.; Guerra-Sa, R.; Leao, V.A.; Teixeira, M.C. Nickel, manganese and copper removal by a mixed consortium of sulfate reducing bacteria at a high COD/sulfate ratio. World J. Microbiol. Biotechnol. 2014, 30, 2171–2180. [Google Scholar] [CrossRef] [PubMed]
- van den Brand, T.P.; Roest, K.; Chen, G.H.; Brdjanovic, D.; van Loosdrecht, M.C. Potential for beneficial application of sulfate reducing bacteria in sulfate containing domestic wastewater treatment. World J. Microbiol. Biotechnol. 2015, 31, 1675–1681. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, Y.T.; Kieu, H.T.; West, S.; Dang, Y.T.; Horn, H. Community structure of a sulfate-reducing consortium in lead-contaminated wastewater treatment process. World J. Microbiol. Biotechnol. 2017, 33, 10. [Google Scholar] [CrossRef] [PubMed]
- Van den Brand, T.; Snip, L.; Palmen, L.; Weij, P.; Sipma, J.; van Loosdrecht, M. Sulfate reducing bacteria applied to domestic wastewater. Water Pract. Technol. 2018, 13, 542–554. [Google Scholar] [CrossRef]
- Kieu, H.T.Q.; Müller, E.; Horn, H. Heavy metal removal in anaerobic semi-continuous stirred tank reactors by a consortium of sulfate-reducing bacteria. Water Res. 2011, 45, 3863–3870. [Google Scholar] [CrossRef]
- Gadd, G.M. Microbial influence on metal mobility and application for bioremediation. Geoderma 2004, 122, 109–119. [Google Scholar] [CrossRef]
- Serio, A.; Fusella, G.C.; Chaves López, C.; Sacchetti, G.; Paparella, A. A survey on bacteria isolated as hydrogen sulfide-producers from marine fish. Food Control 2014, 39, 111–118. [Google Scholar] [CrossRef]
- Roman, P.; Lipińska, J.; Bijmans, M.F.M.; Sorokin, D.Y.; Keesman, K.J.; Janssen, A.J.H. Inhibition of a biological sulfide oxidation under haloalkaline conditions by thiols and diorgano polysulfanes. Water Res. 2016, 101, 448–456. [Google Scholar] [CrossRef]
- Delforno, T.P.; Moura, A.G.; Okada, D.Y.; Sakamoto, I.K.; Varesche, M.B. Microbial diversity and the implications of sulfide levels in an anaerobic reactor used to remove an anionic surfactant from laundry wastewater. Bioresour. Technol. 2015, 192, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Jabari, L.; Gannoun, H.; Khelifi, E.; Cayol, J.-L.; Godon, J.-J.; Hamdi, M.; Fardeau, M.-L. Bacterial ecology of abattoir wastewater treated by an anaerobic digestor. Braz. J. Microbiol. 2016, 47, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.-G.; Wang, A.-J.; Ren, N.-Q. Effect of carbon sources on sulfidogenic bacterial communities during the starting-up of acidogenic sulfate-reducing bioreactors. Bioresour. Technol. 2010, 101, 2952–2959. [Google Scholar] [CrossRef] [PubMed]
