Characterization of Drought-Responsive miRNAs in Peanut Through Integrated Transcriptomic Approaches
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. sRNA Library Construction and DNA Sequencing
2.3. Degradome Library Construction and Sequencing
2.4. Northern Blotting Analysis
2.5. Generation of Transgenic Plants
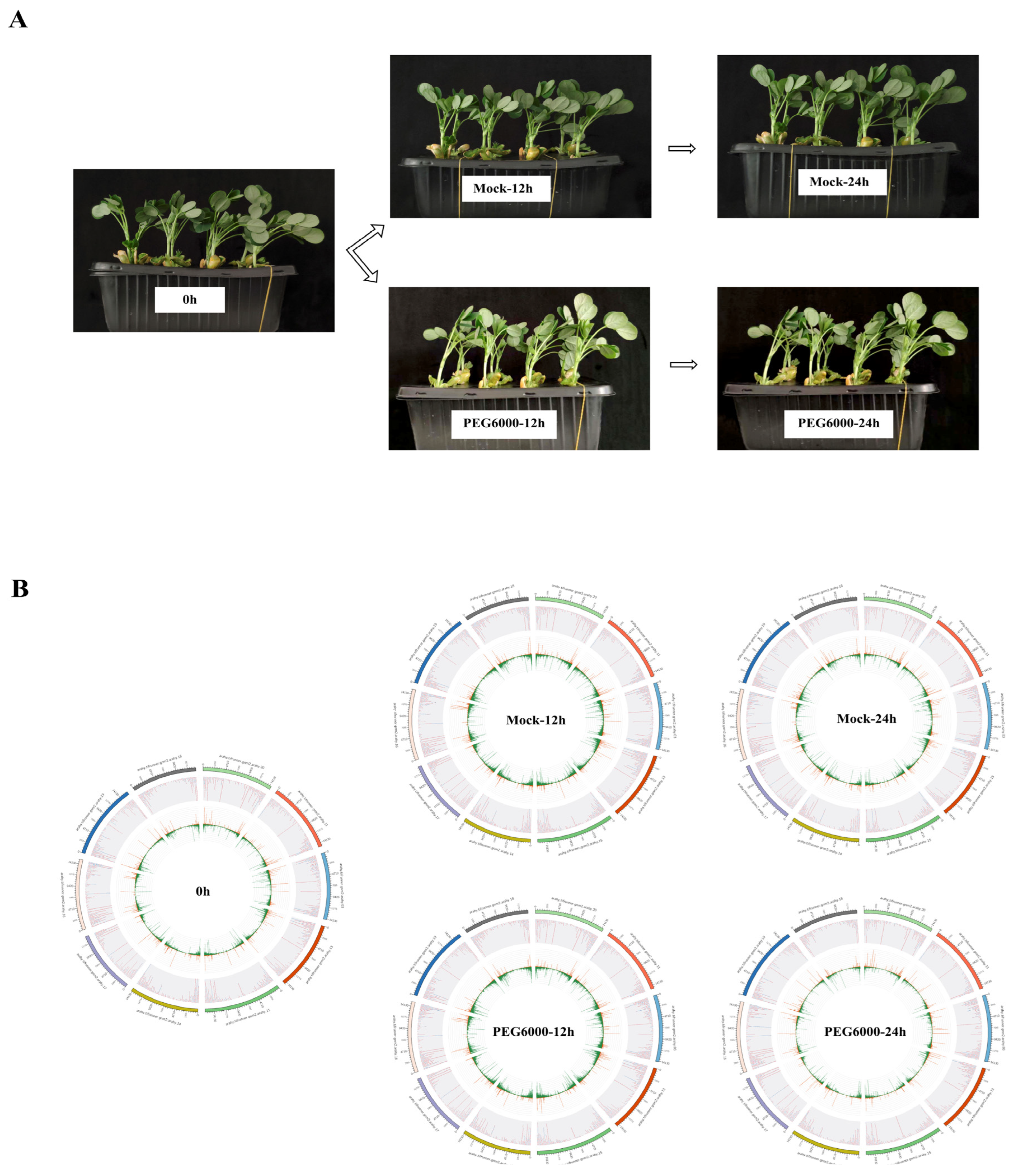
2.6. Drought Stress Treatment
3. Results
3.1. High-Throughput Sequencing and Annotation of Peanut sRNAs
3.2. Identification of Known, Conserved, and Novel Peanut miRNAs
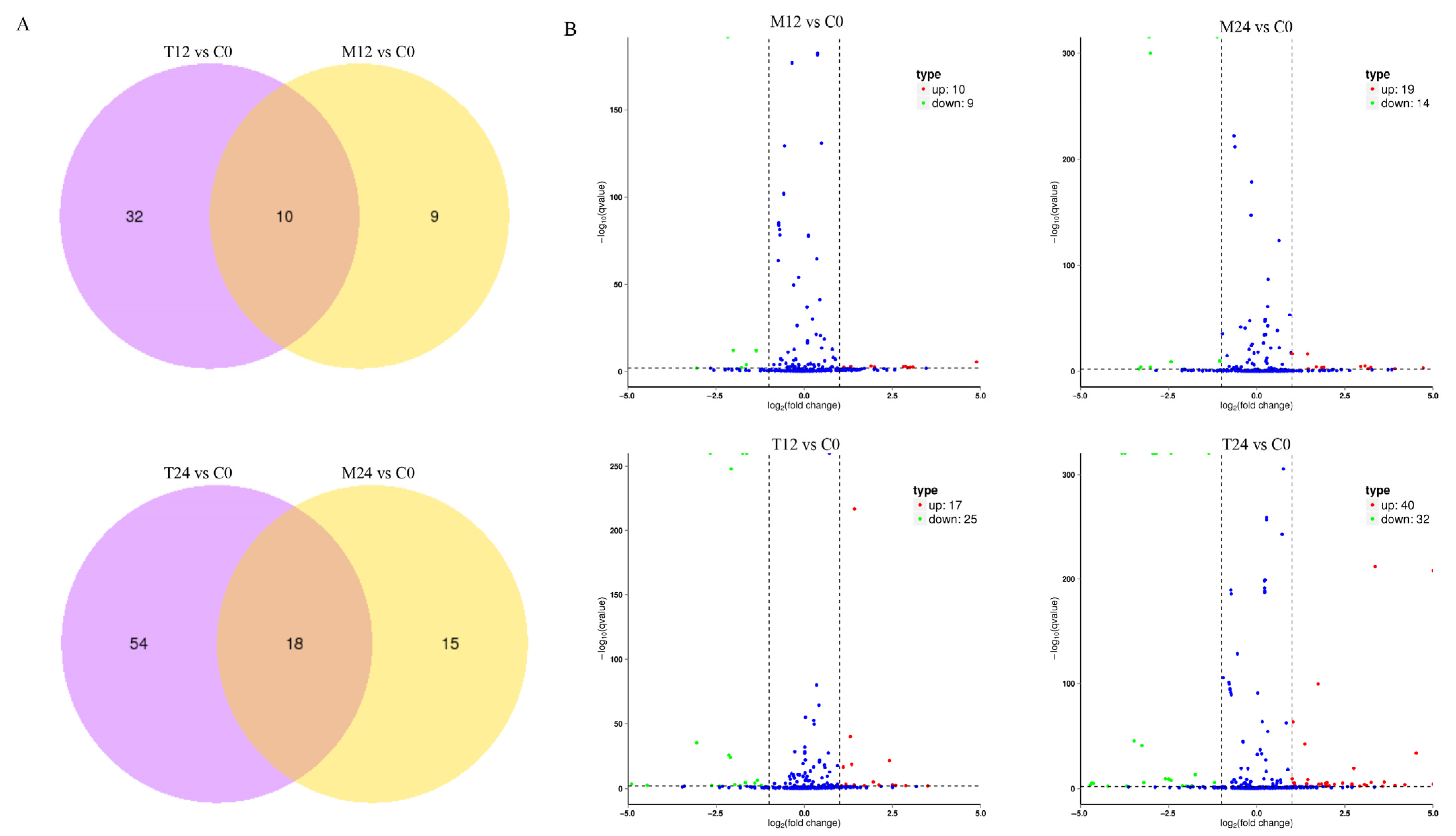
3.3. Revealed Differential Expression Profiling of miRNAs Between PEG6000-Treated and Mock Control Groups
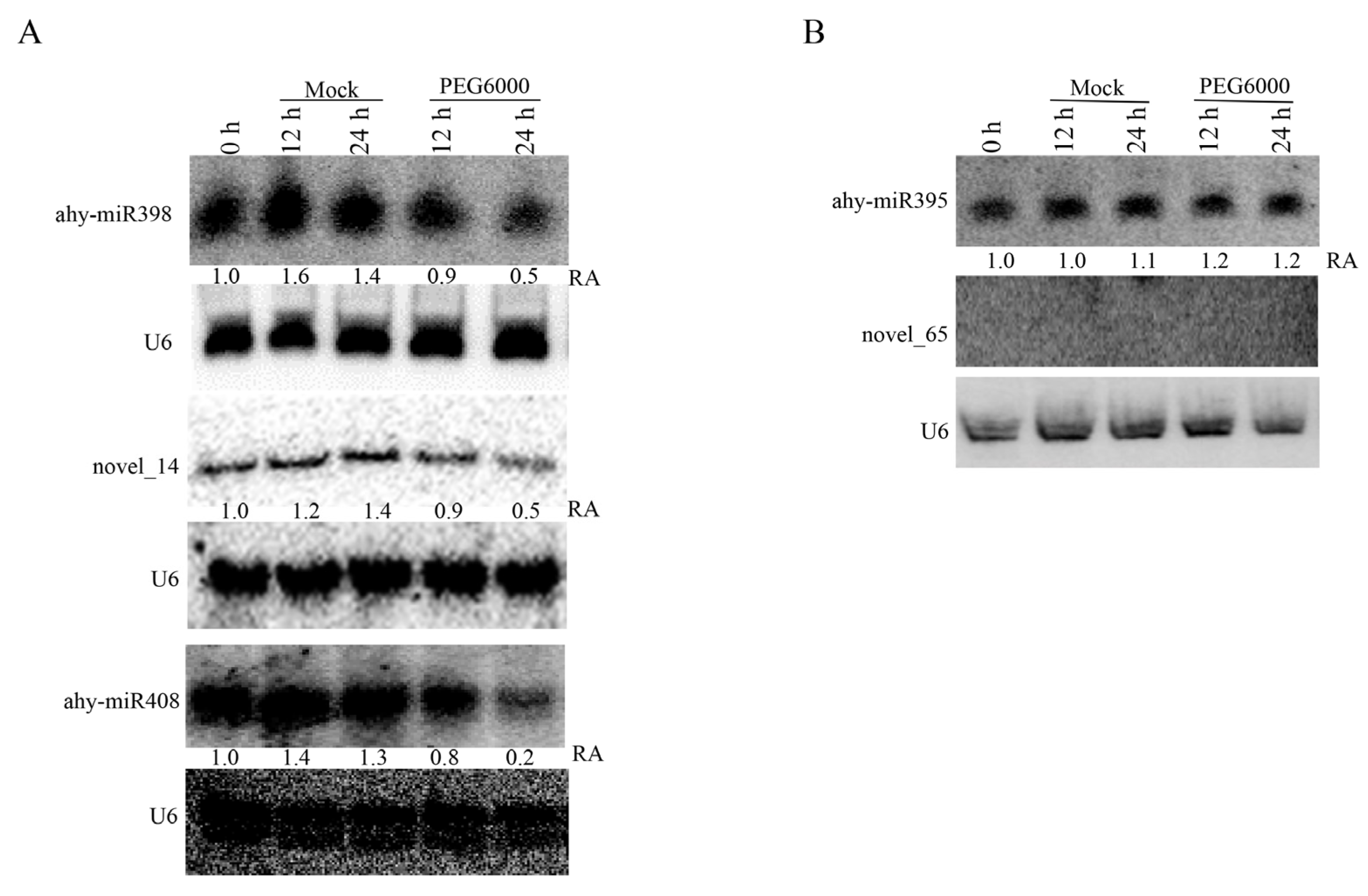
3.4. Validation of Drought-Responsive miRNAs by Northern Blotting
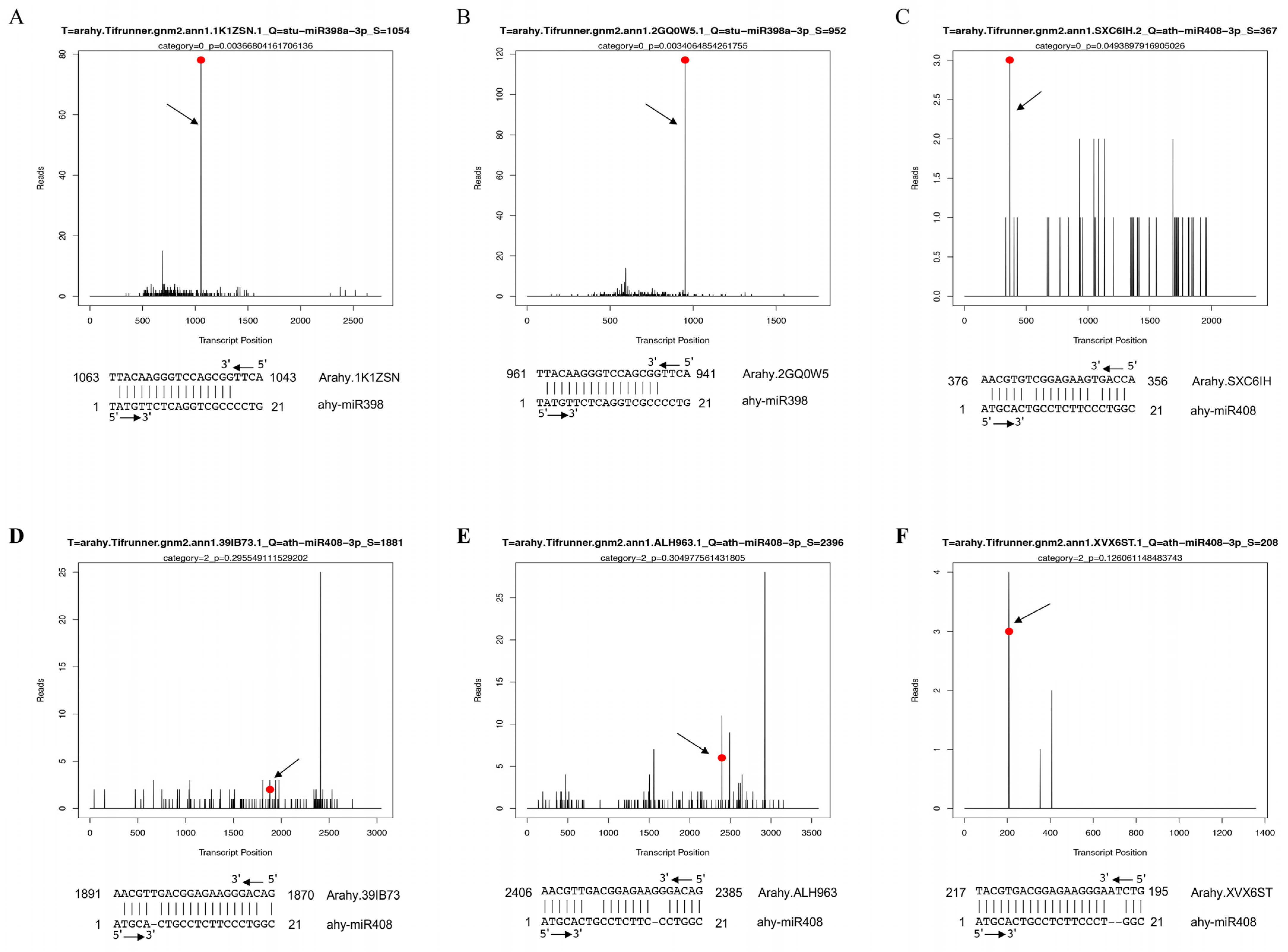
3.5. Target Identification and Validation of Drought-Responsive miRNAs
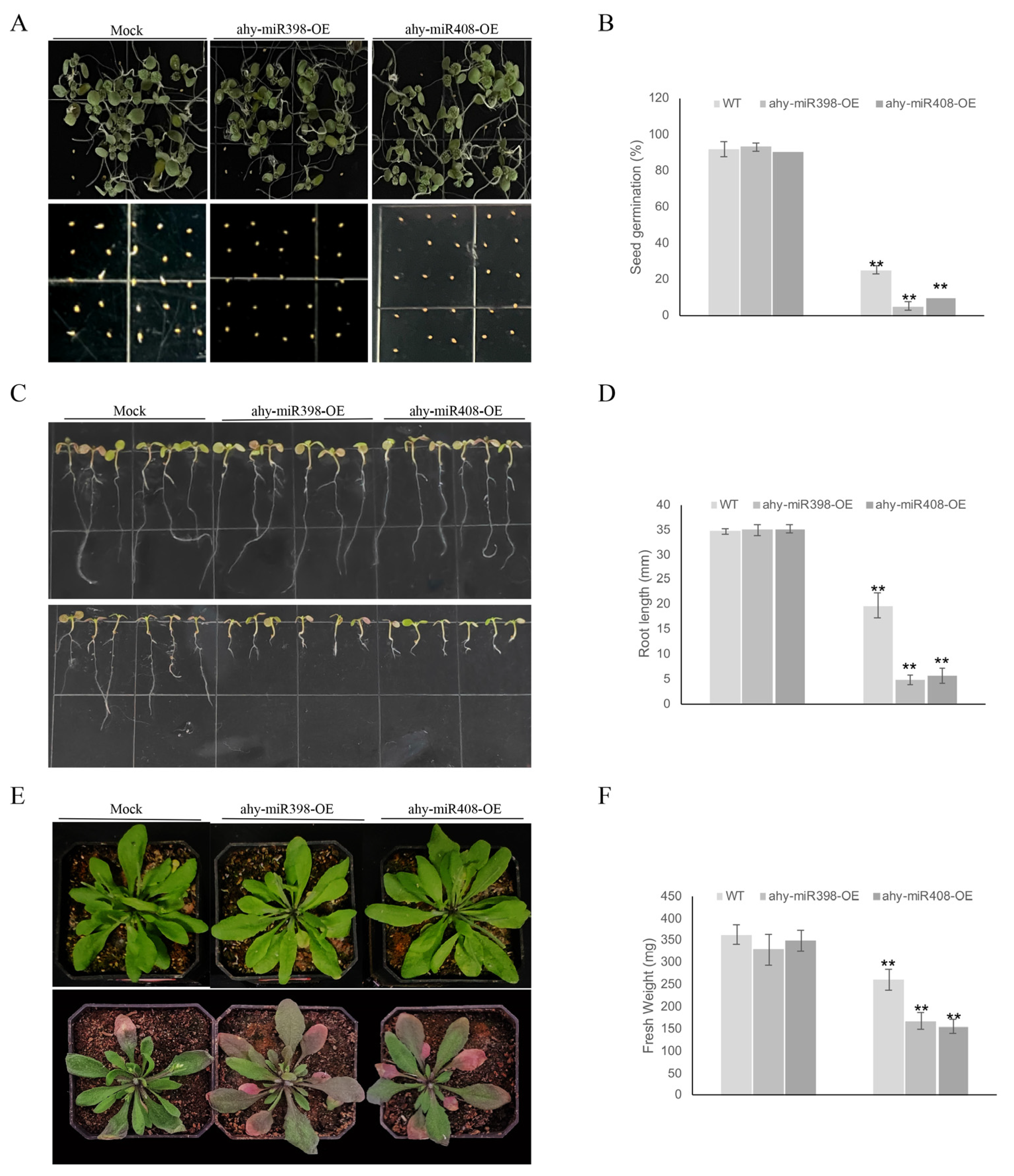
3.6. Overexpression of Ahy-miR398 and Ahy-miR408 Negatively Regulates Drought Tolerance
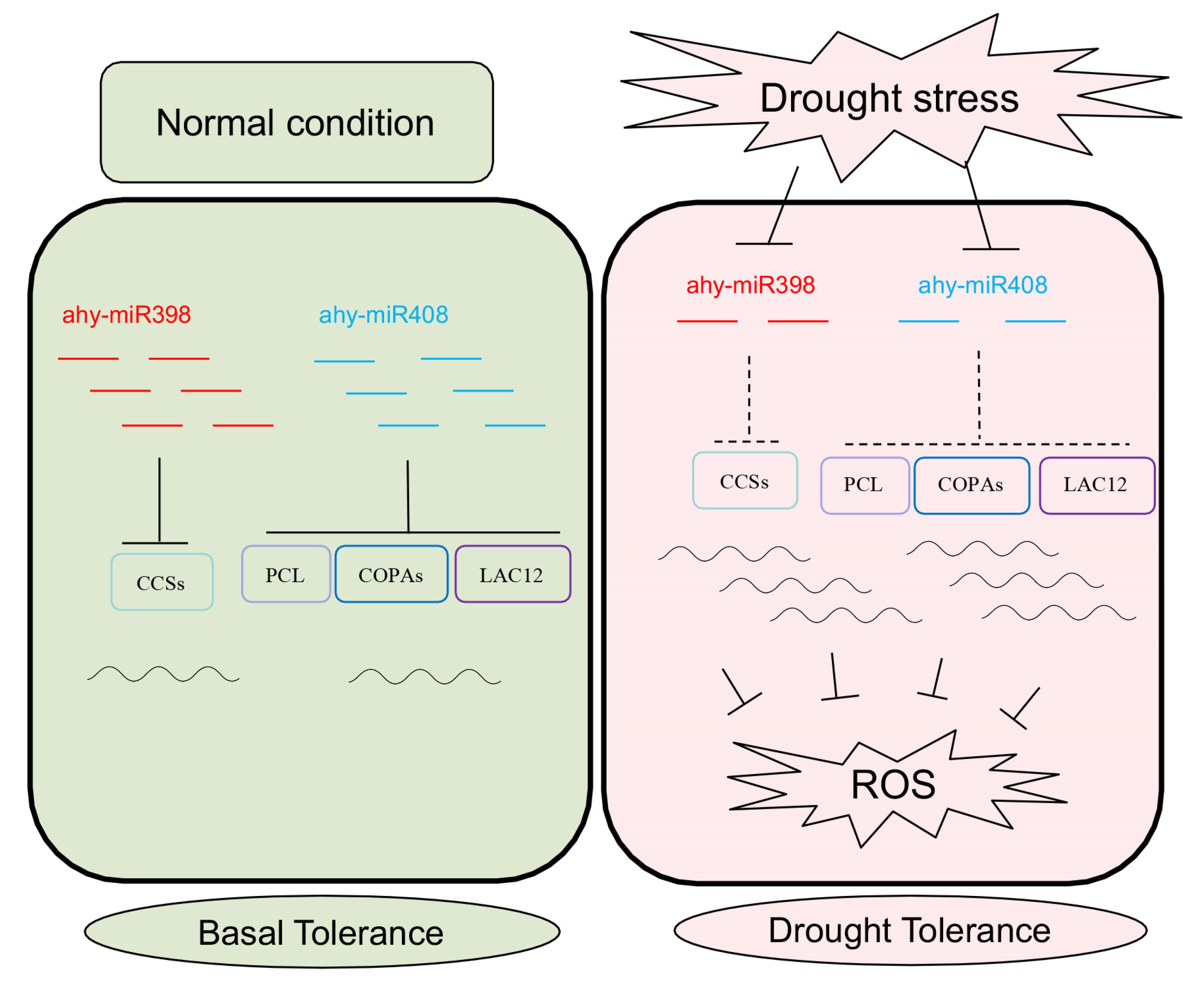
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| TPM | Transcripts per kilobase of exon model per million mapped reads |
| ROS | Reactive oxygen species |
| CCS | Cu chaperone for SOD |
| PLC | Plantacyanin |
| PCL | Blue copper protein-like |
| PCY | Plastocyanin |
| UCL | Uclacyanin |
| LAC | Laccase |
| DEM | Differentially expressed miRNA |
| COPA | P-type ATPase copper transporter |
| PCR | Polymerase chain reaction |
| MS | Murashige and Skoog |
References
