1. Introduction
Given the intensification of livestock production and increasing emphasis on animal welfare, dairy cow health management has become a critical component for ensuring milk yield and quality, and lameness is one of the most severe diseases affecting production performance and welfare levels in cows [
1]. It has been shown that lameness in cows results in an average 23% reduction in milk yield [
2], and treatment costs associated with lameness rank second among common diseases. Without timely intervention, more serious health complications may arise [
3]. Early detection and timely treatment of lame cows are essential for interrupting the chronic cycle associated with this condition [
4]. Therefore, early lameness detection in cows facilitates the prompt identification and treatment of hoof disorders, thereby reducing economic losses.
Manual identification exhibits drawbacks such as low efficiency, high cost, and strong subjectivity [
5]. Computer vision-based detection methods extracted lameness characteristic parameters and constructed diagnostic models through techniques such as foreground detection, image segmentation, keypoint localization, and trajectory tracking. Related research has also been conducted in subtle motion feature extraction [
5], key behavior detection [
6], multi-scale keypoint detection [
7], three-dimensional information from depth cameras [
8], and thermal infrared temperature measurement [
9]. Livestock behavior recognition using computer vision has been found to face substantial challenges in complex, dynamic farm environments due to highly variable scenes, lighting, and background conditions, which hinder reliable performance and limit industrial-scale adoption [
10,
11]. Moreover, accurate operation often requires animals to traverse dedicated alleys under fixed camera mounts, and long-term monitoring may be compromised by dust, humidity, and fluctuating light, making large-scale deployment technically challenging [
12].
Recently, increasing attention has been devoted to monitoring cow behavior and health using wearable devices, particularly through the extraction and modeling of gait data to recognize movement states and early lameness signs. Early studies predominantly employed low-frequency accelerometers (10–50 Hz) combined with traditional machine learning algorithms, with devices deployed on the cow’s neck [
13] or limbs [
14]. Basic features such as activity level and Stride Frequency were extracted to construct binary classification models, achieving sensitivities up to 77% but remaining susceptible to environmental interference [
15]. Limited feature representation capability made differentiation between early lameness and normal gait deviations difficult. To improve detection accuracy, research shifted to high-frequency sampling devices (100–400 Hz) and refined gait analysis, applying methods such as peak detection and time-frequency transforms to extract gait parameters, including Step Amplitude cycle and root mean square values [
16,
17]. However, these approaches depend on high-frequency sensors, which inherently suffer from high power consumption and short battery life, and impose substantial challenges in data storage and transmission. In cows’ activity, actual walking time accounts for only a small proportion; continuous acquisition without filtering generates large volumes of redundant stationary data, further burdening the system. Recent efforts have sought to integrate deep learning to enhance model generalization, such as employing LSTM-Autoencoders to process six-axis sensor time series data [
18] or augmenting time-frequency feature expression via wavelet transforms [
19]. Although these methods can capture subtle gait anomalies, they typically rely on high-quality annotated data and complex offline analysis pipelines, and they lack pre-screening mechanisms for raw data, making them unsuitable for online processing on low-power devices.
With the widespread application of time-series detection methods in fields such as electrocardiography [
20,
21] and industrial equipment monitoring [
22,
23], techniques for online detection and rapid alerting have attracted increasing academic attention. Existing studies have demonstrated that machine-learning-based online ECG classification systems can achieve a recognition accuracy of 90.89% [
24] and that multivariate time-series models exhibit high robustness in the presence of artifacts, effectively reducing false alarms for myocardial infarction [
21]. Furthermore, LSTM networks have been widely employed to construct online anomaly-detection frameworks in smart-grid and IoT scenarios [
25]. These approaches highlight the potential of deep-learning-based time-series models for real-time monitoring in complex environments.
Inspired by these advances, this study sought to introduce mature sequence-modeling techniques into the task of early lameness detection in cows by identifying an architecture suited to the characteristics of dairy cow gait signals.
Comparative analyses indicated that the InceptionTime network, owing to its multi-scale convolutional structure, offers significant advantages in mining temporal patterns and exhibits strong feature-extraction capability and adaptability [
26], making it well suited to handling non-stationary, highly variable gait data [
27]. The network’s ability to automatically learn hierarchical time-domain features has led to excellent performance across various time-series classification tasks. However, the relatively large size of the InceptionTime architecture renders it ill-suited for deployment on resource-constrained edge devices. Moreover, static periods dominate in dairy farms, and walking data are sparse. As a result, a conventional InceptionTime network tends to overfit on the limited samples and struggles to balance detection accuracy with online processing efficiency [
28]. Consequently, it is necessary to optimize the network architecture and integrate additional feature-fusion strategies tailored to the specific demands of cow gait analysis.
To date, early online detection of dairy cow gait abnormalities still faces several technical bottlenecks. First, traditional sensor-based monitoring methods rely heavily on manual data screening, and the collected data typically require offline analysis, which does not satisfy the need for real-time early detection. Second, although deep-learning techniques offer powerful feature-extraction capabilities, their complexity hinders on-site deployment in farm environments; moreover, the limited availability of annotated dairy cow gait samples increases the risk of model overfitting. Third, because static intervals comprise a large proportion of dairy cow gait recordings and motion data are sparse, general time-series models (e.g., LSTM) often fail to perform reliably on such data. These challenges point to the urgent need for a more robust, adaptive method that supports early online lameness detection.
To address these bottlenecks, this study proposes a lightweight online lameness detection model featuring three key innovations. First, a threshold discrimination module based on variance was designed to automatically distinguish between static and motion data windows, thereby filtering out redundant stationary data and enhancing online-analysis efficiency. Second, the InceptionTime architecture was upgraded by replacing standard Conv1D layers with YOLOConv1D and SeparableConv1D modules and inserting Dropout layers after each Inception block, which reduced model parameter count and helped mitigate overfitting. Finally, handcrafted expert features (Stride Frequency, Signal Amplitude, Step Amplitude, and Support Time) were fused with the multi-scale temporal features learned by the deep network, resulting in improved discrimination accuracy on limited gait datasets. Literature review indicates that no prior research has applied an improved InceptionTime network to dairy cow lameness detection.
2. Materials and Methods
2.1. Wearable Sensor Data Acquisition
Data acquisition was carried out in September 2024 at the Beijing Agricultural Machinery Experimental Station. A short habituation period was included to minimize behavioral artifacts associated with unfamiliar equipment. One week before formal data collection, each experimental cow was fitted with the same limb-mounted IMU (LPMS-B2, Alubi, Guangzhou, China) at the intended attachment point once per day. Sensor-wear time was increased progressively—from roughly 2 h on the first 2 days, to 6 h on days 3–5, and to a full working day on the day immediately preceding the trial. Throughout this week, trained stock personnel observed the animals at 15-min intervals and recorded signs of head-shaking, limb-kicking, altered stride length, or changes in feed intake. No abnormal behaviors were noted after the third day, and stride parameters measured during the final acclimation session overlapped the baseline range obtained without sensors. These observations indicate that the cows had adapted to the device before the start of data acquisition in September 2024, reducing the likelihood that novelty effects influenced the recorded gait signals.
