Multi-Trait Phenotypic Analysis and Biomass Estimation of Lettuce Cultivars Based on SFM-MVS
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Materials
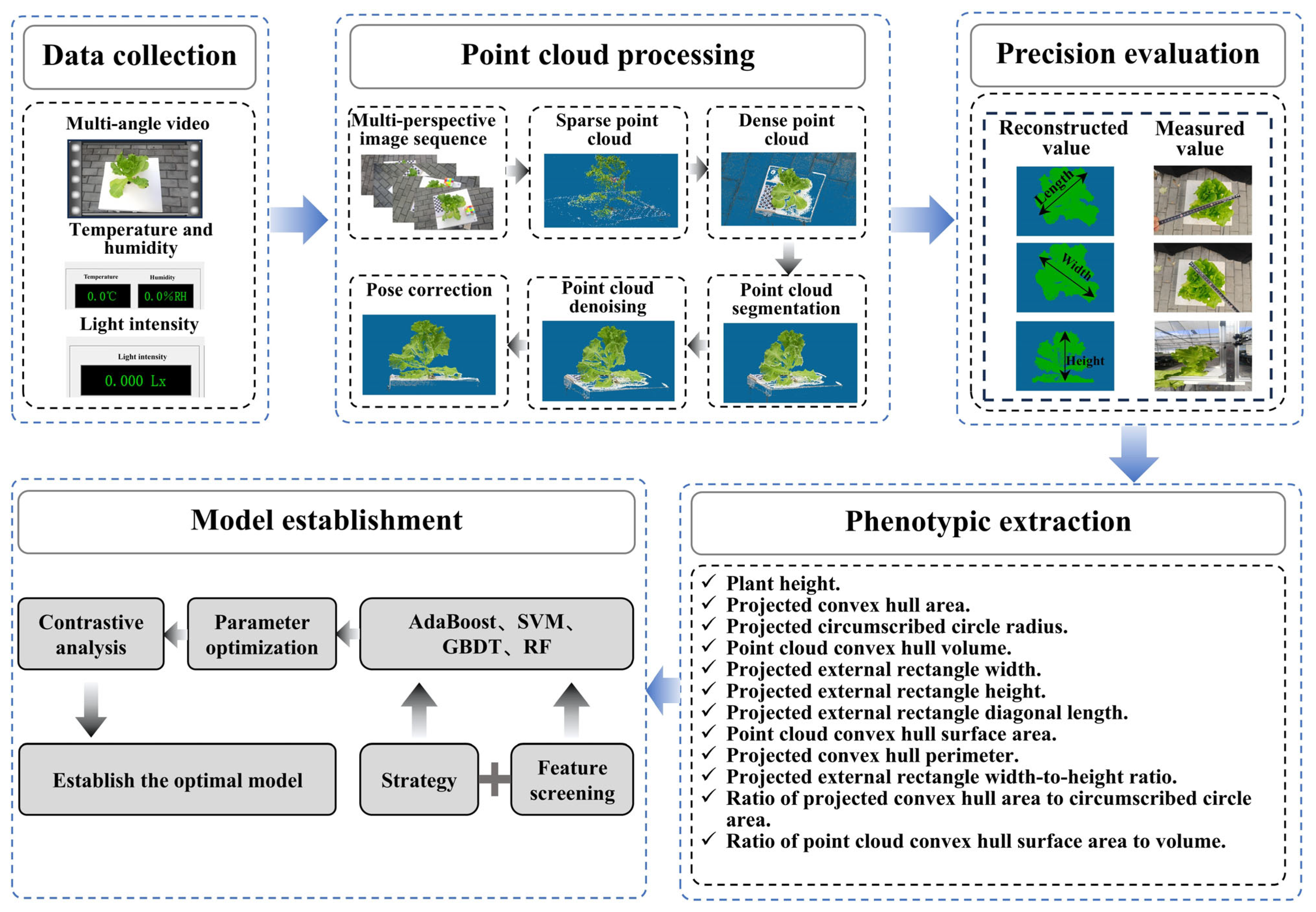
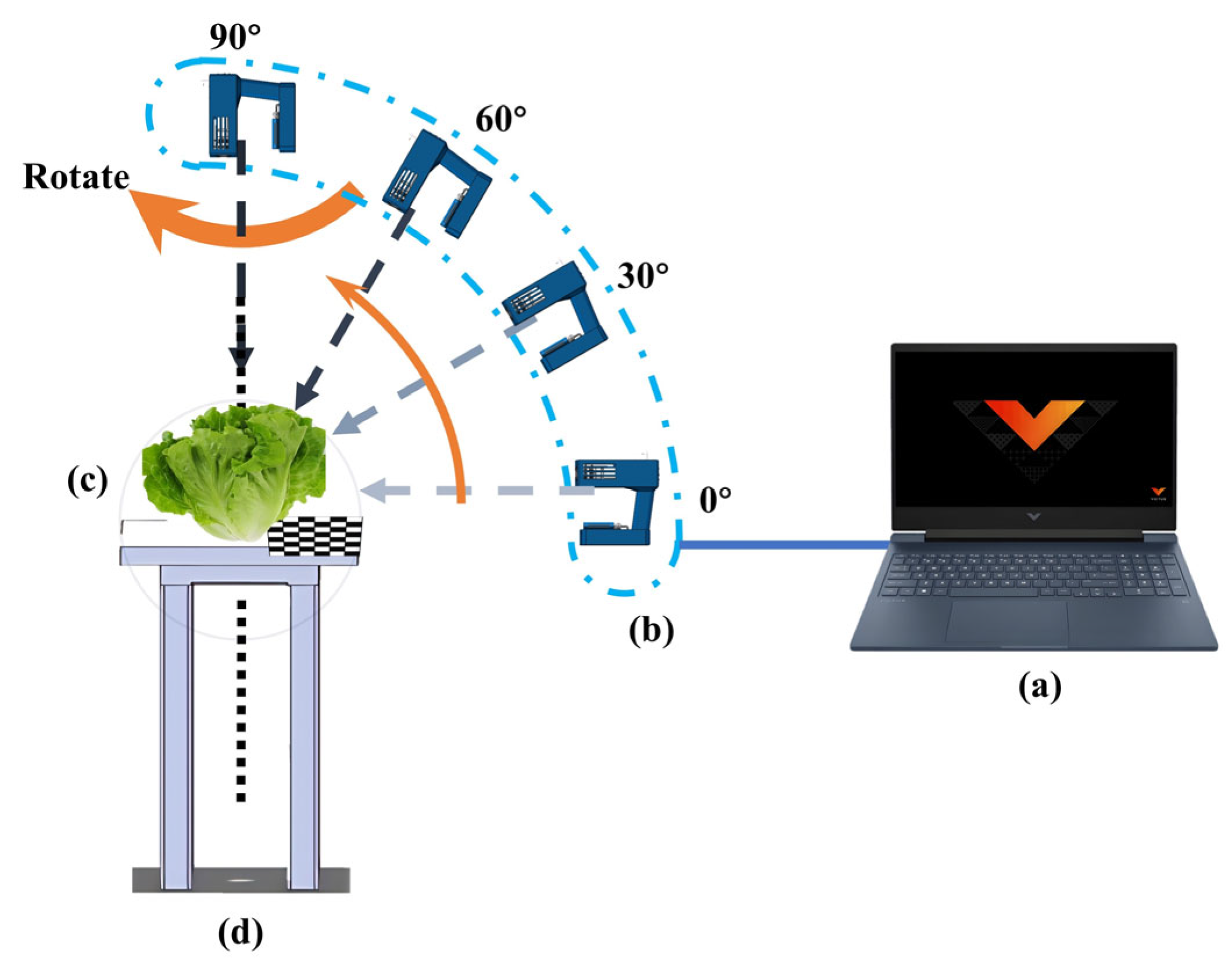
2.2. Overview of the Platform
2.3. Point Cloud Reconstruction
2.3.1. Overview of Point Cloud Processing Workflow
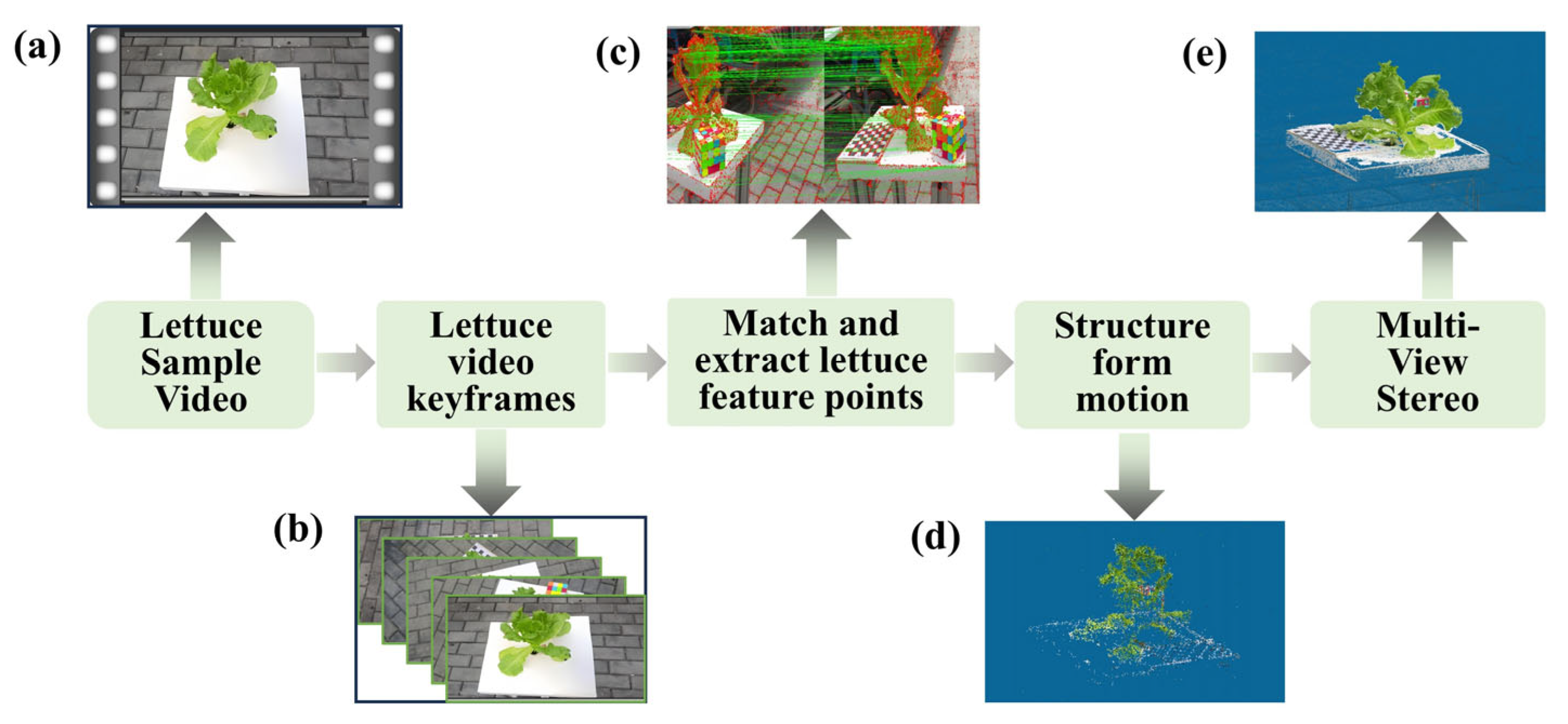
2.3.2. SFM-MVS Reconstruction
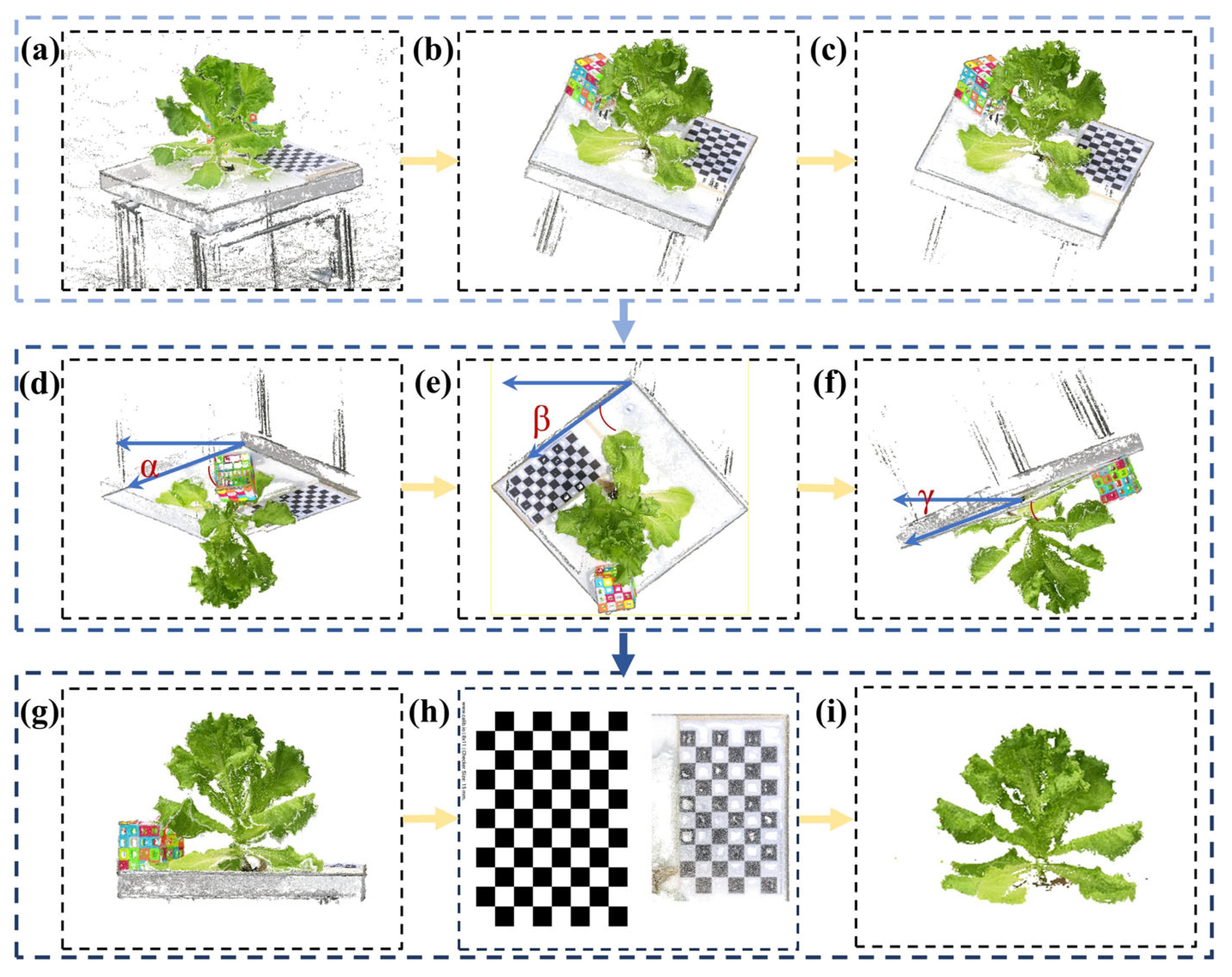
2.3.3. Point Cloud Preprocessing
2.4. Phenotypic Traits Extraction Methods
2.5. Dataset Exploration
2.6. Biomass Estimation Modeling Methods
2.7. Performance Evaluation Metrics
2.8. Parameter Importance Analysis
3. Results
3.1. Comparison of Multi-View Imaging and Light Field Imaging
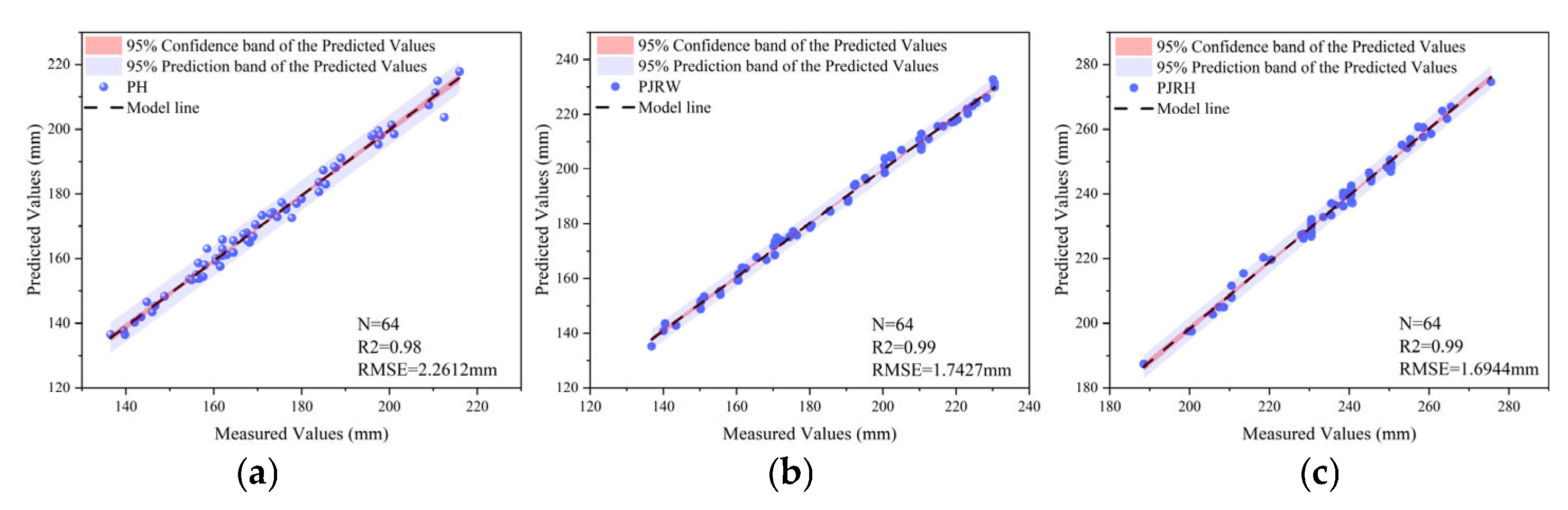
3.2. Evaluation of Phenotypic Data
3.3. SHAP Analysis Results
3.4. Biomass Prediction Based on Machine Learning Algorithms
3.5. Evaluating and Comparing Machine Learning Models for Estimating Aboveground Biomass
4. Discussion
4.1. Relationship Between Morphological Structure and Biomass
4.2. Key Morphological Feature Extraction and Modeling Strategies
4.3. Performance Comparison of Different Machine Learning Models
4.4. Limitations and Future Work
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Diez, Y.; Kentsch, S.; Fukuda, M.; Caceres, M.L.L.; Moritake, K.; Cabezas, M. Deep learning in forestry using UAV-acquired RGB data: A practical review. Remote Sens. 2021, 13, 2837. [Google Scholar] [CrossRef]
- Vázquez-Arellano, M.; Griepentrog, H.W.; Reiser, D.; Paraforos, D.S. 3-D imaging systems for agricultural applications—A review. Sensors 2016, 16, 618. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wen, W.; Wu, S.; Wang, C.; Yu, Z.; Guo, X.; Zhao, C. Maize plant phenotyping: Comparing 3D laser scanning, multi-view stereo reconstruction, and 3D digitizing estimates. Remote Sens. 2018, 11, 63. [Google Scholar] [CrossRef]
- Kim, M.J.; Moon, Y.; Tou, J.C.; Mou, B.; Waterland, N.L. Nutritional value, bioactive compounds and health benefits of lettuce (Lactuca sativa L.). J. Food Compos. Anal. 2016, 49, 19–34. [Google Scholar] [CrossRef]
- Du, H.; Liu, L.; Su, L.; Zeng, F.; Wang, K.; Peng, W.; Zhang, H.; Song, T. Seasonal changes and vertical distribution of fine root biomass during vegetation restoration in a karst area, Southwest China. Front. Plant Sci. 2019, 9, 2001. [Google Scholar] [CrossRef]
- Wei, L.; Yang, H.; Niu, Y.; Zhang, Y.; Xu, L.; Chai, X. Wheat biomass, yield, and straw-grain ratio estimation from multi-temporal UAV-based RGB and multispectral images. Biosyst. Eng. 2023, 234, 187–205. [Google Scholar] [CrossRef]
