Abstract
Animal biodiversity is essential for maintaining the functionality of local food systems and ensuring sustainable livelihoods. Starting in 2000, the Food and Agriculture Organization of the United Nations (F.A.O.) has drawn attention to the decline in cattle populations, including the Transylvanian Pinzgau breed from Romania. Renowned for its hardiness, adaptability, and enhanced resistance to diseases and climate change, the Transylvanian Pinzgau is regarded as an important genetic asset for advancing livestock production. As a result, tracking genetic diversity has become a key focus in breeding programs, particularly for small, endangered local populations that play a vital role in regional agro-ecological systems. This research paper sought to assess the genetic diversity of a group of 24 head of cattle from the Transylvania region by analyzing two mtDNA markers, cytochrome b and D-loop sequences, both widely recognized for their relevance and importance in the analysis of genetic diversity of cattle and phylogenetic studies. The findings, derived through statistical analysis of nucleotide sequences using specialized software, indicated that the analyzed cattle are part of the ancestral T haplogroup, with a direct lineage tracing back to Bos taurus. This information can aid in developing crossbreeding programs focused on conserving essential genetic resources, improving other cattle breeds, and protecting biodiversity.
1. Introduction
Human societies have primarily concentrated on cattle husbandry in the agricultural sector, considering it an essential activity for supporting rural economies [1,2]. This practice was closely linked to the needs of communities, providing not only food sources through meat and milk production but also resources for agricultural labor, transportation, and construction, thereby contributing to the social and economic development of society [1,2,3].
The domestication of cattle, dating back to the dawn of civilization, has represented a crucial influence on agricultural practices and the development of societies throughout history. Indigenous cattle breeds belong to a significant social and cultural identity, highly valued for their remarkable resilience to harsh environmental conditions and climatic challenges, innate resistance to diseases and ectoparasites, and rapid recuperative capacity post-illness [1,2,4,5,6,7,8].
In recent decades, there has been a marked global decline in livestock genetic diversity [8,9], primarily driven by production specialization that prioritizes the most widespread, highly productive breeds. Notably, the rigorous selection process of “elite sires” for the purpose of artificial insemination in extensive cattle populations has further reduced genetic diversity within breeds [6,8,10,11,12,13].
Among the oldest European cattle breeds is the Transylvanian Pinzgau, whose historical origins remain incompletely understood [1,13,14,15]. The Romanian Pinzgau breed, also known as the Transylvanian Pinzgau after its location area in the Transylvania region, is an alpine cattle breed, adapted to growth and exploitation in mountainous altitudes [12,16,17]. This breed was developed between 1670 and 1740 through the interbreeding of native cattle from the Pinzgau region (in the Salzburg area of Austria) with the Bernese spotted cattle (imported from Switzerland, specifically from Tux and Zillertal, between 1690 and 1740). Key attributes of this breed include robust resistance to pathogens and environmental stressors, extended productive longevity, and superior mobility compared to improved breeds. These morphological and functional attributes are crucial for the breed’s persistence under the specific demands of high-altitude regions [1,4,7,12]. Consequently, it is essential to prioritize the preservation and growth of the current Pinzgau nucleus, in addition to preserving the breed’s genetic diversity, especially considering accelerating global climate change [15,16,17,18].
Since 2000, reports from the FAO organization have indicated that the Transylvanian Pinzgau cattle breed faces a risk of extinction due to a significant population decline [19,20].
The genetic composition and lineage relationships between the Transylvanian Pinzgau and other varieties from the Pinzgau breed worldwide reveal varying degrees of kinship and evolutionary divergence [16]. Comparative genetic studies have shown that, while the Transylvanian Pinzgau shares ancestral genetic markers with other Pinzgau populations in Europe, distinct genetic drift and adaptation to local environmental conditions have shaped its unique genetic profile over time [16,17]. By analyzing molecular markers and constructing phylogenetic trees, researchers can assess the genetic distances between these populations, which provide insights into their common heritage and the evolutionary processes that have led to regional genetic variations. Such analyses are essential for understanding both the conservation needs and the genetic uniqueness of the Transylvanian Pinzgau within the broader context of the Pinzgau breed globally (Austrian Pinzgau, German Pinzgau, Slovak Pinzgau, Swiss Pinzgau, etc.) [18]. The common lineage of this cattle breed can be traced to the wild progenitor Bos taurus, which was considered extinct by the 16th century [21,22,23]. From this ancestral stock, cattle have inherited numerous valuable traits [2].
Numerous studies focused on the origin of cattle [14,21,24,25,26], based on mtDNA analysis, have established that all taurine descend from a common ancestor in the area of Near East approximately 10,000 years ago, coinciding with the shift to the Neolithic era. Recent investigations into the mitochondrial genome of Bos taurus have identified macro-haplogroup T [21,27], which comprises two sibling clades, T1’2’3 and T5. The T1’2’3 clade includes the initially characterized haplogroups T1, T2, T3, and T4, with T4 being a subgroup of T3. All T haplogroups are believed to have likely originated in the Fertile Crescent, where domestication occurred. From this region, they disseminated alongside the movement of domestic cattle herds of Bos taurus. The mitochondrial DNA of contemporary cattle is not wholly identified by haplogroups T1–T5 [21,28,29,30].