- Lesk, C.; Rowhani, P.; Ramankutty, N. Influence of extreme weather disasters on global crop production. Nature 2016, 529, 84–87. [Google Scholar] [CrossRef]
- Li, W.X.; Oono, Y.; Zhu, J.; He, X.J.; Wu, J.M.; Iida, K.; Lu, X.Y.; Cui, X.; Jin, H.; Zhu, J.K. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell. 2008, 20, 2238–2251. [Google Scholar] [CrossRef]
- Janila, P.; Pandey, M.K.; Shasidhar, Y.; Variath, M.T.; Sriswathi, M.; Khera, P.; Manohar, S.S.; Nagesh, P.; Vishwakarma, M.K.; Mishra, G.P.; et al. Molecular breeding for introgression of fatty acid desaturase mutant alleles (AhFAD2A and AhFAD2B) enhances oil quality in high and low oil containing peanut genotypes. Plant Sci. 2016, 242, 203–213. [Google Scholar] [CrossRef]
- Hamidou, F.; Halilou, O.; Vadez, V. Assessment of groundnut under combined heat and drought stress. J. Agron. Crop Sci. 2013, 199, 1–11. [Google Scholar] [CrossRef]
- Toomer, O.T. Nutritional chemistry of the peanut (Arachis hypogaea). Crit. Rev. Food Sci. Nutr. 2018, 58, 3042–3053. [Google Scholar] [CrossRef] [PubMed]
- Bertioli, D.J.; Jenkins, J.; Clevenger, J.; Dudchenko, O.; Gao, D.; Seijo, G.; Leal-Bertioli, S.C.M.; Ren, L.; Farmer, A.D.; Pandey, M.K.; et al. The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 2019, 51, 877–884. [Google Scholar] [CrossRef] [PubMed]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006, 57, 19–53. [Google Scholar] [CrossRef]