- Jin, X.; Zarco-Tejada, P.J.; Schmidhalter, U.; Reynolds, M.P.; Hawkesford, M.J.; Varshney, R.K.; Yang, T.; Nie, C.; Li, Z.; Ming, B. High-throughput estimation of crop traits: A review of ground and aerial phenotyping platforms. IEEE Geosci. Remote Sens. Mag. 2020, 9, 200–231. [Google Scholar] [CrossRef]
- Sun, G.; Wang, X. Three-dimensional point cloud reconstruction and morphology measurement method for greenhouse plants based on the kinect sensor self-calibration. Agronomy 2019, 9, 596. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, M.; Li, G.; Li, J.; Zheng, L.; Zhang, M.; Wang, M. Multi-phenotypic parameters extraction and biomass estimation for lettuce based on point clouds. Measurement 2022, 204, 112094. [Google Scholar] [CrossRef]
- Si, C.; Lin, Y.; Luo, S.; Yu, Y.; Liu, R.; Naz, M.; Dai, Z. Effects of LED light quality combinations on growth and leaf colour of tissue culture-generated plantlets in Sedum rubrotinctum. Hortic. Sci. Technol. 2024, 42, 53–67. [Google Scholar] [CrossRef]
- Li, W.; Zhang, C.; Ma, T.; Li, W. Estimation of summer maize biomass based on a crop growth model. Emir. J. Food Agric. (EJFA) 2021, 33, 742–750. [Google Scholar] [CrossRef]
- Tunio, M.H.; Gao, J.; Lakhiar, I.A.; Solangi, K.A.; Qureshi, W.A.; Shaikh, S.A.; Chen, J. Influence of atomization nozzles and spraying intervals on growth, biomass yield, and nutrient uptake of butter-head lettuce under aeroponics system. Agronomy 2021, 11, 97. [Google Scholar] [CrossRef]
- Golbach, F.; Kootstra, G.; Damjanovic, S.; Otten, G.; van de Zedde, R. Validation of plant part measurements using a 3D reconstruction method suitable for high-throughput seedling phenotyping. Mach. Vis. Appl. 2016, 27, 663–680. [Google Scholar] [CrossRef]
- Vulpi, F.; Marani, R.; Petitti, A.; Reina, G.; Milella, A. An RGB-D multi-view perspective for autonomous agricultural robots. Comput. Electron. Agric. 2022, 202, 107419. [Google Scholar] [CrossRef]
- Rebetzke, G.; Jimenez-Berni, J.; Fischer, R.; Deery, D.; Smith, D. High-throughput phenotyping to enhance the use of crop genetic resources. Plant Sci. 2019, 282, 40–48. [Google Scholar] [CrossRef]
- Grzybowski, M.; Wijewardane, N.K.; Atefi, A.; Ge, Y.; Schnable, J.C. Hyperspectral reflectance-based phenotyping for quantitative genetics in crops: Progress and challenges. Plant Commun. 2021, 2, 100209. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Y.; Gu, R. Research status and prospects on plant canopy structure measurement using visual sensors based on three-dimensional reconstruction. Agriculture 2020, 10, 462. [Google Scholar] [CrossRef]