Presently, the Research and Development Station for Cattle Breeding in Târgu-Mureș, Romania, houses 24 Pinzgau individuals included in a nationwide biodiversity conservation program. The cows are not known to be closely related. They were identified in the Transylvanian region and selected based on their phenotypic traits characteristic of the Transylvanian Pinzgau breed, ensuring the representation of the breed’s diversity. This population serves as the biological material utilized for ongoing research efforts, aimed at protecting biodiversity and the key genetic materials of this cow, which represents a sustainable alternative to the increasingly pronounced climate changes to which modern improved cattle are increasingly susceptible [4,12,31].
The population trend of this breed demonstrates a decline from approximately 5% of Romania’s total cattle population in 1937 to 1.35% by 1995 [32,33,34,35]. This breed is distinguished by its unique coat pattern, which is a deep cherry color marked with characteristic white patterns (Figure 1).

Figure 1.
The Transylvanian Pinzgau cattle from the Research and Development Station for Cattle Breeding, Târgu-Mureș, Romania (original photograph).
The Transylvanian Pinzgau reaches an average weight up to 900 kg and a height of approximately 134 cm in males, while females can weigh up to 500 kg and reach a height of 127 cm [35]. Morphologically, Transylvanian Pinzgau cattle are considered descendants of Bos taurus, with cranial features that exhibit notable similarities to certain Spanish breeds, making it valuable in terms of a genetic perspective [34].
Studying the phylogeny of the Transylvanian Pinzgau is crucial for several reasons. First, understanding its genetic lineage and evolutionary history can provide insights into the breed’s unique adaptations and resilience to local environmental conditions [12,31]. Additionally, phylogenetic analysis can reveal the relationships between the Transylvanian Pinzgau and other Pinzgau populations worldwide, enhancing our comprehension of genetic diversity within the breed [10,14,31]. Moreover, examining the phylogeny of the Transylvanian Pinzgau can aid in identifying genetic markers linked to disease resistance and overall fitness, thereby contributing to improved management practices [4,5,18]. Ultimately, this research is essential not only for the conservation of this specific breed but also for the continued sustainability of livestock farming in Romania and beyond.
This study sought to assess the diversity of genetic resources of the Transylvanian Pinzgau cattle population utilizing mitochondrial genes (cytochrome b and D-loop) that are pertinent to investigations of variability, phylogeny lineage, and genetic mapping of phylogeography. In mitochondrial DNA (mtDNA), cytochrome b (cyt b) is a component of the cytochrome bc1 complex (also known as Complex III) in the mitochondrial electron transport chain [17,21,24,29]. This protein plays a crucial role in cellular respiration by facilitating electron transfer and contributing to the generation of the proton gradient used for ATP synthesis. Cytochrome b is encoded by the mitochondrial DNA and is highly conserved across species, making it a popular target in studies of genetic variation, phylogenetics, and species identification [21,24,30]. The D-loop is a non-coding region known for its role in the onset of DNA replication and transcription. It contains control elements essential for regulating the replication and expression of the mitochondrial genome. This region is often used in genetic studies because it has a high mutation rate, making it useful for analyzing genetic diversity, maternal lineage, and evolutionary studies [24,30].
Yan et al., in 2019, assessed the genetic diversity and phylogenetic relationships of Chinese cattle using mtDNA, specifically the cytochrome b marker. The researchers found that the analysis of these mitochondrial markers clarified the evolutionary relationships among different cattle breeds and was essential for the safeguarding and oversight of these populations of animals [11].
Another study, published by Prihandini et al. (2020) on the genetic diversity of the mitochondrial cytochrome b gene in a native Indonesian cattle nucleus, demonstrates that the mitochondrial cytochrome b marker can be effectively used to understand genetic structure and identify the main haplogroups that define different cattle breeds [15].
A study published by Davidescu et al. (2022) focusing on the phylogenetic analysis of endangered Grey Steppe cows shows that cytochrome b was the mt marker highly skilled for accurately assessing the phylogenetic lineage between the studied breed and other cattle breeds in Europe, demonstrating importance for identifying genetic differences. Meanwhile, the D-loop mt gene provided the most reliable topological support [13].
This research aims to investigate the genetic composition and evolutionary links of the endangered Transylvanian Pinzgau cattle, a key resource for biodiversity conservation and the sustainability of livestock production.
2. Materials and Methods
2.1. Collection of Blood Samples
The initial phase in fulfilling the research objectives involved obtaining blood specimens from 24 female Transylvanian Pinzgau cattle, a population housed at the Research and Development Station for Cattle Breeding in Târgu-Mureș, within Romania’s Transylvania region. The average age of the analyzed cows was 52 months, with limits between 12 and 90 months, respectively. Blood samples were acquired via jugular venipuncture, using 2 mL vacutainer tubes containing EDTA (ethylene-diamine-tetra-acetic acid) to prevent blood clotting. The collected blood samples were numbered from tp_01 to tp_24, “tp” being the abbreviation from the name of breed, namely Transylvanian Pinzgau. The blood samples were preserved under ideal storage conditions in a freezer at −20 °C before the stage of DNA isolation and analysis.
2.2. Extraction and Quantification of Genomic DNA from Blood Samples
The DNA was isolated from blood through an automated protocol using the Maxwell™ 16 and 16 MDx systems, with a specific kit supplied by the Promega distributor (Promega Corporation, Madison, WI, USA): the Maxwell 16 LEV Blood DNA Kit (catalog code AS1290) [36,37]. This kit includes 50 LEV cleansing cartridges, 50 collection tubes for elution, 20 mL of cell lysis reagent, two 1 mL solutions of K proteinase, and 20 mL of washing buffer. The Maxwell™ 16 system is capable of processing 16 samples in 40 min, and the extracted DNA can be applied to several techniques, including PCR and agarose gel electrophoresis.