- Zhou, L.G.; Liu, Y.H.; Liu, Z.C.; Kong, D.Y.; Duan, M.; Luo, L.J. Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa. J. Exp. Bot. 2010, 61, 4157–4168. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Gonzalez, N.; Inzé, D.; Dubois, M. Emerging connections between small RNAs and phytohormones. Trends Plant Sci. 2020, 25, 912–929. [Google Scholar] [CrossRef]
- Tiwari, M. Blessing in disguise: A loss of miR159 makes plant drought tolerant and ABA sensitive. Physiol. Plant. 2022, 174, e13763. [Google Scholar] [CrossRef]
- Zhao, J.; Yuan, S.; Zhou, M.; Yuan, N.; Li, Z.; Hu, Q.; Bethea, F.G.; Jr Liu, H.; Li, S.; Luo, H. Transgenic creeping bentgrass overexpressing Osa-miR393a exhibits altered plant development and improved multiple stress tolerance. Plant Biotechnol. J. 2019, 17, 233–251. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Z.; Lian, C.; Han, S.; Huang, M.; Xia, X. PtmiR169o plays a positive role in regulating drought tolerance and growth by targeting the PtNF-YA6 gene in poplar. Environ. Exp. Bot. 2021, 189, 104549. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, Z.; Yu, S.; Li, W.; Li, C. Pu-miR172d regulates stomatal density and water use efficiency via targeting PuGTL1 in poplar. J. Exp. Bot. 2020, 72, 1370–1383. [Google Scholar] [CrossRef]
- Addo-Quaye, C.; Miller, W.; Axtell, M.J. CleaveLand: A pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics 2009, 25, 130–131. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ren, C.; Xue, Y.; Tian, Y.; Zhang, H.; Li, N.; Sheng, C.; Jiang, H.; Bai, D. Small RNA and Degradome Deep Sequencing Reveals the Roles of microRNAs in Peanut (Arachis hypogaea L.) Cold Response. Front. Plant Sci. 2022, 13, 920195. [Google Scholar] [CrossRef]