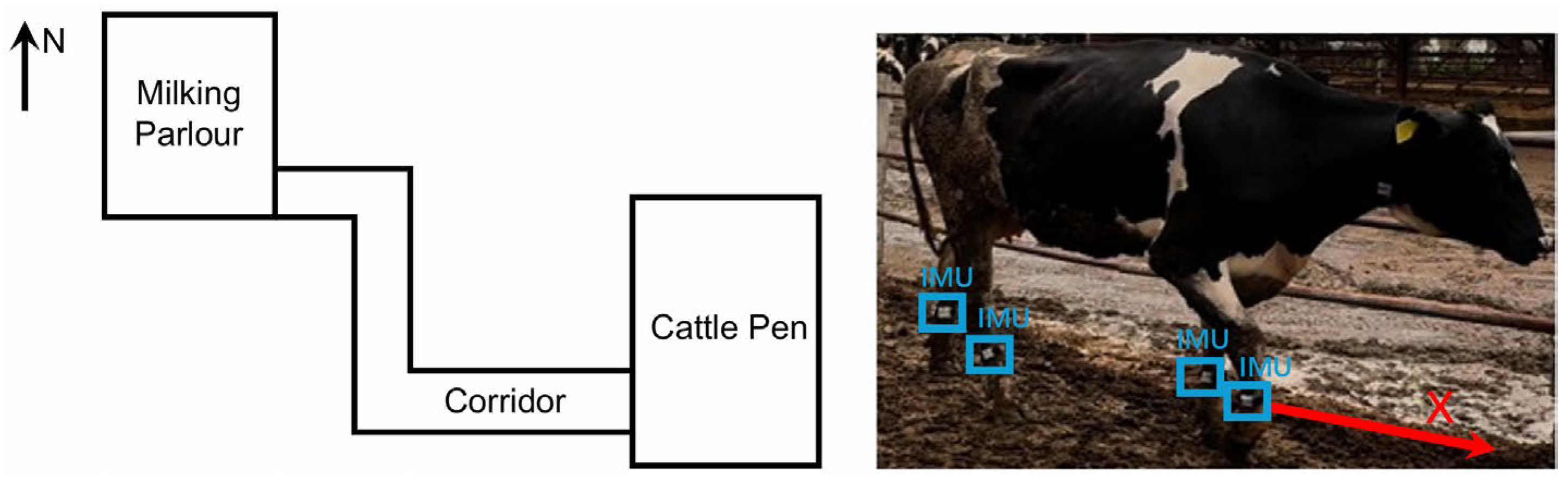
Wearable IMU sensors with a sampling frequency of 30 Hz were employed to capture acceleration signals during cows’ locomotion. To ensure the accuracy and consistency of the collected data, all sensors were rigorously calibrated prior to deployment and mounted on the metatarsal region of each cow’s limb to prevent displacement during walking; the sensor X-axis was aligned parallel to each cow’s walking direction. Video recordings were used for time synchronization.
At the onset of each trial, cows were guided from the milking parlor along a 30 m corridor to the pen (
Figure 1). Continuous back-and-forth walking within the corridor was enforced for at least 20 min to acquire sufficient high-quality walking data. Subsequently, cows entered the pen, and acceleration data were collected for the stationary states (standing and recumbency) without interference with their natural behavior.
2.2. Animal Ethics Approval
The animal study protocol was approved by the Institutional Review Board (or Ethics Committee) of the Agricultural Information Institute of the Chinese Academy of Agricultural Sciences (protocol code AII2021-810, approved on 10 August 2021).
2.3. Dataset Definition
The health status of 36 lactating Holstein cows (36–58 months old) was scored by three experts using the five-point Numerical Rating System (NRS) [
29], where 1 denotes sound and 5 denotes severely lame. Mean scores were used to classify the animals into three groups: healthy (mean < 1.5;
n = 12), early lameness (1.5 ≤ mean ≤ 2.5;
n = 12), and severe lameness (mean > 2.5;
n = 12) [
16]. Because locomotion predominantly occurs along the sensor
X-axis and its acceleration signal most directly reflects key gait-cycle events (acceleration, deceleration, and impact peaks), only the 30 Hz
X-axis acceleration data were retained for analysis to reduce interference from other axes.
Because processing full tri-axial signals on resource-limited edge devices would significantly increase computational load, memory transfers, and energy consumption, and because the sensor is mounted on the distal metatarsus with its X-axis closely aligned to the walking direction—providing clear cyclic patterns of swing, hoof contact, and stride timing—while the Z-axis, being perpendicular to the walking direction, offers no additional discriminative information, and the Y-axis, though capable of capturing limb-lift and impact events, was omitted to simplify the processing pipeline and remain within the embedded platform’s resource budget, only the X-axis acceleration was retained for both variance-threshold screening and deep-learning analysis.
A sliding-window approach (window length
λ = 120, frames ≈ 4 s, step size = 1 frame) was applied to the continuous
X-axis time series, yielding 418,863 windowed samples in total (
Table 1). A total of 94,373 windows corresponded to stationary states and 324,490 to motion states (healthy: 136,790; early: 80,893; severe: 106,807). To avoid data leakage and evaluate generalization on unseen individuals, a stratified split was performed at the cow level: one cow per health category (3 cows total) was randomly held out as an independent test set, contributing 15,840 windows (healthy: 4517; early: 5377; severe: 5946). The remaining 33 cows (308,650 windows) were randomly shuffled and partitioned into a training set (246,920 windows) and a validation set (61,730 windows) in an 8:2 ratio, with class proportions maintained (
Table 1). This ensured that all test samples originated from cows not represented in training or validation, thereby providing a realistic assessment of model performance on “unseen” subjects.