- Kiely, P.D.; Call, D.F.; Yates, M.D.; Regan, J.M.; Logan, B.E. Anodic biofilms in microbial fuel cells harbor low numbers of higher-power-producing bacteria than abundant genera. Appl. Microbiol. Biotechnol. 2010, 88, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Chen, F.; Liu, Q.; Zhao, Y.; Zhou, J.-Z.; Wang, A.-J. Microbial community structure and function in response to the shift of sulfide/nitrate loading ratio during the denitrifying sulfide removal process. Bioresour. Technol. 2015, 197, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zhou, X.; Li, Y.; Xing, J. Bacterial communities in haloalkaliphilic sulfate-reducing bioreactors under different electron donors revealed by 16S rRNA MiSeq sequencing. J. Hazard. Mater. 2015, 295, 176–184. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.-G.; Zheng, Y.; Tian, W.; Bai, J.; Feng, G.; Guo, L.; Gao, M. Enrichment and immobilization of sulfide removal microbiota applied for environmental biological remediation of aquaculture area. Environ. Pollut. 2016, 214, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Marti, E.; Balcázar, J.L. Use of pyrosequencing to explore the benthic bacterial community structure in a river impacted by wastewater treatment plant discharges. Res. Microbiol. 2014, 165, 468–471. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Silveira, J.A.; Comin, F.A.; Aranda-Cirerol, N.; Troccoli, L.; Capurro, L. Coastal water quality assessment in the Yucatan Peninsula: Management implications. Ocean Coast. Manag. 2004, 47, 625–639. [Google Scholar] [CrossRef]
- Walkley, A.; Black, I.A. An examination of the degtjareff method for determining soil organic matter, and a proposed modification of the chromic acid titration method. Soil Sci. 1934, 37, 29–38. [Google Scholar] [CrossRef]
- APHA. Standard Methods for the Examination of Water and Wastewater: 19th Edition Supplement; American Public Health Association: Washington, DC, USA, 1996. [Google Scholar]
- Nagpal, S.; Chuichulcherm, S.; Peeva, L.; Livingston, A. Microbial sulfate reduction in a liquid–solid fluidized bed reactor. Biotechnol. Bioeng. 2000, 70, 370–380. [Google Scholar] [CrossRef]
- Labastida-Nunez, I.; Lazaro, I.; Celis, L.B.; Razo-Flores, E.; Cruz, R.; Briones-Gallardo, R. Kinetic of biogenic sulfide production for microbial consortia isolated from soils with different bioaccessible concentrations of lead. Int. J. Environ. Sci. Tech. 2013, 10, 827–836. [Google Scholar] [CrossRef] [Green Version]