- Zhang, J.X.; Xiao, S.H.; Zhou, S.L.; Dong, X.M.; Jiang, S.; Bao, Y.X.; Jiang, X.L.; Hu, Q.; Duan, Z.Z.; Powell, C.A.; et al. Genome-wide identification of sugarcane SINA family proteins reveals that SsSINA1a positively regulates drought tolerance. Ind. Crop Prod. 2025, 226. [Google Scholar] [CrossRef]
- Li, J.; Song, Q.; Zuo, Z.F.; Liu, L. MicroRNA398: A master regulator of plant development and stress responses. Int. J. Mol. Sci. 2022, 23, 10803. [Google Scholar] [CrossRef]
- Feng, K.; Yu, J.; Cheng, Y.; Ruan, M.; Wang, R.; Ye, Q.; Zhou, G.; Li, Z.; Yao, Z.; Yang, Y.; et al. The SOD gene family in tomato: Identification, phylogenetic relationships, and expression patterns. Front. Plant Sci. 2016, 7, 1279. [Google Scholar] [CrossRef]
- Sunkar, R.; Kapoor, A.; Zhu, J.K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell. 2006, 18, 2051–2065. [Google Scholar] [CrossRef]
- Jovanovic, Z.; Stanisavljevic, N.; Mikic, A.; Radovic, S.; Maksimovic, V. Water deficit down-regulates miR398 and miR408 in pea (Pisum sativum L.). Plant Physiol. Biochem. 2014, 83, 26–31. [Google Scholar] [CrossRef] [PubMed]
- De la Rosa, C.; Covarrubias, A.A.; Reyes, J.L. A dicistronic precursor encoding miR398 and the legume-specific miR2119 coregulates CSD1 and ADH1 mRNAs in response to water deficit. Plant Cell Environ. 2019, 42, 133–144. [Google Scholar] [CrossRef]
- Lu, Y.; Feng, Z.; Bian, L.; Xie, H.; Liang, J. miR398 regulation in rice of the responses to abiotic and biotic stresses depends on CSD1 and CSD2 expression. Funct. Plant Biol. 2010, 38, 44–53. [Google Scholar] [CrossRef]