The DNA quantification was conducted with the spectrophotometer Nanodrop ASP-3700 (ACTGene Inc., Princeton, NJ, USA), measuring absorbance at absorbance wavelengths of A260 nm and A280 nm to determine DNA extraction yield. This spectrophotometric technique is guided by the principle that most biological substances exhibit a characteristic absorbance rate within the ultraviolet (UV) radiation spectrum. Specifically, an absorption rate at 260 nm corresponds to nucleic acids (DNA/RNA), 280 nm to proteins, and 230 nm to various contaminants. Beer–Lambert’s law establishes a linear correlation between the concentration of a compound and its absorbance at a specific wavelength. This principle underlies the measurement of DNA concentration, enabling assessments of purity in relation to protein content [37,38].
2.3. Primer Design, PCR Amplification, and Gene Sequencing
The amplification of mitochondrial markers, specifically cytochrome b and the D-loop, was conducted through polymerase chain reaction (PCR), a molecular technique used to efficiently generate millions to billions of copies of targeted DNA segments using specific amplification primers. In this study, PCR amplification of cytochrome b and the D-loop genes was achieved with two primer pairs (forward primer and reverse primer, detailed in Table 1) designed based on the Bovine Reference Sequence (BRS; GenBank V00654) [39]. The mitochondrial genome in Bos taurus measures 16,341 base pairs (bp) in total length. The cytochrome b marker, approximately 1140 bp in length, and the mitochondrial D-loop control region, about 910 bp long (Figure 2), were amplified using the PCR technique.
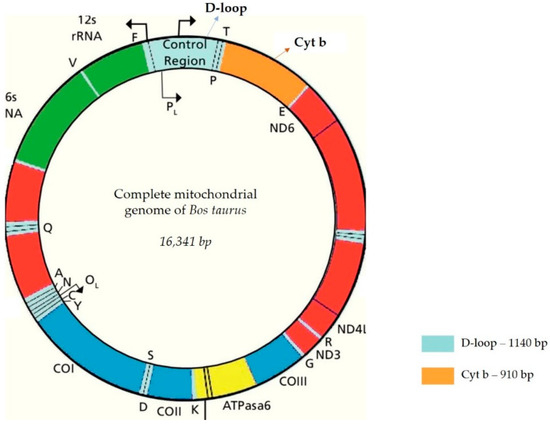
Figure 2.
Total mitochondrial genome of Bos taurus: focus on mt markers analyzed—cytochrome b (1140 bp); D-loop-(910 bp) [13,40,41,42].

Table 1.
Amplification primer pair characteristics for gene sequence analyzed.
Table 1.
Amplification primer pair characteristics for gene sequence analyzed.
| Primer Selection | Primer Specificity Gene | Primer Sequence (5′-3′) | Base Composition G1 + C2(%) | GenBank Accession No. and Genome Position [43] | Amplicon Length (bp) |
|---|---|---|---|---|---|
| BCYT | cytochrome b | Forward sequence primer: TTCTTACATGGAATCTAACCATGA | 33.3 | V00654.1 14,443–14,466 | 1140 |
| cytochrome b | Reverse sequence primer: GGGAGGTTAGTTGTTCTCCTTCTC | 50.0 | V00654.1 473–497 | ||
| BRS | D-loop | Forward sequence primer: CCTAAGACTCAAGGAAGAAACTGC | 45.8 | V00654.1 15,718–15,741 | 910 |
| D-loop | Reverse sequence primer: CAGTGAGAATGCCCTCTAGGTT | 50.0 | V00654.1 496–517 |
“G1”—guanine. “C2”—cytosine.
The DNA extracts that were isolated and purified were processed through amplification using polymerase chain reaction (PCR). The total reaction volume for PCR was set at 25.5 μL, comprising 2 μL of DNA extract, 12.5 μL of GoTaq® Green Master Mix (Promega, Madison, WI, USA), 1.5 μL of both primers, and 7.5 μL of deionized water without nucleases. The reaction was further enhanced with 0.5 μL of MgCl2. For cytochrome b fragment amplification, the primer annealing temperature was adjusted to 62 °C, while for D-loop fragment amplification, it was set to 60 °C. The PCR protocol for amplifying cytochrome b and D-loop sequences comprised 35 cycles, with conditions detailed in Figure 3a,b.
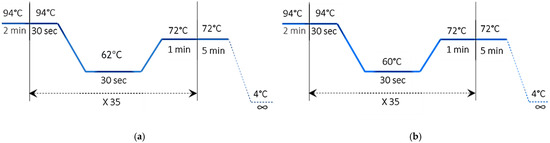
Figure 3.
PCR_Program: (a) cytochrome b; (b) D-loop.
The analyzed markers were successfully replicated, after which the PCR products were submitted to Macrogen Europe Genomic Sequencing Lab in Amsterdam, The Netherlands, for sequencing genes. Sanger sequencing was employed to determine the genetic code sequences of markers, using the primers specified in Table 2.
2.4. Validation of Amplified PCR Products in Agarose Gel Electrophoresis
To assess the degree of purity, molecular size, and potential nonspecific contaminants, the amplicons resulting from the amplification of the gene sequences (cyt b and D-loop) were validated using agarose gel electrophoresis (Figure 4a,b). The PCR amplicons were validated using a 1% agarose gel with 0.5× TBE buffer solution, followed by electrophoretic migration conducted at 100 volts for 30 min. To approximate the length of the PCR fragments, a 100-base-pair molecular weight marker was used. The sequences of interest were effectively amplified, with the specific primers showing strong specificity for the targeted mtDNA markers. The PCR amplicons had lengths of around 1140 base pairs for cytochrome b and about 910 base pairs for the D-loop, aligning with the reported values for the complete Bos taurus mitochondrial genome (GenBank accession no.: V00654).