- Ji, Y.; Chen, Z.; Cheng, Q.; Liu, R.; Li, M.; Yan, X.; Li, G.; Wang, D.; Fu, L.; Ma, Y. Estimation of plant height and yield based on UAV imagery in faba bean (Vicia faba L.). Plant Methods 2022, 18, 26. [Google Scholar] [CrossRef]
- Zhu, W.; Feng, Z.; Dai, S.; Zhang, P.; Wei, X. Using UAV multispectral remote sensing with appropriate spatial resolution and machine learning to monitor wheat scab. Agriculture 2022, 12, 1785. [Google Scholar] [CrossRef]
- Li, S.; Yan, Z.; Guo, Y.; Su, X.; Cao, Y.; Jiang, B.; Yang, F.; Zhang, Z.; Xin, D.; Chen, Q. SPM-IS: An auto-algorithm to acquire a mature soybean phenotype based on instance segmentation. Crop J. 2022, 10, 1412–1423. [Google Scholar] [CrossRef]
- Akhtar, M.S.; Zafar, Z.; Nawaz, R.; Fraz, M.M. Unlocking plant secrets: A systematic review of 3D imaging in plant phenotyping techniques. Comput. Electron. Agric. 2024, 222, 109033. [Google Scholar] [CrossRef]
- Qian, Y.; Xu, Q.; Yang, Y.; Lu, H.; Li, H.; Feng, X.; Yin, W. Classification of rice seed variety using point cloud data combined with deep learning. Int. J. Agric. Biol. Eng. 2021, 14, 206–212. [Google Scholar] [CrossRef]
- Cao, W.; Zhou, J.; Yuan, Y.; Ye, H.; Nguyen, H.T.; Chen, J.; Zhou, J. Quantifying variation in soybean due to flood using a low-cost 3D imaging system. Sensors 2019, 19, 2682. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Teng, P.; Aono, M.; Shimizu, Y.; Hosoi, F.; Omasa, K. 3D monitoring for plant growth parameters in field with a single camera by multi-view approach. J. Agric. Meteorol. 2018, 74, 129–139. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Slaughter, D.C.; Max, N.; Maloof, J.N.; Sinha, N. Structured light-based 3D reconstruction system for plants. Sensors 2015, 15, 18587–18612. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.K.; Park, E.-S.; Lee, H.; Priya, G.L.; Kim, H.; Joshi, R.; Arief, M.A.A.; Kim, M.S.; Baek, I.; Cho, B.-K. Deep learning-based plant organ segmentation and phenotyping of sorghum plants using LiDAR point cloud. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2023, 16, 8492–8507. [Google Scholar] [CrossRef]
- Thapa, S.; Zhu, F.; Walia, H.; Yu, H.; Ge, Y. A novel LiDAR-based instrument for high-throughput, 3D measurement of morphological traits in maize and sorghum. Sensors 2018, 18, 1187. [Google Scholar] [CrossRef]
- He, W.; Ye, Z.; Li, M.; Yan, Y.; Lu, W.; Xing, G. Extraction of soybean plant trait parameters based on SfM-MVS algorithm combined with GRNN. Front. Plant Sci. 2023, 14, 1181322. [Google Scholar] [CrossRef]
- Wang, Y.; Hu, S.; Ren, H.; Yang, W.; Zhai, R. 3DPhenoMVS: A low-cost 3D tomato phenotyping pipeline using 3D reconstruction point cloud based on multiview images. Agronomy 2022, 12, 1865. [Google Scholar] [CrossRef]