2.4. Online Lameness Detection Model Architecture Design
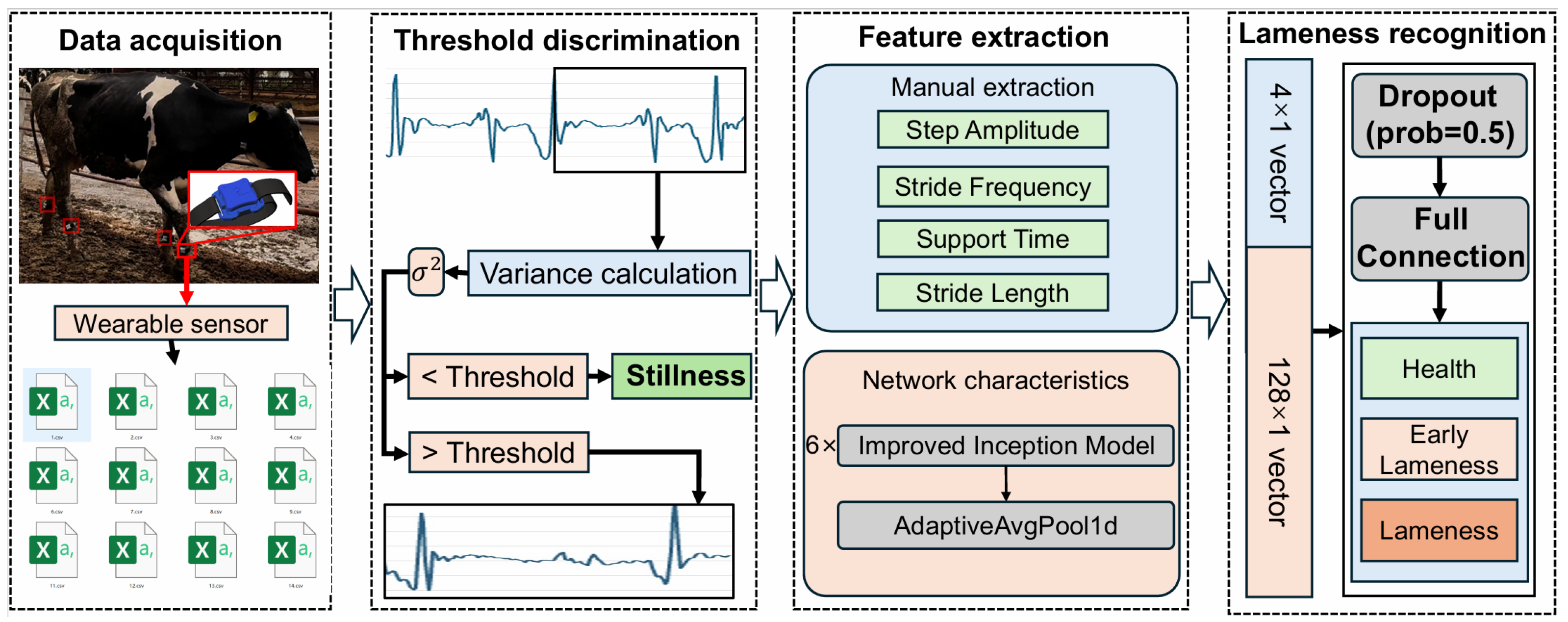
The overall architecture of the proposed Online Early Lameness Detection Model is shown in
Figure 2. It comprised the following modules: sensor-data preprocessing, preliminary state assessment, feature extraction, deep-feature learning, and feature fusion. Acceleration data were segmented into fixed-length windows, and an initial classification was performed by computing the variance of the
X-axis acceleration within each window using a threshold discrimination module. When a window was classified as motion, a dual-branch feature-extraction pipeline was activated to distinguish among healthy gait, early lameness, and severe lameness.
In the handcrafted branch, four key gait features (Stride Frequency, Signal Amplitude, Step Amplitude, and Support Time) were extracted to form a 4 × 1 feature vector. Simultaneously, the raw windowed data were fed into multiple improved Inception modules to automatically learn deep temporal features, yielding a deep-feature vector. The handcrafted and deep feature vectors were concatenated along the channel dimension to create a fused feature vector. This fused vector was then passed through a fully connected layer to produce predicted probabilities for each lameness state (healthy, early lameness, and severe lameness).
To meet the low-latency requirements of edge-device deployment, the entire pipeline (from feature extraction through to final classification) was executed online at short, regular intervals (e.g., one complete inference every 4 s), ensuring rapid response.
2.5. Variance-Threshold Discrimination Module
To enable online differentiation between stationary and motion states, a variance-threshold discrimination module was designed. The module exploits the statistical characteristics of stationary and motion time-series samples in the training set by computing the variance of X-axis acceleration within each fixed-length window.
For each window of length, sensor bias was eliminated by adding an offset of +5 to all acceleration values, ensuring that all processed values, denoted as
, lie in the positive domain. The variance
of a window was then computed as
where
represents the mean acceleration within the window.
During module construction, the variances of all windows in the stationary dataset
and the motion dataset
were computed. If
then the discrimination threshold
T was defined as
For each newly acquired window, its variance was calculated; if , the window was classified as stationary, otherwise as motion. It should be noted that cow motion was predominantly walking, whereas other actions (e.g., turning, standing up) were transient, and their sporadic windows have a negligible impact on overall lameness detection.
2.6. Improved InceptionTime Architecture Design
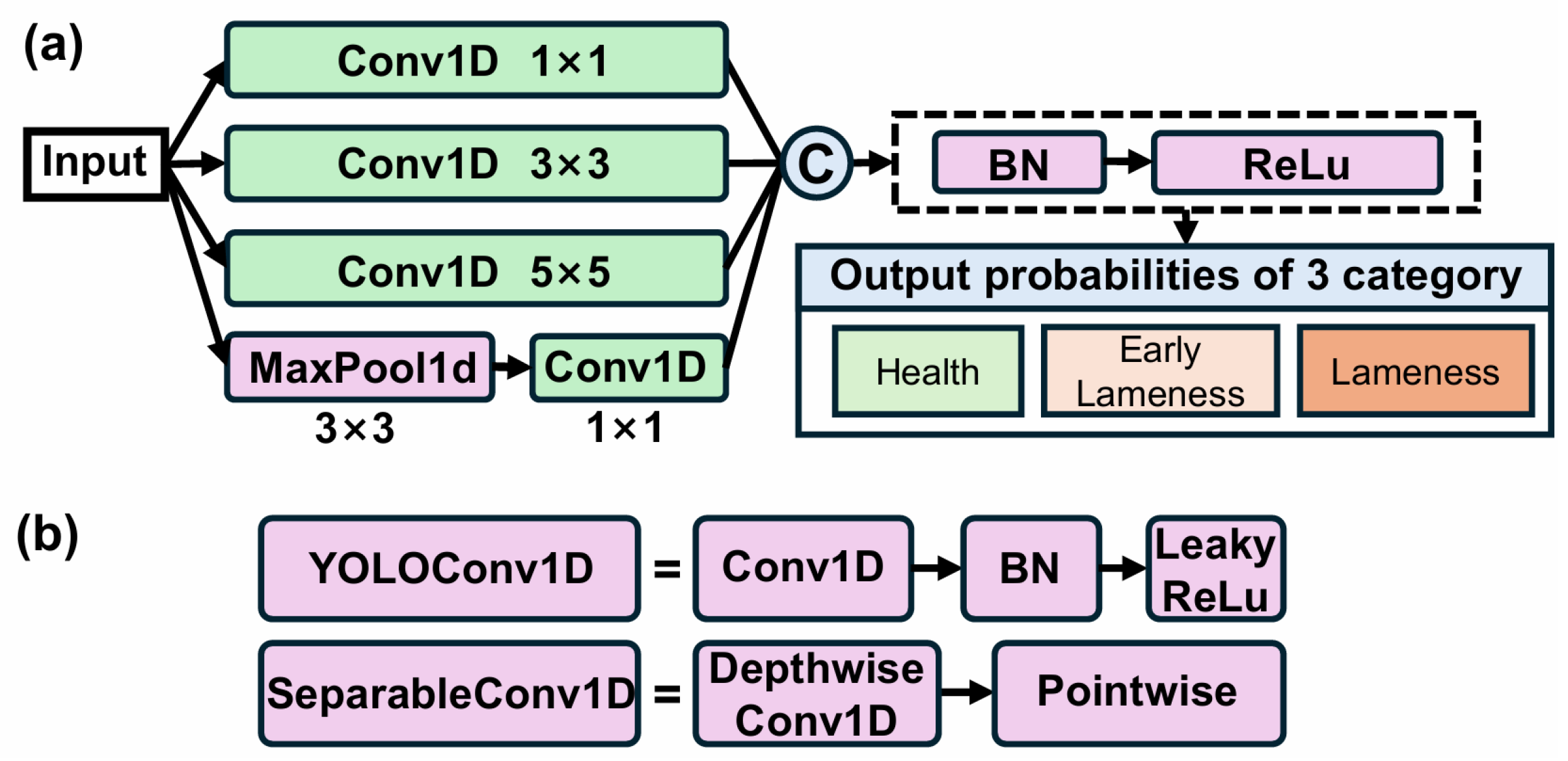
The InceptionTime network, a classical deep neural network in the time-series processing domain [
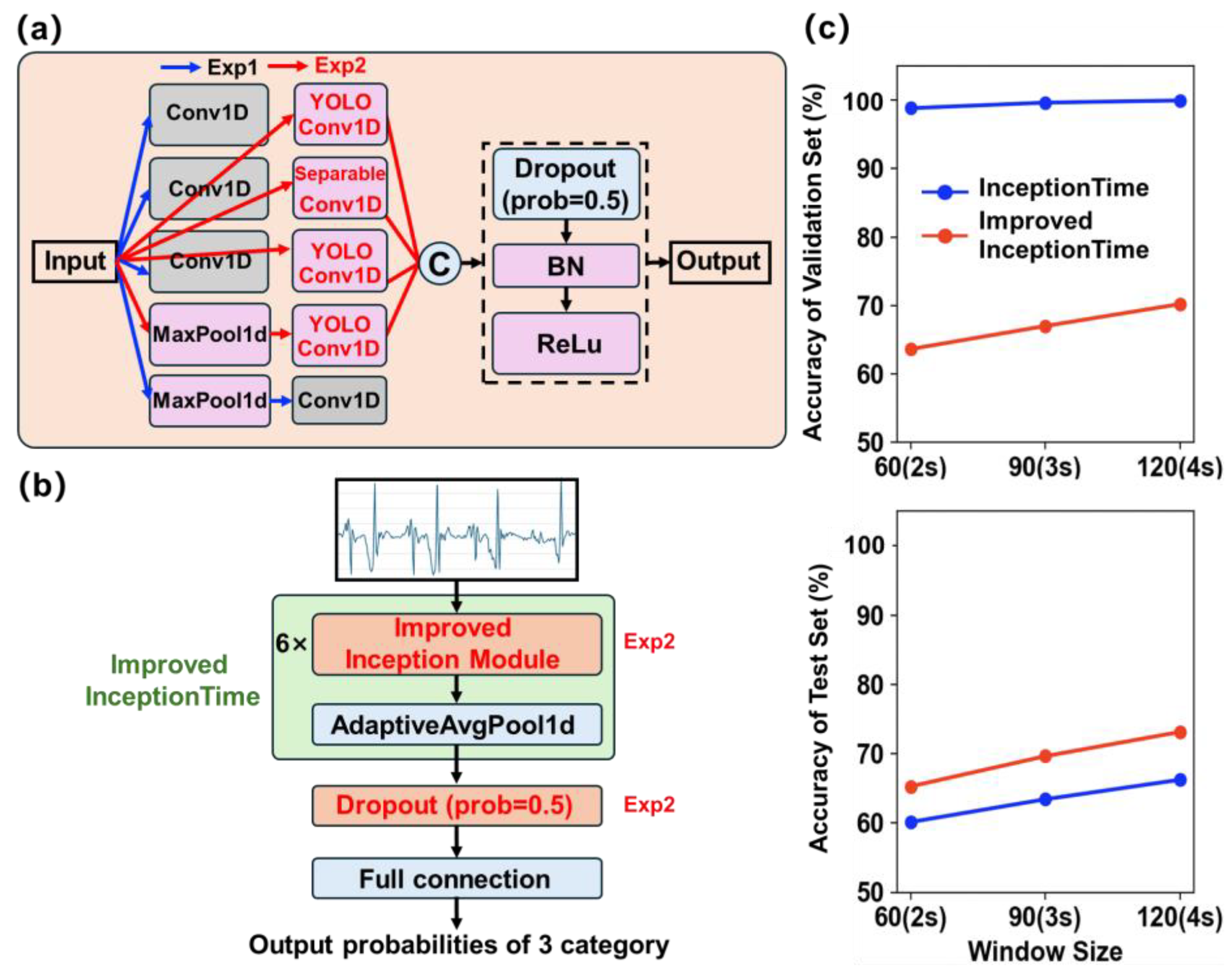
26], employs multiple stacked Inception modules to extract multi-scale features and excels at capturing temporal patterns. A conventional InceptionTime architecture consists of six Inception modules, each comprising four parallel branches (