- Gao, Y.; Feng, B.; Gao, C.; Zhang, H.; Wen, F.; Tao, L.; Fu, G.; Xiong, J. The evolution and functional roles of miR408 and its targets in plants. Int. J. Mol. Sci. 2022, 23, 530. [Google Scholar] [CrossRef]
- Esmaeili, F.; Shiran, B.; Fallahi, H.; Mirakhorli, N.; Budak, H.; Martinez-Gomez, P. In silico search and biological validation of microRNAs related to drought response in peach and almond. Funct. Integr. Genom. 2017, 17, 189–201. [Google Scholar] [CrossRef]
- Candar-Cakir, B.; Arican, E.; Zhang, B. Small RNA and degradome deep sequencing reveals drought-and tissue-specific micrornas and their important roles in drought-sensitive and drought-tolerant tomato genotypes. Plant Biotechnol. J. 2016, 14, 1727–1746. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Burd, S.; Lers, A. miR408 is involved in abiotic stress responses in Arabidopsis. Plant J. 2015, 84, 169–187. [Google Scholar] [CrossRef] [PubMed]
- Ghorecha, V.; Patel, K.; Ingle, S.; Sunkar, R.; Krishnayya, N.S. Analysis of biochemical variations and microRNA expression in wild (Ipomoea campanulata) and cultivated (Jacquemontia pentantha) species exposed to in vivo water stress. Physiol. Mol. Biol. Plants. 2014, 20, 57–67. [Google Scholar] [CrossRef]
- Ghorecha, V.; Zheng, Y.; Liu, L.; Sunkar, R.; Krishnayya, N.S.R. MicroRNA dynamics in a wild and cultivated species of Convolvulaceae exposed to drought stress. Physiol. Mol. Biol. Plants. 2017, 23, 291–300. [Google Scholar] [CrossRef]