- Gu, W.; Wen, W.; Wu, S.; Zheng, C.; Lu, X.; Chang, W.; Xiao, P.; Guo, X. 3D Reconstruction of Wheat Plants by Integrating Point Cloud Data and Virtual Design Optimization. Agriculture 2024, 14, 391. [Google Scholar] [CrossRef]
- Zhu, L.; Jiang, W.; Sun, B.; Chai, M.; Li, S.; Ding, Y. Three-dimensional Reconstruction and Phenotype Parameters Acquisition of Seeding Vegetables Based on Neural Radiance Fields. Trans. Chin. Soc. Agric. Mach. 2024, 55, 391. [Google Scholar]
- Iglhaut, J.; Cabo, C.; Puliti, S.; Piermattei, L.; O’Connor, J.; Rosette, J. Structure from motion photogrammetry in forestry: A review. Curr. For. Rep. 2019, 5, 155–168. [Google Scholar] [CrossRef]
- Hui, F.; Zhu, J.; Hu, P.; Meng, L.; Zhu, B.; Guo, Y.; Li, B.; Ma, Y. Image-based dynamic quantification and high-accuracy 3D evaluation of canopy structure of plant populations. Ann. Bot. 2018, 121, 1079–1088. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Sun, L.; Zhang, J.; Fan, X.; Zhang, D.; Ren, T.; Liu, M.; Zhang, Z.; Ma, W. Comprehensive Analysis of Phenotypic Traits in Chinese Cabbage Using 3D Point Cloud Technology. Agronomy 2024, 14, 2506. [Google Scholar] [CrossRef]
- Wu, S.; Wen, W.; Wang, Y.; Fan, J.; Wang, C.; Gou, W.; Guo, X. MVS-Pheno: A portable and low-cost phenotyping platform for maize shoots using multiview stereo 3D reconstruction. Plant Phenomics 2020, 2020, 1848437. [Google Scholar] [CrossRef]
- Bhuyan, B.P.; Tomar, R.; Singh, T.; Cherif, A.R. Crop type prediction: A statistical and machine learning approach. Sustainability 2022, 15, 481. [Google Scholar] [CrossRef]
- Castillo-Botón, C.; Casillas-Pérez, D.; Casanova-Mateo, C.; Ghimire, S.; Cerro-Prada, E.; Gutierrez, P.A.; Deo, R.C.; Salcedo-Sanz, S. Machine learning regression and classification methods for fog events prediction. Atmos. Res. 2022, 272, 106157. [Google Scholar] [CrossRef]
- Paidipati, K.K.; Chesneau, C.; Nayana, B.; Kumar, K.R.; Polisetty, K.; Kurangi, C. Prediction of rice cultivation in India—Support vector regression approach with various kernels for non-linear patterns. AgriEngineering 2021, 3, 182–198. [Google Scholar] [CrossRef]
- Apergis, N.; Manickavasagam, J.; Visalakshmi, S. A novel hybrid approach to forecast crude oil futures using intraday data. Technol. Forecast. Soc. Change 2020, 158, 120126. [Google Scholar]
- Schonberger, J.L.; Frahm, J.-M. Structure-from-motion revisited. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 4104–4113. [Google Scholar]
- Schönberger, J.L.; Zheng, E.; Frahm, J.-M.; Pollefeys, M. Pixelwise view selection for unstructured multi-view stereo. In Proceedings, Part III 14, Proceedings of the Computer Vision–ECCV 2016: 14th European Conference, Amsterdam, The Netherlands, 11–14 October 2016; Springer: Cham, Switzerland, 2016; pp. 501–518. [Google Scholar]