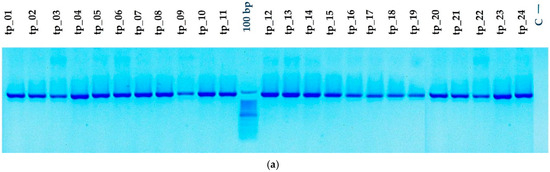
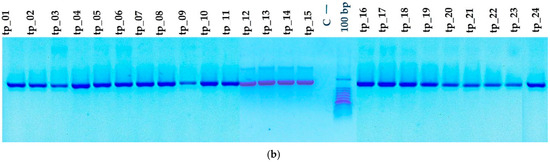
Figure 4.
PCR amplification of mtDNA genes: (a) Sequence cytochrome b (M, 100 bp molecular marker; C—negative sample control; tp_1–tp_24—sample numbers). (b) Sequence D-loop (M, 100 bp molecular marker; C—negative sample control; tp_1–tp_24—sample numbers).
2.5. Quantitative Assessment of Nitrogenous Base Proportions in Cyt b and D-loop Sequences from Transylvanian Pinzgau
The complete mitochondrial DNA (mtDNA) cytochrome b sequence, measuring 1140 base pairs [24], was successfully obtained for all 24 samples analyzed. The nucleotide composition was determined to be as follows: 30.5% adenine, 25.4% thymine, 29.9% cytosine, and 14.2% guanine. In addition, the nucleotide composition of the D-loop marker, which spans 910 base pairs [24], was characterized as follows: 32.6% adenine, 29.1% thymine, 24.5% cytosine, and 13.8% guanine. These results demonstrate the success of the sequencing (Figure 5).
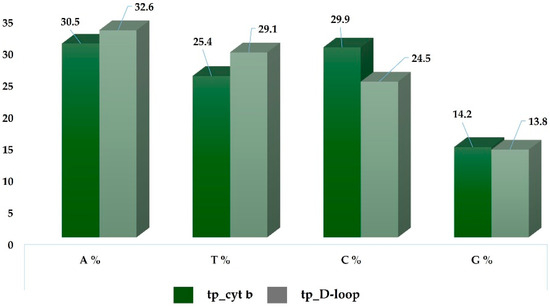
Figure 5.
Frequency distribution of DNA nucleotides (A: adenine; T: thymine; C: cytosine; G: guanine) in the sequences of cytochrome b and the D-loop.
The values are very similar to those obtained in analyses of sequences specific to the Pinzgau breed from other studies, with only slight variation [16,17]. This indicates a balanced proportion among the four DNA nucleotides.
2.6. The Coefficient of Specificity
The specificity coefficient was determined using the frequencies of the nucleotide bases, represented by the A + T/C + G ratio, which illustrates the variations among individuals of a species. This parameter is utilized in molecular phylogenetic research to assess variability in nucleotide structure. Typically, it exhibits values ranging from 1.2 to 1.5 [21], which are characteristic of most animal species. When considering the nitrogenous dominant bases in frequency, two distinct types of DNA can be identified: AT-type DNA (where A + T > C + G) and GC-type DNA (where A + T < C + G). In the analysis of the cytochrome b nucleotide sequences, all individuals examined displayed a characteristic AT-type DNA profile (A + T > C + G). A comparable conclusion was reached regarding the nucleotide sequences of the D-loop region. The coefficient of specificity for the two combined nucleotide sequences averaged 1.43, a value consistent with the typical range observed in animal organisms.
2.7. Data Analysis
Data analysis was conducted using a series of specialized software programs (Table 2). Initial analysis of the sequences, including fluorogram generation and correction, was performed with DNABaser (DNA assembly programs). To examine the phylogenetic lineage between the Transylvanian Pinzgau cattle and their wild ancestor, Bos taurus, we aligned the total mitochondrial genome sequences of the species, alongside the specific cytochrome b genes and mitochondrial fragments (D-loop) of the Pinzgau cows, obtained from database GenBank-NCBI [43]. For every sample, the sequences forward and reverse were combined based on the sequences generated by the two primers, then trimmed to produce a single, unique sequence. ClustalW was used to align all sequences from the samples [44], implemented in the MegaAlignment module. Sequence alignment and phylogenetic tree generation were performed using the MEGA X 11.0.10 software (Center for Evolutionary Medicine and Informatics, Tempe, AZ, USA, providing Molecular Evolutionary Genetics Analysis).

Table 2.
Data analysis tools and software used.
Table 2.
Data analysis tools and software used.
| Software No. | Specification of the Software Program Utilized | Type of Data Analysis | Reference |
|---|---|---|---|
| 1. |  | PCR_amplicon sequencing ✓→Sanger sequencing | [45] |
| 2. |  | Chromatogram alignment and adjustment of sequences ✓→DNA Baser 5.15 | [46] |
| 3. |  | DNA sequence alignment ✓→Mega X 11.0.10 | [46,47] |
| 4. |  | Evaluation of the optimal substitution model ✓→jModelTest 2.1.10 | [48,49] |
| 5. |  | Assembly of the haplotype network ✓→PopArt 1.7 | [49,50,51] |
| 6. |  | Phylogenetic tree build ✓→SeaView/PhyML 5.0.5 | [52,53,54] |
| 7. |  | Evaluation of nucleotide sequence diversity ✓→DnaSP 5.10.1 | [51,55,56] |
The sequence diversity under examination was assessed using LaunchDnaSP version 4.50.3 program [52]. Median-joining network analysis was conducted utilizing the PopART 1.7 software. The neighbor-joining tree was constructed employing the Kimura 2-parameter model, incorporating the following parameters: 1000 bootstrap replicates, a gamma distribution (+G) utilizing five rate categories and an evolutionary invariability model (+I).