- Sun, M.; Yang, J.; Cai, X.; Shen, Y.; Cui, N.; Zhu, Y.; Jia, B.; Sun, X. The opposite roles of OsmiR408 in cold and drought stress responses in Oryza sativa. Mol. Breed. 2018, 10, 12. [Google Scholar] [CrossRef]
- Zhang, J.P.; Yu, Y.; Feng, Y.Z.; Zhou, Y.F.; Zhang, F.; Yang, Y.W.; Lei, M.Q.; Zhang, Y.C.; Chen, Y.Q. MiR408 regulates grain yield and photosynthesis via a phytocyanin protein. Plant Physiol. 2017, 175, 1175–1185. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Niu, J.; Cao, X. Heterologous expression of Salvia miltiorrhiza microRNA408 enhances tolerance to salt stress in Nicotiana benthamiana. Int. J. Mol. Sci. 2018, 19, 3985. [Google Scholar] [CrossRef] [PubMed]
| Types | Normalized Reads | ||||
|---|---|---|---|---|---|
| Mock | PEG6000 | ||||
| C0 (0 h) | M12 (12 h) | M24 (24 h) | T12 (12 h) | T24 (24 h) | |
| Total sRNA | 8,318,209 | 12,456,065 | 14,081,185 | 11,767,476 | 14,303,309 |
| Mapped sRNA | 8,216,527 | 12,298,867 | 13,877,512 | 11,602,168 | 14,101,597 |
| unique reads | 1,800,233 | 2,923,698 | 2,787,519 | 2,625,305 | 3,199,519 |
| Rfam | 2,971,490 | 4,180,176 | 4,808,352 | 4,528,665 | 5,440,521 |
| rRNA | 2,850,733 | 4,006,316 | 4,612,913 | 4,330,644 | 5,219,816 |
| tRNA | 105,892 | 151,185 | 173,190 | 177,060 | 196,697 |
| snRNA | 795 | 1306 | 1416 | 921 | 1401 |
| snoRNA | 14,070 | 21,369 | 20,833 | 20,040 | 22,607 |
| Repeat | 910,921 | 1,300,354 | 1,723,883 | 1,287,662 | 1,349,558 |
| Peanut small RNA | 4,334,116 | 6,818,337 | 7,345,277 | 5,785,841 | 4,365,027 |
| Conserved_miRNA (Total) | 701,945 | 787,691 | 1,068,334 | 589,234 | 643,141 |