- Jin, S.; Bi, H.; Feng, J.; Xu, W.; Xu, J.; Zhang, J. Research on 4-D Imaging of Holographic SAR Differential Tomography. Remote Sens. 2023, 15, 3421. [Google Scholar] [CrossRef]
- Wu, S.; Wen, W.; Gou, W.; Lu, X.; Zhang, W.; Zheng, C.; Xiang, Z.; Chen, L.; Guo, X. A miniaturized phenotyping platform for individual plants using multi-view stereo 3D reconstruction. Front. Plant Sci. 2022, 13, 897746. [Google Scholar] [CrossRef]
- Combs, T.P.; Didan, K.; Dierig, D.; Jarchow, C.J.; Barreto-Muñoz, A. Estimating Productivity Measures in Guayule Using UAS Imagery and Sentinel-2 Satellite Data. Remote Sens. 2022, 14, 2867. [Google Scholar] [CrossRef]
- Guo, Y.; Schöb, C.; Ma, W.; Mohammat, A.; Liu, H.; Yu, S.; Jiang, Y.; Schmid, B.; Tang, Z. Increasing water availability and facilitation weaken biodiversity–biomass relationships in shrublands. Ecology 2019, 100, e02624. [Google Scholar] [CrossRef] [PubMed]
- Vermeiren, J.; Villers, S.L.; Wittemans, L.; Vanlommel, W.; van Roy, J.; Marien, H.; Coussement, J.R.; Steppe, K. Quantifying the importance of a realistic tomato (Solanum lycopersicum) leaflet shape for 3-D light modelling. Ann. Bot. 2020, 126, 661–670. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Han, W.; Zhang, H.; Zhang, L.; Chen, H. Estimating maize plant height using a crop surface model constructed from UAV RGB images. Biosyst. Eng. 2024, 241, 56–67. [Google Scholar] [CrossRef]
- Jiao, Y.; Niklas, K.J.; Wang, L.; Yu, K.; Li, Y.; Shi, P. Influence of leaf age on the scaling relationships of lamina mass vs. area. Front. Plant Sci. 2022, 13, 860206. [Google Scholar] [CrossRef]
- Dieleman, J.A.; De Visser, P.H.; Meinen, E.; Grit, J.G.; Dueck, T.A. Integrating morphological and physiological responses of tomato plants to light quality to the crop level by 3D modeling. Front. Plant Sci. 2019, 10, 839. [Google Scholar] [CrossRef]
- Nasim, W.; Ahmad, A.; Belhouchette, H.; Fahad, S.; Hoogenboom, G. Evaluation of the OILCROP-SUN model for sunflower hybrids under different agro-meteorological conditions of Punjab—Pakistan. Field Crops Res. 2016, 188, 17–30. [Google Scholar] [CrossRef]
- Dhawi, F.; Ghafoor, A.; Almousa, N.; Ali, S.; Alqanbar, S. Predictive modelling employing machine learning, convolutional neural networks (CNNs), and smartphone RGB images for non-destructive biomass estimation of pearl millet (Pennisetum glaucum). Front. Plant Sci. 2025, 16, 1594728. [Google Scholar] [CrossRef]