3. Results
3.1. Dynamics of the Evolutionary Rate of mtDNA Markers
To evaluate variation in the genetic makeup of D-loop sequences, the analysis included aligning the 24 sequences to identify haplotypes and nucleotide variation sites, using the reference sequence encoded V00654 in the GenBank database. The haplogroup T3 exhibited the most frequent occurrence, consistent with predictions for European cattle [57,58]. Also, the observed genetic variation in haplotypes was measured at 0.908 ± 0.005. Utilizing the gene sequences of the mitochondrial markers studied, the demographic and territorial expansion of the Transylvanian Pinzgau cattle was evaluated through the model for calculating mismatch distribution [59,60]. This model illustrates the distribution of nucleotide differences observed among haplotype pairs. Generally, the distribution can take a unimodal form, indicative of populations that have experienced recent expansion in population or area, characterized by significant migration, or multimodal, typical of populations in a stable demographic equilibrium. Concerning the population investigated, the analysis of both mitochondrial markers yielded a multimodal distribution (Figure 6a,b).
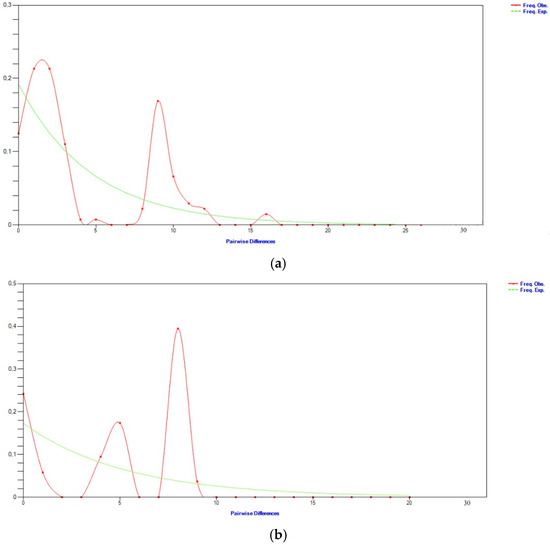
Figure 6.
Demographic expansion of the Transylvanian Pinzgau cattle: (a) cytochrome b; (b) D-loop.
The gene sequences were aligned using the ClustalW method via the MegAlign module. This process involved merging both forward and reverse sequences for each animal through alignment with the respective primer sequences, followed by the separation of the two strands and their subsequent combination into a singular sequence. A total of 23 variable sites (1.98%) were identified, among which 17 were classified as informative sites (1.51%). In the analysis of D-loop sequences, 22 variable sites (2.44%) were recorded, with 15 of these being informative sites (1.61%).
The occurrence rates of the nitrogenous bases were 30.5% A, 25.4% T, 14.3% G, and 29.9% C (cytochrome b). A<->G- and T<->C-type transversions exhibited values of 4.6481 and 0.5065, respectively (Figure 7a), with their proportion being 0.637 (Table 3).
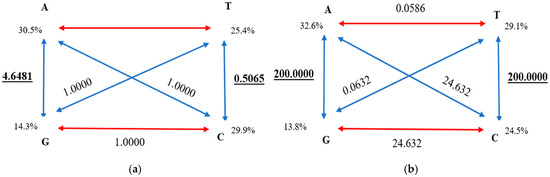
Figure 7.
Rates of nucleotide substitution calculated using jModelTest: (a) sequences of cytochrome b; (b) sequences of the D-loop.

Table 3.
Count of nucleotide sites and Ti/Tv ratio for cyt b and D-loop mtDNA characteristic of the Transylvanian Pinzgau.
3.2. Haplotype Frequency Evaluation
Through the analysis and interpretation of the DNA sequences of the two mitochondrial markers (cyt b and D-loop), three haplotypes with varying frequencies were identified (T1, T2, and T3), into which the 24 cows examined were grouped (Figure 8). The molecular diversity assessment revealed that contemporary taurine mitochondrial genomes cluster within several closely related lineages, designated as T haplogroup, respectively T1, T2, T3, and T4, exhibiting distinct geographical structuring: T1 is predominantly located in Africa; T2 is believed to have originated in the Near East and Western Asia; T3 is primarily found in Europe and is originating from the growth of a small cattle population domesticated in the Middle East; T4 was identified as a derived subclade within T3, likely disseminated throughout East Asia due to a founder effect resulting from the eastward migration of cattle [21,40].
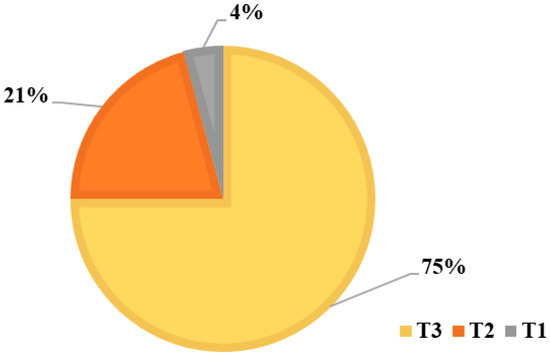
Figure 8.