| Conserved_miRNA (Unique) | 1396 | 1530 | 1642 | 1508 | 1542 |
| Novel_miRNA (Total) | 65,970 | 75,236 | 100,879 | 56,351 | 63,170 |
| Novel_miRNA (Unique) | 1477 | 1814 | 1895 | 1689 | 1667 |
| TAS (Total) | 2170 | 2775 | 3041 | 1929 | 2045 |
| TAS (Unique) | 257 | 342 | 329 | 288 | 302 |
| NAT (Total) | 704,741 | 1,025,461 | 1,204,380 | 936,825 | 1,145,790 |
| NAT (Unique) | 163,853 | 239,479 | 239,611 | 221,336 | 249,808 |
| exon:+ | 188,098 | 261,121 | 327,090 | 222,604 | 73,051 |
| exon:− | 114,204 | 168,497 | 195,878 | 134,026 | 40,458 |
| intron:+ | 174,486 | 301,486 | 296,639 | 258,942 | 117,667 |
| intron:− | 107,808 | 190,269 | 182,374 | 167,246 | 89,658 |
| other | 2,274,694 | 4,005,801 | 3,966,662 | 3,418,684 | 2,190,047 |
| miRNA Identifier | Normalized Reads (reads/mgs) | Fold-Change | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mock | PEG6000 | Mock | PEG6000 | |||||||
| C0 | M12 | M24 | T12 | T24 | M12 | M24 | T12 | T24 | ||
| ahy-miR408 | ppt-miR408b | 4641.21 | 11,839.82 | 8686.84 | 2212.68 | 1375.16 | 2.55 | 1.87 | 0.48 | 0.30 |
| gma-miR408d | 4634.50 | 11,837.44 | 8682.56 | 2210.28 | 1373.68 | 2.55 | 1.87 | 0.48 | 0.30 | |
| ath-miR408-3p | 4633.16 | 11,837.44 | 8682.56 | 2210.28 | 1373.68 | 2.55 | 1.87 | 0.48 | 0.30 | |
| osa-miR408-3p | 1467.48 | 3723.15 | 2688.65 | 681.56 | 523.45 | 2.54 | 1.83 | 0.46 | 0.36 | |
| ahy-novel_14 | novel_14 | 663.99 | 1271.90 | 794.63 | 153.59 | 248.42 | 1.92 | 1.20 | 0.23 | 0.37 |
| ahy-novel_65 | novel_65 | 268.28 | 473.40 | 534.03 | 191.99 | 102.03 | 1.76 | 1.99 | 0.72 | 0.38 |