Figure 3a). In branch 1, a 1 × 1 convolution is used for inter-channel feature fusion and dimensionality reduction; branch 2 applies a 3 × 3 convolution to capture local temporal variations; branch 3 employs a 5 × 5 convolution to gather information over a larger receptive field; and branch 4 performs 3 × 3 max-pooling followed by a 1 × 1 convolution to extract robust local statistical features. The outputs of all branches are concatenated along the channel dimension, then passed through Batch Normalization and a ReLU activation to form the module output. Finally, global average pooling aggregates all temporal features across the network, and a fully connected layer maps to the three lameness classes (healthy, early lameness, severe lameness).
The strength of the traditional InceptionTime lies in its multi-scale convolutional structure, which effectively captures subtle variations in time-series data, and has the ability to enhance feature representation through module stacking [
30]. However, the large model size and high parameter count hinder direct deployment on resource-constrained edge devices and can exacerbate overfitting issues [
31].
An improved InceptionTime network was constructed by replacing the standard Conv1D layers with YOLOConv1D and SeparableConv1D modules, as illustrated in
Figure 3b. The detailed architecture parameters are provided in
Table 2. In the YOLOConv1D branch, Batch Normalization and LeakyReLU were applied immediately after each convolution, which enhanced training stability [
32] and alleviated the vanishing-gradient issue [
33]. The SeparableConv1D branch employed a combination of Depthwise and Pointwise convolutions, substantially reducing the model’s parameter count while preserving its capacity for local temporal-feature extraction [
34]. Finally, a Dropout layer (node-retention probability set to 0.5) was inserted after the concatenation of each Inception module to mitigate overfitting and bolster the model’s generalization ability [
35].
In the improved InceptionTime architecture, the network comprises six stacked enhanced Inception modules. Temporal features are aggregated via an adaptive global average pooling layer, then passed through a Dropout layer and a fully connected layer to produce the three-class output.
2.7. Lameness Feature Extraction Module
Lameness features were extracted from each fixed-length signal window by means of a dedicated feature-extraction module. This module was designed to leverage expert knowledge to derive physically meaningful gait indicators from the preprocessed acceleration signal, thereby compensating for potential deficiencies in feature representation by the deep neural network and enabling their subsequent fusion. The extraction process consisted of two stages: data preprocessing and handcrafted feature computation.
For data preprocessing, raw acceleration signals were first filtered using a fourth-order Butterworth band-pass filter with a low cutoff frequency of 0.5 Hz and a high cutoff frequency of 5 Hz. The filter’s transfer function was defined as
where
denotes the cutoff angular frequency (calculated from the sampling rate) and
is the filter order. This filtering step removed low-frequency drift and high-frequency noise, retaining only the frequency components relevant to the cow gait cycle (0.5–5 Hz).