Distribution of haplotype frequencies within the sampled animals of the Transylvanian Pinzgau population.
The T3 haplotype had the highest frequency, occurring in 18 of the analyzed animals (75%). The T2 haplotype was found in five animals, representing 21% and one animal individual was included in the T1 haplotype, which showed the lowest occurrence, at only 4% (Table 4).

Table 4.
Identified haplotypes and their representative individuals within the Transylvanian Pinzgau population.
3.3. Analysis of Haplotype Networks and Construction of Phylogenetic Trees
The gene sequences were examined by utilizing the Network 10.2.0.0 software, producing the haplotype design (Figure 9). Three primary haplotypes (T1, T2, and T3) were identified, each with distinct connection patterns. All haplotypes were associated with a specific number of animals.
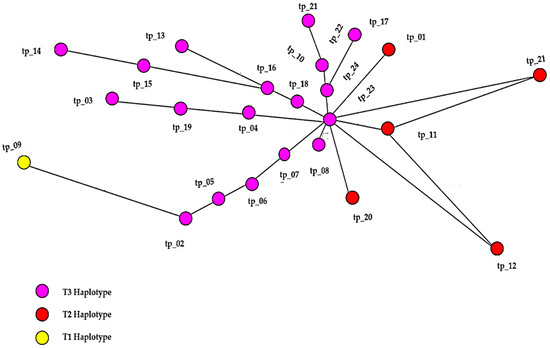
Figure 9.
Haplotype network of Transylvanian Pinzgau cattle.
The analysis of the distribution of the three haplotypes revealed that the T3 haplotype had the highest frequency, found in 18 of the 24 individuals studied. The genetic divergence between haplotypes T1, T2, and T3 ranged from one to four nucleotide sites. The identification of T-derived haplotypes indicates that the population has undergone demographic expansion and confirms that the cattle analyzed are directly descended from Bos taurus. This is a crucial finding for genetic conservation programs aimed at preserving this valuable breed (Figure 10).
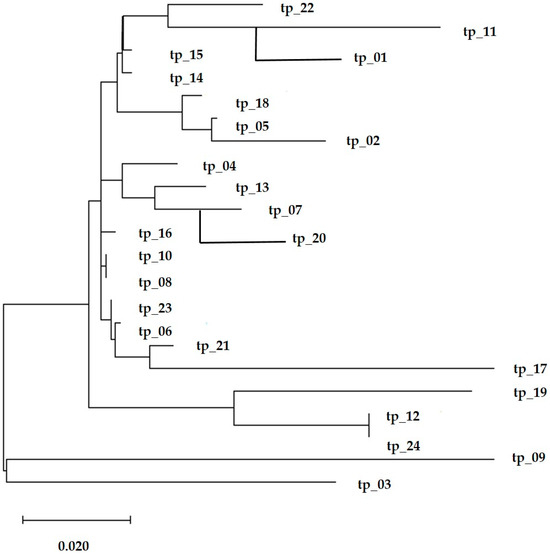
Figure 10.
Phylogenetic tree constructed based on the sequence analysis of the Transylvanian Pinzgau cattle.
4. Discussion
Until now, only a limited number of studies have been conducted regarding the genetic structure of endangered Romanian cattle breeds in comparison to other cattle breeds globally. Most of these studies have been restricted to a limited number of breeds and primarily concentrated on the nuclear DNA. This research presents, for the first time, the genetic diversity and phylogenetic characteristics of native Transylvanian Pinzgau cattle, derived from the sequencing of mitochondrial DNA genes (cytochrome b and D-loop).
Numerous studies show that the haplotypes identified in the Transylvanian Pinzgau were found to be identical to those documented in Austrian Pinzgau, German Pinzgau, Slovak Pinzgau, and Swiss Pinzgau populations [17,57,58].
Recent investigations [17,55,57,61,62,63] have demonstrated that nearly all taurine cattle are assigned to the T macro-haplogroup, with a predicted divergence time of approximately 16,000 years, suggesting a significant evolutionary bottleneck in the taurine lineage within the Bos taurus genus. The T macro-haplogroup is further subdivided into two related subclades (T5 and T1/T2/T3), with T1/T2/T3 being the most common subclade. Over time, T4 has been incorporated into T3 [17,64].
The phylogeny of the Transylvanian Pinzgau cattle breed, when analyzed alongside other Pinzgau breeds in Europe and beyond, reveals both shared ancestry and unique genetic distinctions shaped by regional breeding practices and geographic isolation. Mitochondrial DNA (mtDNA) studies, often leveraging markers like the D-loop region, trace maternal lineage and suggest that the Transylvanian Pinzgau shares a close ancestry with other European Pinzgau populations, particularly the Austrian Pinzgau [18,32,34,35]. This connection reflects historical breed movements across central and eastern Europe, although some genetic markers show divergence, possibly due to limited gene flow in isolated regions such as Transylvania [18,32,34,64,65,66]. Genomic studies indicate that while Transylvanian Pinzgau cattle maintain a unique genetic profile, they are also closely linked to broader European cattle populations, shaped by shared domestication and migration patterns [64,65,66]. Understanding these phylogenetic relationships is crucial for conservation, as it highlights the variability of genetic traits within the breed and underscores the need to preserve this unique lineage.