- Cord-Ruwisch, R. A quick method for the determination of dissolved and precipitated sulfides in cultures of sulfate-reducing bacteria. J. Microbiol. Methods 1985, 4, 33–36. [Google Scholar] [CrossRef]
- Rittmann, B.; McCarthy, P. Environmental Biotechnology: Principles and Applications; McGraw Hill: New York, NY, USA, 2001. [Google Scholar]
- Schloss, P.D.; Gevers, D.; Westcott, S.L. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS ONE 2011, 6, e27310. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 2014, 30, 614–620. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Meyer, F.; Paarmann, D.; D’Souza, M.; Olson, R.; Glass, E.M.; Kubal, M.; Paczian, T.; Rodriguez, A.; Stevens, R.; Wilke, A.; et al. The metagenomics RAST server—A public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinform. 2008, 9, 386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Team, R.D.C. R: A Language and Environment for Statistical Computing; Foundation for Statistical Computing: Vienna, Austria, 2006. [Google Scholar]
- Cao, J.; Zhang, G.; Mao, Z.; Fang, Z.; Yang, C. Precipitation of valuable metals from bioleaching solution by biogenic sulfides. Miner. Eng. 2009, 22, 289–295. [Google Scholar] [CrossRef]
- Bertolino, S.M.; Rodrigues, I.C.; Guerra-Sá, R.; Aquino, S.F.; Leão, V.A. Implications of volatile fatty acid profile on the metabolic pathway during continuous sulfate reduction. J. Environ. Manag. 2012, 103, 15–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lynd, L.R.; Weimer, P.J.; Zyl, W.H.v.; Pretorius, I.S. Microbial cellulose utilization: Fundamentals and biotechnology. Microbiol. Molec. Biol. Rev. 2002, 66, 739. [Google Scholar] [CrossRef] [Green Version]
- Purdy, K.J.; Embley, T.M.; Nedwell, D.B. The distribution and activity of sulphate reducing bacteria in estuarine and coastal marine sediments. Antonie Leeuwenhoek 2002, 81, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.-H.; Chang, Y.-J.; Chung, W.-C.; Hsieh, P.-H.; Lin, C.-Y.; Ho, J.-M. Subset selection of high-depth next generation sequencing reads for de novo genome assembly using MapReduce framework. BMC Genom. 2015, 16, S9. [Google Scholar] [CrossRef] [Green Version]
- So, C.M.; Young, L.Y. Isolation and characterization of a sulfate-reducing bacterium that anaerobically degrades alkanes. Appl. Environ. Microbiol. 1999, 65, 2969–2976. [Google Scholar] [CrossRef] [Green Version]
- Fauque, G.D. Ecology of sulfate-reducing bacteria. In Sulfate-Reducing Bacteria; Barton, L.L., Ed.; Plenum Press: New York, NY, USA, 1995; pp. 217–241. [Google Scholar]