| ahy-miR395a | ath-miR395a | 52.31 | 67.63 | 71.20 | 167.99 | 1148.93 | 1.29 | 1.36 | 3.21 | 21.96 |
| ahy-miR398 | zma-miR398a-3p | 104.63 | 215.94 | 166.62 | 52.80 | 16.27 | 2.06 | 1.59 | 0.50 | 0.16 |
| ppe-miR398b | 79.14 | 169.67 | 98.26 | 28.80 | 29.57 | 2.14 | 1.24 | 0.36 | 0.37 | |
| stu-miR398a-3p | 57.68 | 129.33 | 89.72 | 24.00 | 11.83 | 2.24 | 1.56 | 0.42 | 0.21 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Zhang, R.; Chen, Z.; Zhang, X.; Zhang, X.; Tian, Y.; Xue, Y.; Zhang, H.; Li, N.; Bai, D. Characterization of Drought-Responsive miRNAs in Peanut Through Integrated Transcriptomic Approaches. Agriculture 2025, 15, 2190. https://doi.org/10.3390/agriculture15212190
Zhang X, Zhang R, Chen Z, Zhang X, Zhang X, Tian Y, Xue Y, Zhang H, Li N, Bai D. Characterization of Drought-Responsive miRNAs in Peanut Through Integrated Transcriptomic Approaches. Agriculture. 2025; 15(21):2190. https://doi.org/10.3390/agriculture15212190
Chicago/Turabian StyleZhang, Xin, Rui Zhang, Zhenbo Chen, Xiaoyu Zhang, Xiaoji Zhang, Yuexia Tian, Yunyun Xue, Huiqi Zhang, Na Li, and Dongmei Bai. 2025. "Characterization of Drought-Responsive miRNAs in Peanut Through Integrated Transcriptomic Approaches" Agriculture 15, no. 21: 2190. https://doi.org/10.3390/agriculture15212190
APA StyleZhang, X., Zhang, R., Chen, Z., Zhang, X., Zhang, X., Tian, Y., Xue, Y., Zhang, H., Li, N., & Bai, D. (2025). Characterization of Drought-Responsive miRNAs in Peanut Through Integrated Transcriptomic Approaches. Agriculture, 15(21), 2190. https://doi.org/10.3390/agriculture15212190