Current research in bovine genomics emphasizes the importance of maintaining genetic diversity in isolated breeds like the Transylvanian Pinzgau to ensure the resilience and sustainability of global cattle genetics [23,24,67].
The Transylvanian Pinzgau cattle, like many European cattle breeds, are primarily associated with mitochondrial haplogroup T3, which is common across Europe and reflects maternal lineage tracing back to Near Eastern domestication events that later spread into Europe. Haplogroup T3 is widely prevalent in central and eastern European breeds, including the Pinzgau lineage, due to shared ancestry and historical cattle movement within Europe [40,64,68]. Although rarer, minor occurrences of other haplogroups such as T1—more common in African cattle—have occasionally been identified in European cattle, likely due to ancient trade and migration influences, though it is less common in the Pinzgau [21,69]. These haplogroups, studied through mitochondrial DNA analysis, highlight the genetic distinctions and evolutionary history of the Transylvanian Pinzgau relative to other Pinzgau and European breeds, offering valuable insights for conservation and genetic diversity assessments [21,24,67].
The Transylvanian Pinzgau cattle population analyzed shows a strong presence of haplogroup T3 (18 out of 24 individuals), which is typical of European cattle breeds and mirrors findings in other Pinzgau populations, such as the Austrian Pinzgau, where T3 also predominates due to shared ancestry from early European domesticated cattle [21,67]. The presence of haplogroups T2 (in five individuals) and T1 (in one individual) adds diversity to the Transylvanian Pinzgau’s lineage, with T2 being relatively common in Near Eastern and European breeds and reflecting ancient domestication patterns that reached Europe. The single T1 individual is intriguing, as T1 is typically associated with African cattle and likely entered Europe through historic trade or migration routes, as occasionally seen in Mediterranean breeds [21]. These findings align with genomic studies on European cattle that show clustering but with distinct genetic variations based on geographic isolation, as seen in the Transylvanian Pinzgau, which may have undergone genetic bottlenecks due to isolation in Transylvania [23,70]. This diversity highlights the conservation value of the Transylvanian Pinzgau, as preserving unique haplotypes within isolated populations contributes to overall genetic resilience in cattle breeds [7].
Studies of mitochondrial DNA (mtDNA) across various Pinzgau cattle populations, including Austrian, German, Slovak, and Swiss breeds, reveal a strong predominance of haplogroup T3, which is the dominant lineage in European cattle [23,70]. In Austrian Pinzgau, haplogroup T3 accounts for nearly all identified mtDNA haplotypes, consistent with its ancestry from early domestication events in the Near East. Similarly, German Pinzgau cattle predominantly exhibit haplogroup T3, with occasional instances of haplogroup T2, reflecting historical breeding practices and limited genetic mixing from neighboring regions. The Slovak Pinzgau also primarily features haplogroup T3, with rare occurrences of T2 and even more infrequently, T1, which may indicate low-level gene flow from ancient trade routes [71,72,73,74,75,76,77]. Likewise, Swiss Pinzgau cattle show a similar pattern, predominantly carrying haplogroup T3, with T2 appearing sporadically [78]. Collectively, these findings highlight the genetic homogeneity within Pinzgau breeds, largely shaped by shared ancestry and geographical isolation, with haplogroup T3 serving as a marker of their common European lineage [79].
The occurrence of haplotypes derived from haplogroup T indicates a demographic expansion model for these individuals and signifies their lineage within Bos taurus, along with domestication in the Fertile Crescent during the Neolithic era [80,81,82].
5. Conclusions
This research highlights the importance of conserving the genetic diversity and unique phylogenetic lineage of the endangered Transylvanian Pinzgau cattle. Using mitochondrial DNA markers, we identified strong genetic links to other European Pinzgau populations, alongside distinct variations shaped by geographical isolation. The detection of haplogroups T3 and T1 emphasizes shared ancestry and ancient gene flow, contributing to the breed’s genetic richness.
These findings underscore the need for targeted conservation efforts to preserve isolated breeds like the Transylvanian Pinzgau, which enhance global cattle resilience. Additionally, maintaining unique haplotypes supports sustainable breeding practices, ensuring adaptability to future environmental and agricultural challenges.
Author Contributions
Conceptualization, M.-A.D.; methodology, M.-A.D., C.P., R.-M.R.-R. and B.-M.M.; software, M.-A.D., Ș.C. and M.G.D.; validation, M.-A.D. and C.S.; formal analysis, M.-A.D., Ș.C. and A.C.; investigation, M.-A.D., A.U., R.-M.R.-R. and B.-M.M.; data curation, C.P.; writing—original draft preparation, M.-A.D.; writing—review and editing, M.-A.D., A.U., M.G.D. and A.C.; supervision, M.-A.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
In this research, blood samples were obtained from 24 head of cattle, in compliance with the EU regulation UE 2016/679 concerning the “general data protection regulation.” The samples were collected via jugular vein puncture, following the procedures outlined in the EU Directive 2010/63/EU regarding the “protection of animals used for scientific purposes.” No experimental procedures were applied to the animals.