- Garcia, G.P.P.; Diniz, R.C.O.; Bicalho, S.K.; Franco, V.; Pereira, A.D.; Brandt, E.F.; Etchebehere, C.; Chernicharo, C.A.L.; de Araujo, J.C. Microbial community and sulphur behaviour in phototrophic reactors treating UASB effluent under different operational conditions. Int. Biodeteriorat. Biodegradat. 2016, 119, 486–498. [Google Scholar] [CrossRef]
- Rabbani, M.; Dincer, I.; Naterer, G.F. Experimental investigation of hydrogen production in a photo-electrochemical chloralkali processes reactor. Int. J. Hydrogen Energy 2016, 41, 7766–7781. [Google Scholar] [CrossRef]
- Ipsakis, D.; Kraia, T.; Marnellos, G.E.; Ouzounidou, M.; Voutetakis, S.; Dittmeyer, R.; Dubbe, A.; Haas-Santo, K.; Konsolakis, M.; Figen, H.E.; et al. An electrocatalytic membrane-assisted process for hydrogen production from H2S in Black Sea: Preliminary results. Int. J. Hydrogen Energy 2015, 40, 7530–7538. [Google Scholar] [CrossRef]
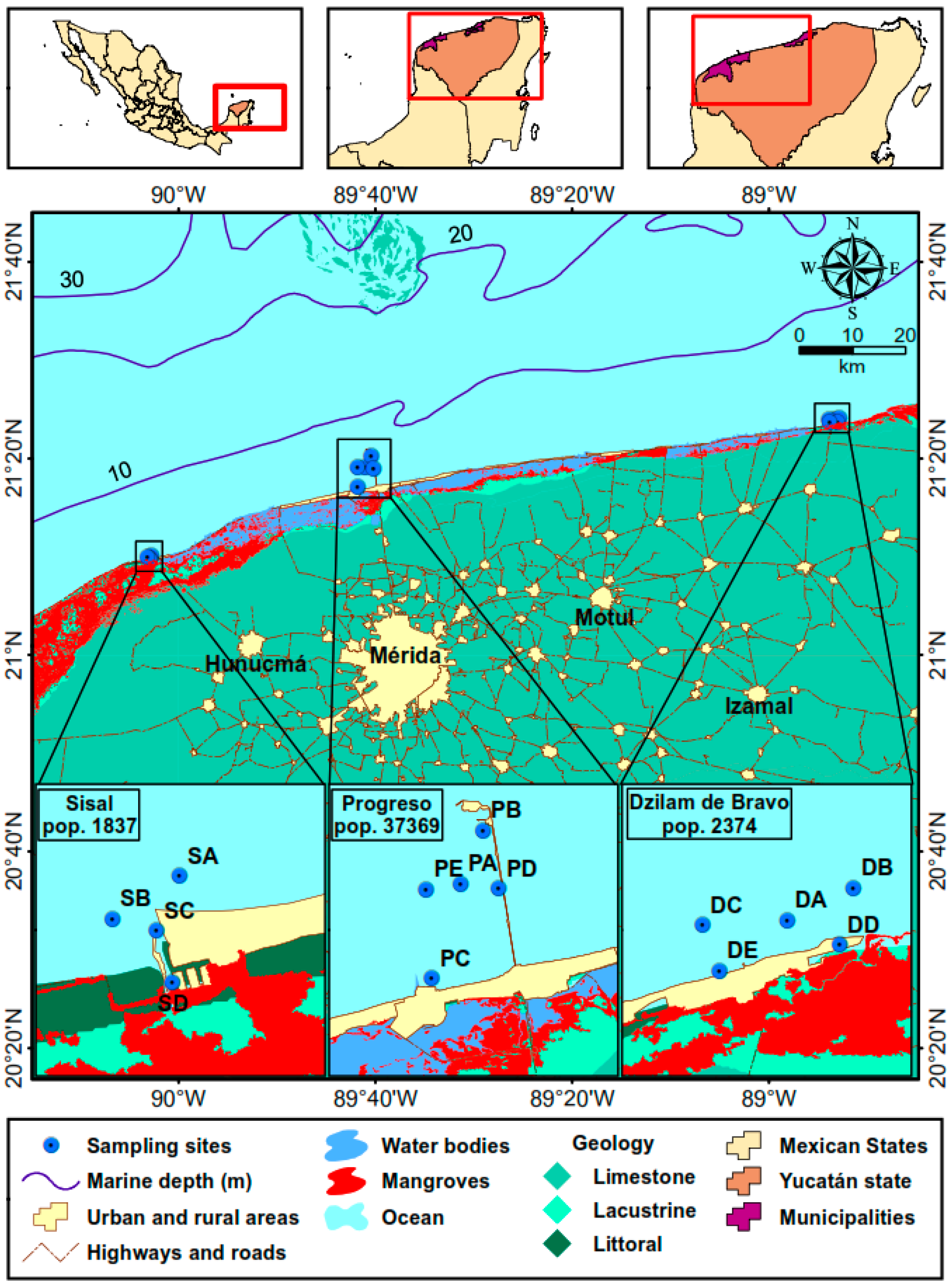


| Surficial Sediment (−40 cm) | |||||||||||
| Port | Site 1 | T (°C) | pH | Eh (mV) | H2S (mg/L) | TOC (%) | TN (µM) | TP (µM) | GPS Location | Depth (m) | |
| Sisal | SA | 28.8 | 7.9 | 28 | 0.8 | 0.4 | 19.9 | 5.3 | 21°10.100′ N | 90°02.817′ O | 4.1 |
| SB | 30.3 | 6.7 | −279 | OR | 3.3 | 339.5 | 6.3 | 21°09.917′ N | 90°03.17′ O | 2.3 | |
| SC | 30.5 | 6.8 | −251 | 469 | 1.9 | 158.6 | 5.9 | 21°09.850′ N | 90°02.950′ O | 1.7 | |
| SD | 31.3 | 6.8 | −256 | 636 | 4.0 | 354.6 | 7.8 | 21°09.600′ N | 90°02.883′ O | 2.5 | |
| SE | 31.2 | 6.8 | −273 | OR | 2.8 | 190.9 | 7.2 | 21°09.617′ N | 90°02.733′ O | 2.8 | |
| Progreso | PA | 29.4 | 8.0 | −107 | 29 | 0.9 | 91.8 | 1.3 | 21°19.135′ N | 89°41.044′ O | 7.0 |
| PB | 30.2 | 7.7 | −166 | 129.8 | 2.9 | 263.7 | 1.8 | 21°20.254′ N | 89°40.455′ O | 8.0 | |
| PC | 27.6 | 6.9 | −361 | OR | 4.6 | 530.3 | 6.8 | 21°17.117′ N | 89°41.833′ O | 2.8 | |
| PD | 26.0 | 6.6 | −368 | 102.2 | 2.4 | 359.3 | 6.8 | 21°18.983′ N | 89°40.167′ O | 6.4 | |
| PE | 27.6 | 7.6 | −306 | 31.3 | 1.1 | 110.8 | 5.4 | 21°19.050′ N | 89°41.850′ O | 3.3 | |
| Dzilam de Bravo | DA | 28.2 | 7.6 | −90 | DL | 0.9 | 74.7 | 5.0 | 21.3980° N | 88.8889° O | 1.8 |
| DB | 28.3 | 7.6 | −162 | 0.1 | 1.6 | 120.8 | 5.5 | 21.4008° N | 88.8809° O | 1.4 | |
| DC | 27.0 | 7.5 | −110 | 0.1 | 1.0 | 62.1 | 4.9 | 21.3978° N | 88.8987° O | 1.5 | |
| DD | 29.3 | 7.3 | −238 | 0.1 | 5.6 | 380.2 | 7.5 | 21.3951° N | 88.8830° O | 1.0 | |
| DE | 29.4 | 7.4 | −44 | 0.1 | 1.9 | 83.5 | 6.3 | 21.2931° N | 88.8971° O | 0.8 | |
| Water Column Close to Surficial Sediment | |||||||||||