Subsequently, the filtered signal was smoothed using a Savitzky-Golay filter. A second-order polynomial with a window length of five was fitted within each sliding window to attenuate noise while preserving signal peaks and waveform morphology. The smoothed signal was then detrended by subtracting its mean, producing a zero-mean signal that removed static offsets arising from sensor bias or environmental factors, thus ensuring that subsequent feature extraction focused solely on dynamic variations. These preprocessing steps preserved only the gait-related frequency components, reduced noise interference, and yielded signals with more stable statistical properties.
From each zero-mean window, four lameness-related features were extracted. First, the signal amplitude
, which reflects the dynamic range within the window, was calculated as
where
denotes the preprocessed, zero-mean signal.
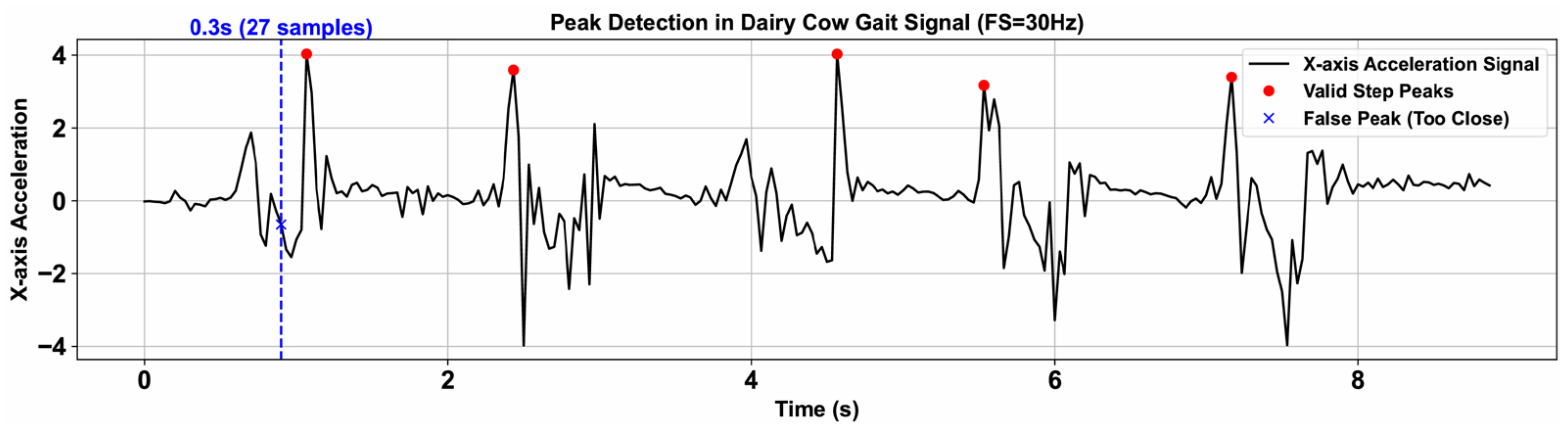
Stride Frequency denotes the number of gait cycles completed by the cow per unit time. Local maxima were identified in the preprocessed, zero-mean signal using a peak-detection algorithm. To prevent multiple detections within a single gait cycle, a minimum peak-to-peak interval of 0.3 s was enforced (
samples, where
). Additionally, to exclude spurious peaks caused by non-cyclic signal fluctuations, a prominence threshold of 3 was applied. As illustrated in
Figure 4, this procedure reliably isolates peaks corresponding to true gait cycles.
Let
denote the number of detected peaks and
the window length (in samples). The window duration is
seconds, converted to minutes (denoted as
) as
The Stride Frequency
is then calculated as
Support Time reflects the duration during which the signal remained near zero (i.e., ), typically corresponding to the phase of foot-ground contact. The signal was first scanned to count all frames satisfying . Thereafter, the frame count of each contiguous interval was divided by the sampling rate FS to obtain its duration. Finally, the mean duration across all intervals was computed to yield the Support Time .
An approximately linear relationship between Step Amplitude and Stride Frequency was assumed to simplify computation. The time required for a single step (i.e., the gait cycle) was first calculated as follows
The gait-cycle duration
was then multiplied by a calibration coefficient
to derive the Step Amplitude
. In this study,
was set to 0.5 based on the walking speeds of healthy and lame cows (0.52–1.37 m·s
−1) and the regression slope of dairy cow gait data [
36], yielding
The four handcrafted features were assembled into a feature vector and concatenated with the deep temporal features automatically extracted by the improved InceptionTime network to form the final feature representation.
All processing and analysis were performed on a workstation equipped with an Intel i7-12650H @ 2.30 GHz CPU, 32 GB RAM, and an NVIDIA GeForce RTX 4060 GPU, running with Pytorch 2.7.0 as the deep-learning framework.
3. Results and Discussion
3.1. Variance-Threshold Discrimination Module Distinguished Cow Motion State Quickly
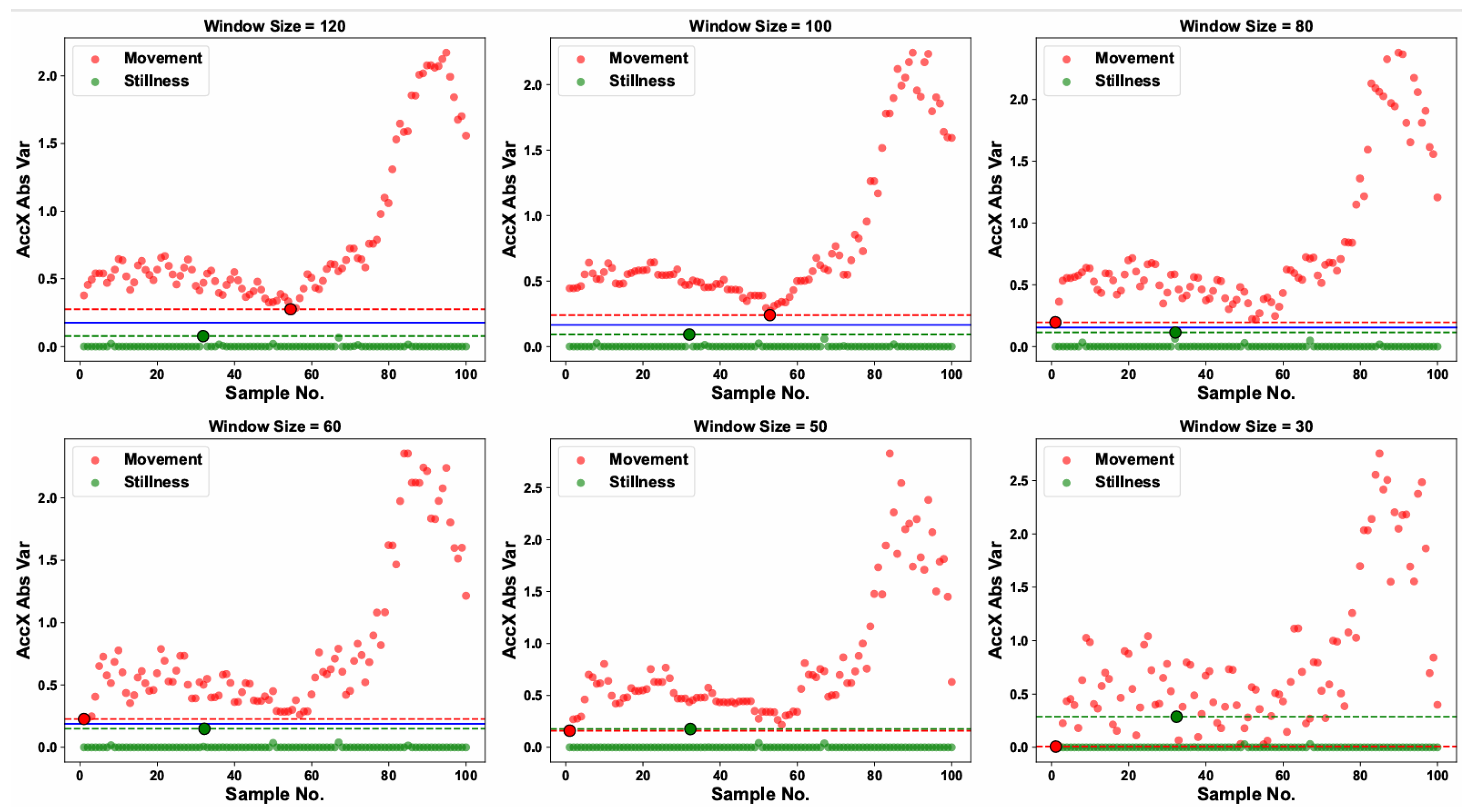
Six window lengths (30, 50, 60, 80, 100, and 120 frames) were selected to generate six datasets of windowed time-series samples for stationary and motion states, each of which was split into training and validation subsets. The threshold discrimination module was applied to the training sets under each window length to compute the variance of every time-window sample. As shown in
Figure 5, red and green dots denote the variance values of individual motion and stationary samples, respectively, while the blue horizontal line indicates the discrimination threshold determined for each window length.