- Parsa, A.B.; Movahedi, A.; Taghipour, H.; Derrible, S.; Mohammadian, A.K. Toward safer highways, application of XGBoost and SHAP for real-time accident detection and feature analysis. Accid. Anal. Prev. 2020, 136, 105405. [Google Scholar] [CrossRef]
- Lim, B.; Zohren, S. Time-series forecasting with deep learning: A survey. Philos. Trans. R. Soc. A 2021, 379, 20200209. [Google Scholar] [CrossRef]
- Santoši, Ž.; Budak, I.; Stojaković, V.; Šokac, M.; Vukelić, Đ. Evaluation of synthetically generated patterns for image-based 3D reconstruction of texture-less objects. Measurement 2019, 147, 106883. [Google Scholar] [CrossRef]
- Li, Y.; Liu, J.; Zhang, B.; Wang, Y.; Yao, J.; Zhang, X.; Fan, B.; Li, X.; Hai, Y.; Fan, X. Three-dimensional reconstruction and phenotype measurement of maize seedlings based on multi-view image sequences. Front. Plant Sci. 2022, 13, 974339. [Google Scholar] [CrossRef] [PubMed]
- Tatsumi, K.; Igarashi, N.; Mengxue, X. Prediction of plant-level tomato biomass and yield using machine learning with unmanned aerial vehicle imagery. Plant Methods 2021, 17, 77. [Google Scholar] [CrossRef]
- Dhakal, R.; Maimaitijiang, M.; Chang, J.; Caffe, M. Utilizing spectral, structural and textural features for estimating oat above-ground biomass using UAV-based multispectral data and machine learning. Sensors 2023, 23, 9708. [Google Scholar] [CrossRef] [PubMed]
- Cardenas-Gallegos, J.S.; Lacerda, L.N.; Severns, P.M.; Peduzzi, A.; Klimeš, P.; Ferrarezi, R.S. Advancing biomass estimation in hydroponic lettuce using RGB-depth imaging and morphometric descriptors with machine learning. Comput. Electron. Agric. 2025, 234, 110299. [Google Scholar] [CrossRef]
- Yu, D.; Zha, Y.; Sun, Z.; Li, J.; Jin, X.; Zhu, W.; Bian, J.; Ma, L.; Zeng, Y.; Su, Z. Deep convolutional neural networks for estimating maize above-ground biomass using multi-source UAV images: A comparison with traditional machine learning algorithms. Precis. Agric. 2023, 24, 92–113. [Google Scholar] [CrossRef]
- Viljanen, N.; Honkavaara, E.; Näsi, R.; Hakala, T.; Niemeläinen, O.; Kaivosoja, J. A novel machine learning method for estimating biomass of grass swards using a photogrammetric canopy height model, images and vegetation indices captured by a drone. Agriculture 2018, 8, 70. [Google Scholar] [CrossRef]
- Xiao, S.; Chai, H.; Shao, K.; Shen, M.; Wang, Q.; Wang, R.; Sui, Y.; Ma, Y. Image-based dynamic quantification of aboveground structure of sugar beet in field. Remote Sens. 2020, 12, 269. [Google Scholar] [CrossRef]
- Maimaitijiang, M.; Sagan, V.; Sidike, P.; Hartling, S.; Esposito, F.; Fritschi, F.B. Soybean yield prediction from UAV using multimodal data fusion and deep learning. Remote Sens. Environ. 2020, 237, 111599. [Google Scholar] [CrossRef]