| Port | Site | T (°C) | pH | Eh (mV) | DO (mg L−1) | NH4− (µM) | NO2− (µM) | NO3− (µM) | TN (mg m−3) | TP (mg m−3) | |
| Sisal | SA | 29.0 | 8.5 | −27.7 | 6.3 | 3.2 | 0.1 | 0.3 | 48.0 | 3.7 | |
| SB | 29.0 | 8.4 | −45 | 6.2 | 4.1 | 0.4 | 4.2 | 121.0 | 0.6 | ||
| SC | 29.2 | 8.4 | −57.1 | 8.4 | 12.0 | 0.7 | 17.3 | 419.3 | 2.8 | ||
| SD | 29.1 | 8.2 | −71.9 | 6.7 | 2.7 | 0.1 | 1.1 | 54.7 | 1.5 | ||
| SE | 29.5 | 7.9 | −44.6 | 6.4 | 29.9 | 2.0 | 15.0 | 656.0 | 7.1 | ||
| Progreso | PA | 29.1 | 8.6 | −44 | 6.7 | 1.5 | 0.1 | 0.1 | 23.0 | 2.8 | |
| PB | 29.7 | 8.7 | −46.1 | 6.6 | 0.9 | 0.1 | 0.2 | 16.5 | 3.7 | ||
| PC | 25.6 | 8.9 | −49.3 | 7.2 | 3.1 | 0.2 | 0.8 | 57.5 | 5.6 | ||
| PD | 26.5 | 9.0 | −100 | 6.9 | 1.8 | 0.1 | 0.3 | 29.8 | 2.8 | ||
| PE | 26.5 | 8.9 | −51.5 | 6.4 | 1.8 | 0.2 | 1.0 | 42.3 | 4.6 | ||
| Dzilam de Bravo | DA | 26.4 | 8.2 | −6.4 | 6.2 | 1.4 | 0.3 | 2.5 | 59.1 | 5.3 | |
| DB | 26.5 | 8.1 | −8.5 | 6.1 | 4.4 | 0.6 | 4.6 | 133.6 | 8.1 | ||
| DC | 27.1 | 8.0 | −21.2 | 5.8 | 1.5 | 0.3 | 2.6 | 60.1 | 1.9 | ||
| DD | 27.3 | 7.9 | −9.7 | 6.1 | 2.4 | 0.5 | 2.4 | 75.5 | 2.8 | ||
| DE | 27.2 | 8.3 | −26.8 | 6 | 1.1 | 0.5 | 4.2 | 81.3 | 5.6 | ||
| Reaction Number | Reduced-Oxidized Compounds | Half Chemical Reaction | ∆G° (kJ/mol) |
|---|---|---|---|
| 1 | Lactate | −387.48 | |
| 2 | Acetate | −219.2 | |
| 3 | Propionate | −386.82 | |
| 4 | Sulfate | 166.8 | |
| 5 | Sulfate | 117 | |
| 6 | Cell synthesis | - |
| Kinetic Parameters | Consortia | ||
|---|---|---|---|
| SC469 | SD636 | PD102 | |
| [SO42−]max consumption (mmol g−1 VSS) | 242 ± 2 a | 197.3 ± 2 a | 200 ± 10 a |
| [H2S]max production (mmol g−1 VSS) | 246 ± 28 a | 132 ± 8 b | 110 ± 3 b |
| −rmax, sulfate (mmol g−1 VSS d−1) | 100 ± 23 a | 144 ± 38 b | 22 ± 2 c |
| rmax, sulfid (mmol g−1 VSS d−1) | 113 ± 50 a | 95 ± 30 a | 144 ± 89 a |
| R2, sulfate | 0.97 | 0.99 | 0.94 |
| R2, sulfide | 0.99 | 0.91 | 0.92 |
| Reaction Number | Lactate Biodegradation Pathway | Reaction | ∆G° (kJ/mol) | Involved Consortium |
|---|---|---|---|---|
| 7 | Total sulfate-reducing metabolism | −137.28 | CS459 | |
| 8 | Lactate fermentation | −58.68 | CS459 | |
| 9 | Total sulfate reducing metabolism | −160.1 | SD636 PD102 | |
| 10 | Lactate fermentation | −55.92 | SD636 PD102 | |
| 11 | Partial sulfate-reducing metabolism | −66 | SD636 PD102 | |
| 12 | Cell synthesis | - | CS459 SD636 PD102 |
| Inoculum | Lactate | Sulfate | Mass Products (mmol) | Ratio [OCS/OCL] | fs | Biomass Net Yield (Yn) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| OCL (mmol) | TMD (mmol) | OCS (mmol) | ||||||||