For a window length of 120 frames, the variance range for stationary windows was 1.33 × 10
−5 to 0.076, and for motion windows, 0.275 to 2.18; accordingly, a threshold of 0.175 was defined. Smaller window lengths correspond to faster model response and stronger online detection capability. As illustrated in
Figure 5, decreasing the window length narrowed the gap between the minimum variance of motion windows and the maximum variance of stationary windows. When the window length was reduced to 50 frames, the minimum motion variance exceeded the maximum stationary variance, making threshold determination infeasible. Consequently, the threshold discrimination module was effective for window lengths
frames, corresponding to a minimum supported discrimination interval of 2 s.
The algorithm was then evaluated on the test set, which comprised 94 373 stationary windows and 15,630 motion windows. When the window length was reduced to 60 frames (2 s), the misclassification rate for motion windows was only 1.2%; extending the window to 90 frames (3 s) reduced this rate to 0.1%. Furthermore, at a window length of 120 frames, the variance-threshold module achieved a stationary-state detection accuracy of 100% (no false positives) and a motion-state detection accuracy of 99.7% (false-negative rate of 0.3%), with a single-window inference latency of just 0.8 ms on the CPU used.
Unlike other approaches, the variance-threshold module proposed here enables preliminary filtering of invalid data before model inference, effectively removing redundant information from stationary periods and substantially reducing both data storage requirements and communication load. In future deployments, this mechanism is expected to alleviate data backlog issues associated with high-frequency sensors and to enhance the system’s online processing efficiency and response speed.
3.2. Cross-Model Evaluation Revealed Superior Performance of InceptionTime
Firstly, the InceptionTime modules were employed in the network architecture to train a lameness detection model (Exp 1,
Figure 6a). Experiments were conducted using three window lengths (60 frames, 90 frames, and 120 frames) to investigate the impact of time-window size on model performance. The training results for each window length are presented in
Figure 6c. Both validation and test accuracies increased as the window length grew. However, regardless of window size, the validation accuracy approached nearly 100%, indicating overfitting. Moreover, the InceptionTime-based lameness detection model exceeded 5 MB in size and contained millions of parameters, making it unsuitable for deployment on lightweight hardware such as microcontrollers.
To assess InceptionTime’s performance relative to prevalent industry models, TCN, FNet, Transformer, and LSTM architectures were selected as baselines, with results collated in
Table 3. Both TCN and LSTM achieved >99% accuracy on the validation set, but their test-set accuracies declined to 63.7% and 58.8%, respectively, indicating the most severe overfitting. FNet and Transformer demonstrated mediocre performance under small-sample conditions, with test accuracies below 62%. The InceptionTime baseline, leveraging its multi-scale convolutional structure, obtained the highest test accuracy of 66.24% among the evaluated models; however, it also possessed the largest model size and parameter count and exhibited overfitting, thereby motivating subsequent architectural enhancements.
3.3. YOLOConv1D and SeparableConv1D Lightened Network Sharply
To reduce model size and mitigate overfitting, the standard Conv1D layers within each Inception module were replaced with YOLOConv1D (1 × 1, 1 × 5) and SeparableConv1D (3 × 3) in Experiment 2 (
Figure 3a), and a Dropout layer (
p = 0.5) was appended to the InceptionTime backbone (
Figure 3b), resulting in the improved InceptionTime architecture.
An ablation study was designed (
Table 4) to assess the impact of these modifications on model compactness and generalization. InceptionTime-YOLO1D involved replacing only the Conv1D layers with YOLOConv1D (1 × 1, 1 × 5). InceptionTime-YoloSep further substituted the remaining Conv1D layers with SeparableConv1D (3 × 3, expansion = 2) on top of the YOLOConv1D replacements. Although the lameness-detection model built on the original InceptionTime architecture attained higher accuracy on the validation set, its performance declined sharply on the test set, revealing pronounced overfitting. The architectural refinements in the improved InceptionTime effectively mitigated this problem.
Across all sliding-window lengths, the baseline InceptionTime exhibited consistently higher validation accuracy than its improved counterpart, but substantially lower test accuracy. At a window size of 120 frames, test accuracy rose from 66.24% to 73.14% (an improvement of 6.90%), while model size shrank from 5.39 MB to 0.72 MB and parameter count fell from 1.40 M to 0.17 M. These findings demonstrate that the Improved InceptionTime effectively reduced overfitting on the validation set and enhanced generalization to unseen cows. Furthermore, the structural optimizations greatly decreased both parameter count and computational complexity, producing a lightweight model that not only shortened inference time but is also more suitable for deployment on edge devices or in resource-constrained environments.
YOLOConv1D augments the standard Conv1D layer with Batch Normalization and LeakyReLU, which improves training stability and mitigates the vanishing-gradient problem. Because LeakyReLU is less susceptible than ReLU to “dying” neurons, it also helps preserve fine-grained details in the feature maps. SeparableConv1D, which first applies a depthwise convolution (one filter per input channel; parameters = in_channels × kernel_size), and a pointwise 1 × 1 convolution that mixes channels (parameters = in_channels × out_channels) drastically reduces the total parameter count relative to a conventional convolution (in_channels × out_channels × kernel_size). The savings become more pronounced as the kernel size grows. As a result, SeparableConv1D yields a lighter model without sacrificing its ability to extract local temporal features, thereby maintaining robust detection performance.