- Wan, L.; Cen, H.; Zhu, J.; Zhang, J.; Zhu, Y.; Sun, D.; Du, X.; Zhai, L.; Weng, H.; Li, Y. Grain yield prediction of rice using multi-temporal UAV-based RGB and multispectral images and model transfer—A case study of small farmlands in the South of China. Agric. For. Meteorol. 2020, 291, 108096. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, X.; Sun, J.; Yu, T.; Cai, Z.; Zhang, Z.; Mao, H. Low-cost lettuce height measurement based on depth vision and lightweight instance segmentation model. Agriculture 2024, 14, 1596. [Google Scholar] [CrossRef]
- Cutler, D.R.; Edwards, T.C., Jr.; Beard, K.H.; Cutler, A.; Hess, K.T.; Gibson, J.; Lawler, J.J. Random forests for classification in ecology. Ecology 2007, 88, 2783–2792. [Google Scholar] [CrossRef]
- Zhai, W.; Li, C.; Cheng, Q.; Mao, B.; Li, Z.; Li, Y.; Ding, F.; Qin, S.; Fei, S.; Chen, Z. Enhancing wheat above-ground biomass estimation using UAV RGB images and machine learning: Multi-feature combinations, flight height, and algorithm implications. Remote Sens. 2023, 15, 3653. [Google Scholar] [CrossRef]
- Wengert, M.; Piepho, H.-P.; Astor, T.; Graß, R.; Wijesingha, J.; Wachendorf, M. Assessing spatial variability of barley whole crop biomass yield and leaf area index in silvoarable agroforestry systems using UAV-borne remote sensing. Remote Sens. 2021, 13, 2751. [Google Scholar] [CrossRef]


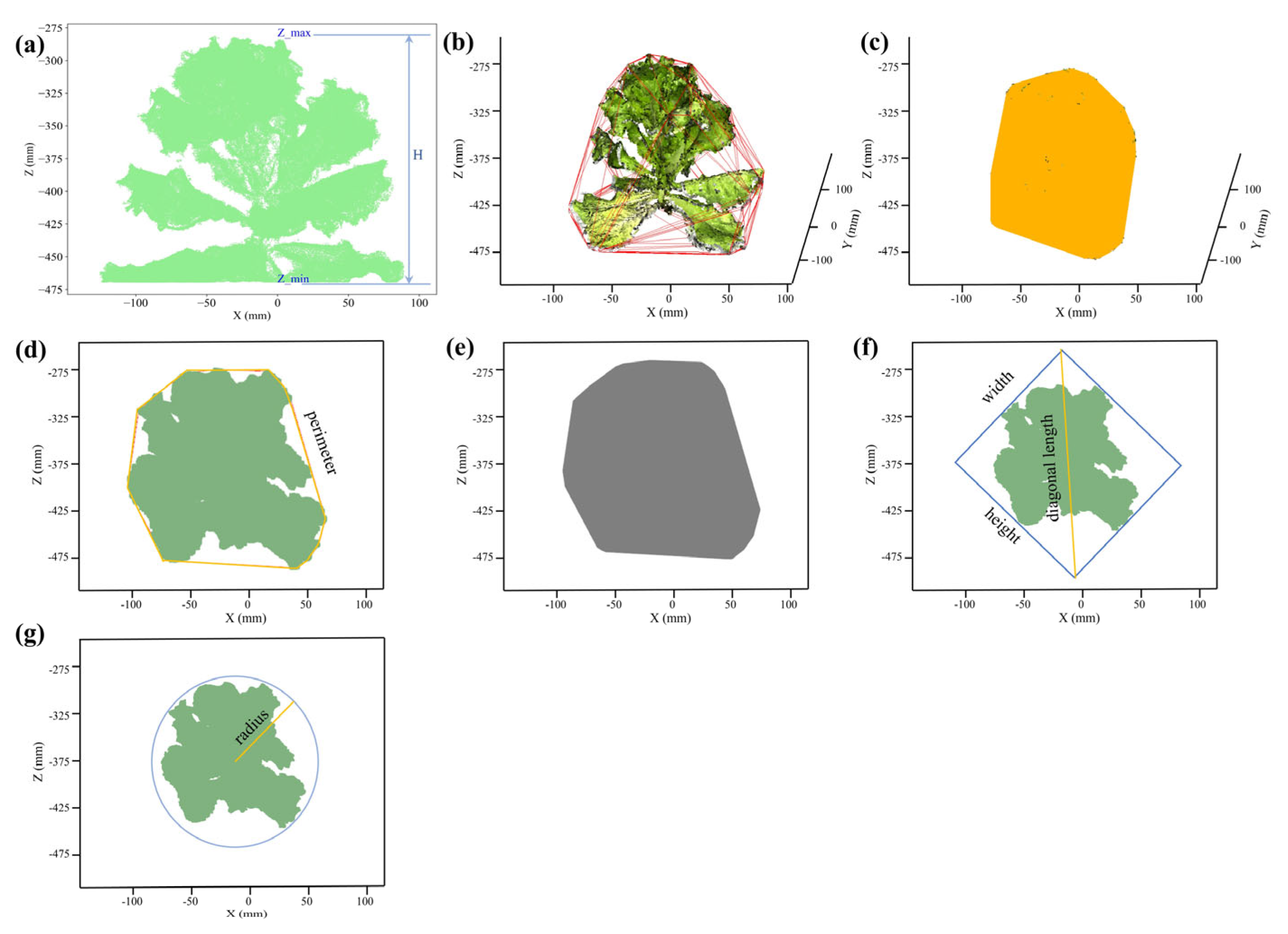




| Phenotypic Parameters | Abbreviations |
|---|---|
| Plant height | PH |
| Projected convex hull area | PCHA |
| Projected circumscribed circle radius | PCCR |
| Point cloud convex hull volume | PCCHV |
| Projected external rectangle width | PER-W |
| Projected external rectangle height | PER-H |
| Projected external rectangle diagonal length | PER-DL |
| Point cloud convex hull surface area | PCCHSA |
| Projected convex hull perimeter | PCHP |
| Projected external rectangle width-to-height ratio | PER-WHR |
| Ratio of projected convex hull area to circumscribed circle area | PCHA/CCA |
| Ratio of point cloud convex hull surface area to volume | PCCHSA/PCCHV |
| ML Method | Features | List of Hyperparameters and Their Optimal Value |
|---|---|---|
| RFR | 3 features | {‘max_depth’: None, ‘max_features’: None, ‘min_samples_leaf’: 2, ‘min_samples_split’: 3, ‘n_estimators’: 200} |
| 6 features | {‘max_depth’: 5, ‘max_features’: None, ‘min_samples_leaf’: 1, ‘min_samples_split’: 3, ‘n_estimators’: 200} | |
| 12 features | {‘max_depth’: 5, ‘max_features’: None, ‘min_samples_leaf’: 1, ‘min_samples_split’: 2, ‘n_estimators’: 50} | |
| GBDT | 3 features | {‘learning_rate’: 0.05, ‘max_depth’: 5, ‘max_features’: ‘sqrt’, ‘n_estimators’: 100, ‘subsample’: 0.7} |