| Acetate | Propionate | Biomass | Sulfide | |||||||
| SC469 | 1.84 | 1.8 | 1.46 | 0.80 | 0.0 | 0.30 | 1.19 | 0.79 | 0.11 | 0.16 |
| SD636 | 2.64 | 2.15 | 1.21 | 0.57 | 0.10 | 0.27 | 0.79 | 0.46 | 0.07 | 0.10 |
| PD102 | 2.16 | 1.93 | 1.09 | 0.60 | 0.0 | 0.36 | 0.66 | 0.5 | 0.11 | 0.17 |
| Inoculum | NEDS (meq e−) | TED (meq e−) | IEA (meq e−) | Electron-acceptor products (meq e−) | ∑(EAP) (meq e−) | % EA-SRMP | % loss electrons | |||
| SC469 | 22.1 | 21.6 | 12.2 | 7.44 | 0.0 | 2.4 | 9.52 | 19.35 | 87.6 | 12.4 |
| SD636 | 31.7 | 25.8 | 9.7 | 6.73 | 0.82 | 2.12 | 6.35 | 16.03 | 50.6 | 49.4 |
| PD102 | 25.9 | 23.2 | 7.9 | 5.36 | 0.0 | 2.85 | 5.34 | 13.56 | 52.3 | 47.7 |
| Samples | OTU Observed | Chao1 | Shannon (H′) |
|---|---|---|---|
| PD102-1 | 698 | 897.9303 | 3.563574 |
| PD102-2 | 903 | 1150.779 | 3.808334 |
| PD102-3 | 761 | 1099.942 | 3.828718 |
| SC469-1 | 590 | 1008.361 | 3.40248 |
| SC469-2 | 1009 | 1137.648 | 5.626442 |
| SC469-3 | 427 | 591.5 | 2.403016 |
| SD636-1 | 745 | 938.7 | 4.673162 |
| SD636-2 | 783 | 1306.837 | 4.643861 |
| SD636-3 | 1070 | 1384.204 | 5.619174 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Briones-Gallardo, R.; González-Muñoz, M.; García-Bautista, I.; Valdés-Lozano, D.; Toledano-Thompson, T.; Polanco-Lugo, E.; Rivera-Madrid, R.; Valdez-Ojeda, R. Hydrogen Sulfide Production with a Microbial Consortium Isolated from Marine Sediments Offshore. J. Mar. Sci. Eng. 2022, 10, 436. https://doi.org/10.3390/jmse10030436
Briones-Gallardo R, González-Muñoz M, García-Bautista I, Valdés-Lozano D, Toledano-Thompson T, Polanco-Lugo E, Rivera-Madrid R, Valdez-Ojeda R. Hydrogen Sulfide Production with a Microbial Consortium Isolated from Marine Sediments Offshore. Journal of Marine Science and Engineering. 2022; 10(3):436. https://doi.org/10.3390/jmse10030436
Chicago/Turabian StyleBriones-Gallardo, Roberto, Muriel González-Muñoz, Itza García-Bautista, David Valdés-Lozano, Tanit Toledano-Thompson, Erik Polanco-Lugo, Renata Rivera-Madrid, and Ruby Valdez-Ojeda. 2022. "Hydrogen Sulfide Production with a Microbial Consortium Isolated from Marine Sediments Offshore" Journal of Marine Science and Engineering 10, no. 3: 436. https://doi.org/10.3390/jmse10030436
APA StyleBriones-Gallardo, R., González-Muñoz, M., García-Bautista, I., Valdés-Lozano, D., Toledano-Thompson, T., Polanco-Lugo, E., Rivera-Madrid, R., & Valdez-Ojeda, R. (2022). Hydrogen Sulfide Production with a Microbial Consortium Isolated from Marine Sediments Offshore. Journal of Marine Science and Engineering, 10(3), 436. https://doi.org/10.3390/jmse10030436