Window length did not affect model size or parameter count. Based on overall accuracy, real-time responsiveness, and resource usage, a window size of 120 frames (4 s per inference) was identified as the optimal compromise: it yielded the highest test accuracy while satisfying the storage and computational constraints of edge devices.
As shown by
Table 3 and
Table 4, the improved InceptionTime network achieved an 86% reduction in model size and a 6.9% increase in test accuracy, surpassing mainstream architectures such as TCN, LSTM, FNet, and Transformer. Thus, the proposed model strikes an optimal balance between accuracy and lightweight design, rendering it well-suited for deployment on microcontrollers and other resource-constrained platforms.
3.4. Feature Extraction Further Enhanced the Detection Accuracy of Lameness
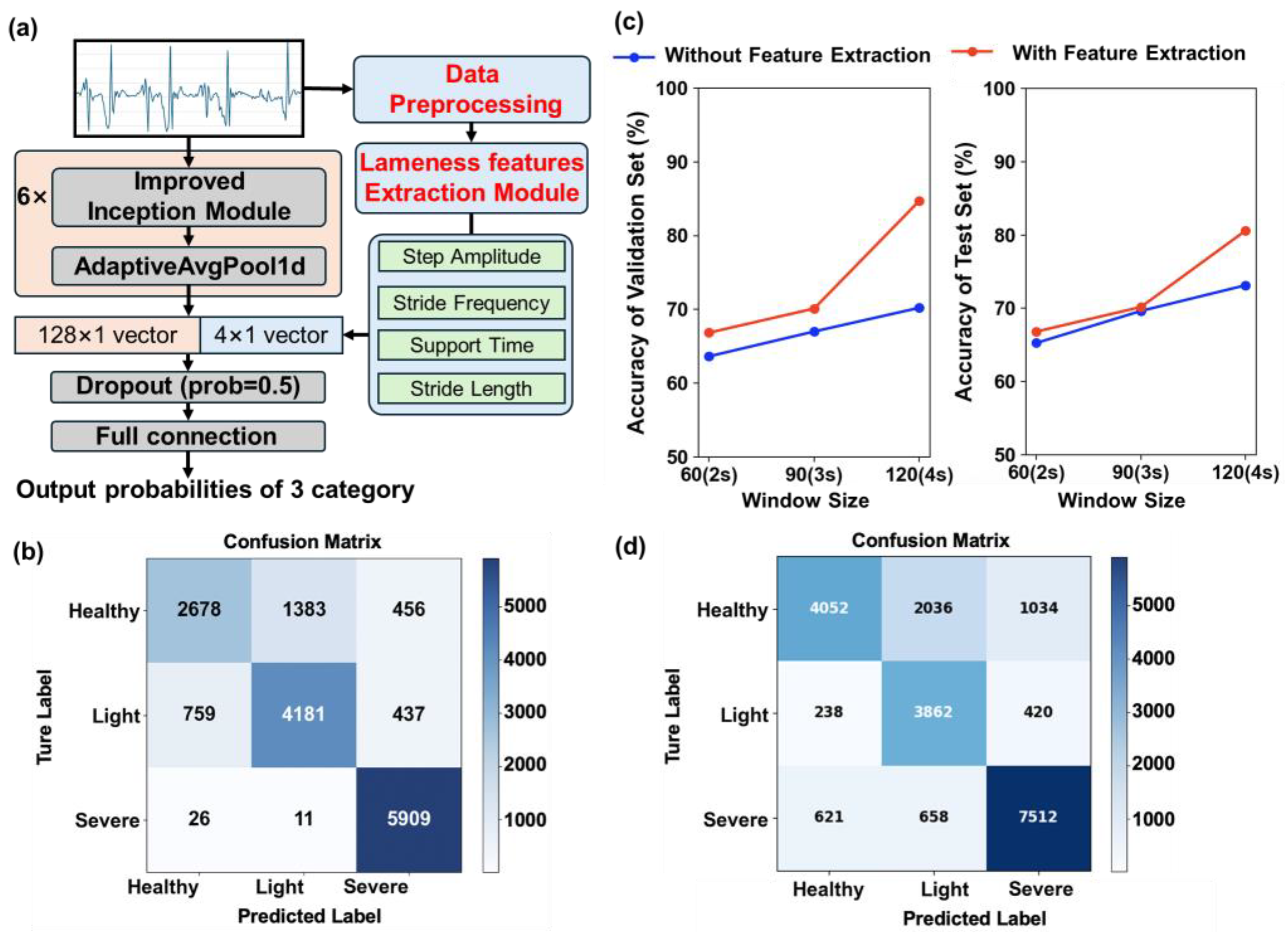
After the improved InceptionTime achieved a 6.9% increase in test accuracy, its overall performance surpassed existing models; however, a gap remained relative to production-level requirements, and end-to-end convolutional networks risked under-extracting key physiological gait features (e.g., Stride Frequency, Step Amplitude). To address this, handcrafted gait features were incorporated to create a “feature-fusion” version of the improved InceptionTime (
Figure 7a).
First, raw acceleration signals were preprocessed using a fourth-order Butterworth band-pass filter and a Savitzky-Golay filter to retain only gait-related frequency components (0.5–5 Hz), reducing noise interference and facilitating the extraction of Stride Frequency, Step Amplitude, Signal Amplitude, and Support Time. Next, each preprocessed window was passed through the feature-extraction module to derive these four gait indicators, which were assembled into a 4 × 1 handcrafted feature vector. Concurrently, the improved InceptionTime branch produced a 128 × 1 deep-feature vector from the same window. These two vectors were concatenated and fed through a fully connected layer to yield the final predicted probabilities for each lameness state.
After the feature-extraction module was incorporated, the training and inference results for each window size are presented in
Figure 7c. Across all window sizes, the lameness detection model augmented with handcrafted features achieved further improvements in accuracy, with the largest gain observed at a window size of 120 frames: validation and test accuracies increased to 84.71% and 80.6%, respectively (
Table 4). Validation accuracy rose by 20.63% relative to the model without the feature-extraction module, while neither model size nor parameter count changed. The confusion matrix (
Figure 7b) demonstrates high accuracy in classifying both early and severe lameness, indicating that lameness symptoms (light and severe) at these stages can be distinguished reliably, producing a recall of 89.11%.
End-to-end deep-learning modules often rely solely on data-driven feature learning, which can be hindered by noise and limited samples, resulting in a failure to capture subtle gait variations. By introducing an expert-knowledge-based feature-extraction module, four handcrafted gait indicators were explicitly computed from the preprocessed signals. These features not only provided physically interpretable representations of dairy cow gait but also supplied additional discriminative cues. In the feature-fusion model, the 128-dimensional deep-feature vector was concatenated with the four-dimensional handcrafted feature vector, enabling the integration of data-driven and knowledge-driven information. Automatically learned features captured complex patterns and nonlinear relationships, while handcrafted features offered clear, physically meaningful metrics. Their complementary effects allowed the model to preserve both local detail and overall trends across different data distributions (validation vs. test), thereby markedly enhancing detection accuracy. Moreover, the presence of handcrafted features reduced dependence on deep layers to extract all discriminative information, mitigating overfitting and substantially improving generalization on unseen data.