| 6 features | {‘learning_rate’: 0.1, ‘max_depth’: 3, ‘max_features’: ‘sqrt’, ‘n_estimators’: 200, ‘subsample’: 0.7} | |
| 12 features | {‘learning_rate’: 0.05, ‘max_depth’: 5, ‘max_features’: ‘sqrt’, ‘n_estimators’: 100, ‘subsample’: 0.7} | |
| SVR | 3 features | {‘C’: 100, ‘degree’: 2, ‘epsilon’: 0.2, ‘gamma’: ‘scale’, ‘kernel’: ‘linear’} |
| 6 features | {‘C’: 10, ‘degree’: 2, ‘epsilon’: 0.5, ‘gamma’: 0.1, ‘kernel’: ‘rbf’} | |
| 12 features | {‘C’: 100, ‘degree’: 2, ‘epsilon’: 0.2, ‘gamma’: ‘scale’, ‘kernel’: ‘linear’} | |
| AdaBoost | 3 features | {‘learning_rate’: 1, ‘loss’: ‘square’, ‘n_estimators’: 50} |
| 6 features | {‘learning_rate’: 0.1, ‘loss’: ‘square’, ‘n_estimators’: 50} | |
| 12 features | {‘learning_rate’: 0.01, ‘loss’: ‘linear’, ‘n_estimators’: 50} |
| Features | ML Model | R2 (Train) | RMSE (g) (Train) | RMSEn (%) (Train) | R2 (Test) | RMSE (g) (Test) | RMSEn (%) (Test) |
|---|---|---|---|---|---|---|---|
| 3 features | RFR | 0.97 | 1.42 | 4.94 | 0.90 | 2.63 | 9.53 |
| GBDT | 0.99 | 0.25 | 0.88 | 0.88 | 2.82 | 10.20 | |
| SVR | 0.90 | 2.62 | 9.10 | 0.91 | 2.40 | 8.68 | |
| AdaBoost | 0.97 | 1.37 | 4.76 | 0.89 | 2.79 | 10.09 | |
| 6 features | RFR | 0.98 | 1.07 | 3.70 | 0.90 | 2.58 | 9.34 |
| GBDT | 0.99 | 0.02 | 0.06 | 0.88 | 2.81 | 10.19 | |
| SVR | 0.94 | 2.01 | 6.99 | 0.92 | 2.32 | 8.40 | |
| AdaBoost | 0.98 | 1.25 | 4.34 | 0.89 | 2.73 | 9.87 | |
| 12 features | RFR | 0.98 | 1.16 | 4.01 | 0.90 | 2.63 | 9.53 |
| GBDT | 0.99 | 0.17 | 0.58 | 0.93 | 2.11 | 7.63 | |
| SVR | 0.92 | 2.39 | 8.29 | 0.90 | 2.59 | 9.38 | |
| AdaBoost | 0.98 | 1.23 | 4.27 | 0.89 | 2.69 | 9.74 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, T.; Zhang, Y.; Hu, L.; Zhao, Y.; Cai, Z.; Yu, T.; Zhang, X. Multi-Trait Phenotypic Analysis and Biomass Estimation of Lettuce Cultivars Based on SFM-MVS. Agriculture 2025, 15, 1662. https://doi.org/10.3390/agriculture15151662
Li T, Zhang Y, Hu L, Zhao Y, Cai Z, Yu T, Zhang X. Multi-Trait Phenotypic Analysis and Biomass Estimation of Lettuce Cultivars Based on SFM-MVS. Agriculture. 2025; 15(15):1662. https://doi.org/10.3390/agriculture15151662
Chicago/Turabian StyleLi, Tiezhu, Yixue Zhang, Lian Hu, Yiqiu Zhao, Zongyao Cai, Tingting Yu, and Xiaodong Zhang. 2025. "Multi-Trait Phenotypic Analysis and Biomass Estimation of Lettuce Cultivars Based on SFM-MVS" Agriculture 15, no. 15: 1662. https://doi.org/10.3390/agriculture15151662
APA StyleLi, T., Zhang, Y., Hu, L., Zhao, Y., Cai, Z., Yu, T., & Zhang, X. (2025). Multi-Trait Phenotypic Analysis and Biomass Estimation of Lettuce Cultivars Based on SFM-MVS. Agriculture, 15(15), 1662. https://doi.org/10.3390/agriculture15151662