3.5. External-Site Validation Under Different Farm and Environmental Conditions
To examine whether the proposed pipeline generalizes beyond the original experimental site, an additional dataset was recorded at Dongxin Dairy Farm, Kaifeng, Henan Province, on 25 June 2025. Six Holstein cows were instrumented with the same metatarsal-mounted IMU sensors and driven along the same 40 m concrete lane used in the primary study. Although breed and attachment protocol were identical, the validation environment differed in several respects: (i) early-summer ambient temperature instead of late-autumn conditions, (ii) lower hygiene scores for the walking surface, and (iii) distinct housing and feeding management. These factors introduce realistic variation in substrate cleanliness, hoof condition, and background limb motion.
After standard preprocessing (30 Hz sampling, 120-frame windows, variance-threshold filtering), 20,433 windows were obtained: 7122 healthy, 4520 mildly lame, and 8791 severely lame. The frozen model trained on the primary site was applied directly without any parameter update. The resulting confusion matrix is shown in
Figure 7d; diagonal elements denote correctly classified windows.
Overall accuracy reached 75.5%. When the mildly and severely lame classes were pooled as a single “lameness” category, the recall for lameness was 85.4%, only 4 percentage points lower than the in-site score (89.1%). Most errors arose from misclassifying healthy windows as lame, likely reflecting greater gait variability caused by higher temperature and less uniform flooring. Nevertheless, the results demonstrate that the learned feature representations retain substantial discriminative power under a new farm, season, and hygiene context, even without retraining. Future work will incorporate this and other external datasets into the training schedule and explore lightweight fine-tuning strategies to further close the domain gap, thereby enhancing robustness across breeds, climates, and management systems.
3.6. X-Axis–Only Signal Selection Balanced Embedded Constraints
All signal preprocessing and model inference in the current system used the forward-aligned acceleration component from the limb-mounted IMU. The sensor was strapped at the distal metatarsus such that its
X-axis was oriented as closely as practical to the direction of limb progression during walking. Under this mounting, the forward component provided a clear cyclic structure related to swing, hoof contact, and stride timing, and therefore served as the input to both the variance-threshold screening (
Section 3.1) and the subsequent deep learning stages (
Section 3.2,
Section 3.3 and
Section 3.4). The study was designed for long-term, real-time monitoring on embedded edge hardware; processing full tri-axial data would substantially increase computational load, memory movement, and energy consumption, which is unfavorable for device miniaturization and operating duration. For these reasons, we prioritized a computationally efficient, low-power input configuration.
Data from the other axes were not incorporated into the present analysis. The sensor’s Z-axis (approximately perpendicular to the walking direction in the limb reference frame) showed no clear additional contribution to gait discrimination in our preliminary inspection. The Y-axis (nominally vertical) could contain information related to limb lift and impact events, but was excluded in order to control experimental variables, simplify the modeling pipeline, and remain within the resource budget of the embedded platform. This forward-axis focus may reduce sensitivity to gait adaptations that manifest outside the sagittal plane—such as lateral displacement, abduction/adduction, or asymmetric weight bearing across contralateral limbs. Future work will evaluate the incremental value of incorporating the Y-axis and, where feasible, full tri-axial signals, together with orientation handling and lightweight feature-fusion strategies, to improve robustness and early detection sensitivity under field conditions.
3.7. Lightweight Design and Data Structure Support Integration with Commercial Herd-Monitoring Systems
Commercial precision-livestock platforms such as AfiAct leg sensor/AfiTag family, CowManager ear-sensor networks, and SenseHub (formerly SCR) collar or tag systems (Afimilk Ltd., Kibbutz Afikim, Israel; CowManager B.V., Harmelen, Netherlands; SCR Engineers Ltd., Netanya, Israel) are widely deployed on dairy farms to generate continuous activity, rumination/ingestion, reproductive, and general health alerts at the individual-cow and group levels. These systems already provide on-farm wireless data acquisition, device addressing, and herd-management software environments into which additional sensor data streams can be ingested. Leveraging such existing infrastructure could substantially reduce the incremental hardware and management burden associated with rolling out specialized lameness analytics.
The proposed early-lameness pipeline produces low-bandwidth, window-level summary outputs (variance screening, feature vectors, class probabilities) rather than requiring continuous high-rate waveform upload, which aligns well with the communication and battery constraints typical of commercial tag networks. In practical deployment, this early-lameness detection pipeline can be embedded directly within a limb-mounted module analogous to current leg-activity tags to preprocess the raw forward-axis acceleration signal and apply variance-threshold screening; after motion is identified, the system transmits only downsampled or event-flagged window-level summaries via the farm’s existing wireless base-station network; finally, the resulting lameness risk scores are integrated seamlessly into herd-management dashboards alongside existing activity, rumination, health, and reproductive alerts to support decision-making.
4. Conclusions
An online early lameness detection method for cows based on an improved InceptionTime architecture and feature fusion was first proposed. Acceleration data were acquired using wearable IMU sensors, downsampled, and segmented into sliding windows to construct a library of dairy cow locomotion time-series samples. To achieve a lightweight model, low-latency processing, and high accuracy, three strategies were implemented. Firstly, a threshold discrimination module based on X-axis acceleration variance was designed to rapidly distinguish stationary from motion states within each fixed-length window. On the test set, this module achieved 100% accuracy for stationary windows and 99.7% accuracy for motion windows, with a per-window inference time of only 0.8 ms, thereby alleviating data-storage burdens under high-frequency sampling. Secondly, within the standard InceptionTime backbone, ordinary Conv1D layers were replaced with YOLOConv1D and SeparableConv1D modules, and Dropout (p = 0.5) was added after each Inception block. These modifications reduced model size and parameter count, helped mitigate overfitting, and markedly enhanced generalization performance on the test set. Finally, four handcrafted features (Signal Amplitude, Stride Frequency, Step Amplitude, and Support Time) were extracted based on dairy cow kinematic characteristics and fused with the 128-dimensional deep features automatically obtained by the improved InceptionTime network. This fusion further improved early-lameness detection, yielding an recall of 89.11% for early and severe lameness with a model size of only 0.72 MB. This approach offers a low-cost, easily deployable solution for real-time early-lameness monitoring and intervention in large-scale dairy farming.
Despite the strong performance demonstrated on both the primary research station and the independent Henan farm, two practical constraints remain. First, the current pipeline processes only the fore-aft (X-axis) acceleration signal, which may limit its ability to detect asymmetric or lateral gait impairments; when hardware resources permit, future work will incorporate multi-axis IMU data to capture three-dimensional movement and improve sensitivity to complex step deviations. Second, although the external-site validation broadens geographic coverage, the overall dataset still represents a single breed and a limited set of housing and management conditions; forthcoming studies will therefore include recordings from multiple commercial farms with varied climates, flooring materials, and production systems, complemented by targeted data augmentation to further enhance generalization. These enhancements are expected to strengthen the pipeline’s robustness across diverse production settings while preserving its low-latency, low-bandwidth operation.