Data Availability Statement
The data present in this study are available on request from the first author and corresponding authors.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Medugorac, I.; Medugorac, A.; Russ, I.; Veit-Kensch, C.E.; Taberlet, P.; Luntz, B.; Mix, H.M.; Förster, M. Genetic diversity of European cattle breeds highlights the conservation value of traditional unselected breeds with high effective population size. Mol. Ecol. 2009, 18, 3394–3410. [Google Scholar] [CrossRef] [PubMed]
- Koyun, H.; Koncagül, S.; Karakuş, K. Significance of Genetic Diversity in Farm Animal Production. Türk Bilimsel Derlemeler Derg. 2016, 1, 82–83. [Google Scholar]
- Simeanu, D.; Radu-Rusu, R.M. Animal nutrition and productions. Agriculture 2023, 13, 943. [Google Scholar] [CrossRef]
- Maciuc, V. Cattle Breeding Management; Alfa: Iasi, Romania, 2006; pp. 76–78. ISBN 973-8953-18-9. [Google Scholar]
- Kasarda, R.; Jamborová, L.; Moravcíková, N. Genetic diversity and production potential of animal food resources. Acta Fytotech. Zootech. 2020, 23, 102–108. [Google Scholar] [CrossRef]
- Mădescu, B.M.; Lazăr, R.; Ciobanu, M.M.; Boișteanu, P.C. Morpfo-Productive Characteristics of Aubrac Cattle Breed: A Sistematic Review. Sci. Pap. Ser. D Anim. Sci. 2021, 64, 260–265. [Google Scholar]
- Groeneveld, L.F.; Lenstra, J.A.; Eding, H.; Toro, M.A.; Scherf, B.; Pilling, D. Genetic diversity in farm animals—A review. Anim. Genet. 2010, 11, 6–31. [Google Scholar] [CrossRef]
- Usturoi, A.; Avarvarei, B.V.; Nistor, C.E.; Simeanu, C.; Davidescu, M.A.; Usturoi, M.G. The Quality of Some Acid Dairy Products Obtained in the Traditional System. Sci. Pap. Anim. Sci. Biotechnol. 2024, 57, 187–193. [Google Scholar]
- Kasarda, R.; Vostrý, L.; Vostrá-Vydrová, H.; Candráková, K.; Moravčíková, N. Food Resources Biodiversity: The Case of Local Cattle in Slovakia. Sustainability 2021, 13, 1296. [Google Scholar] [CrossRef]
- Barnes, K.; Collins, T.; Dion, S.; Reynolds, H.; Riess, S.; Stanzyk, A.; Wolfe, A.; Lonergan, S.; Boettcher, P.; Charrondiere, U.R.; et al. Importance of cattle biodiversity and its influence on the nutrient composition of beef. Anim. Front. 2012, 2, 54–60. [Google Scholar] [CrossRef][Green Version]
- Yan, L.; She, Y.; Elzo, M.A.; Zhang, C.; Fang, X.; Chen, H. Exploring genetic diversity and phylogenic relationships of Chinese cattle using gene mtDNA 16S rRNA. Arch. Anim. Breed. 2019, 62, 325–333. [Google Scholar] [CrossRef]
- Davidescu, M.A.; Grădinaru, A.C.; Creangă, Ș. Endangered romanian cattle breeds–between traditional breeding and genetic conservation. Sci. Pap. Anim. Sci. Ser. Sci. Pap. Anim. Husb. Ser. 2021, 75, 66–75. [Google Scholar]
- Davidescu, M.-A.; Simeanu, D.; Gorgan, D.-L.; Ciorpac, M.; Creanga, S. Analysis of Phylogeny and Genetic Diversity of Endangered Romanian Grey Steppe Cattle Breed, a Reservoir of Valuable Genes to Preserve Biodiversity. Agriculture 2022, 12, 2059. [Google Scholar] [CrossRef]
- Upadhyay, M.R.; European Cattle Genetic Diversity Consortium; Chen, W.; Lenstra, J.A.; Goderie, C.R.J.; MacHugh, D.E.; Park, S.D.E.; Magee, D.A.; Matassino, D.; Ciani, F.; et al. Genetic origin, admixture and population history of aurochs (Bos primigenius) and primitive European cattle. Heredity 2017, 118, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Prihandini, P.W.; Primasari, A.; Luthfi, M.; Efendy, J.; Pamungkas, D. Genetic Diversity of Mitochondrial DNA Cytochrome b in Indonesian Native and Local Cattle Populations. J. Ilmu Ternak Dan Vet. 2020, 25, 39–47. [Google Scholar] [CrossRef]
- Küttel, L.; Letko, A.; Häfliger, I.M.; Signer-Hasler, H.; Joller, S.; Hirsbrunner, G.; Mészáros, G.; Sölkner, J.; Flury, C.; Leeb, T.; et al. A complex structural variant at the KIT locus in cattle with the Pinzgauer spotting pattern. Anim. Genet. 2019, 50, 423–429. [Google Scholar] [CrossRef]
- Kasarda, R.; Moravcikova, N.; Sidlova, V.; Krupova, Z.; Krupa, E.; Kadlecik, O. Progress in evaluation of Diversity in Pinzgau cattle based on molecular markers. Arch. Zootech. 2016, 19, 37–44. [Google Scholar]
- Ciocan-Alupii, M.; Radu-Rusu, R.M.; Pânzaru, C.; Nistor-Anton, M.; Bilkevich, V.; Maciuc, V. Study of productive performance in the Pinzgau breed exploited in the Dornelor Basin, Suceava County. Sci. Pap. Ser. D Anim. Sci. 2022, LXV, 298–303. [Google Scholar]
- Food and Agriculture Organization of the United Nations. Available online: https://www.fao.org (accessed on 12 January 2024).
- Scherf, B.D. World Watch List for Domestic Animal Diversity, 3rd ed.; FAO: Rome, Italy, 2000; Available online: https://www.fao.org/docrep/009/x8750e/x8750e00.htm (accessed on 11 October 2023).
- Bonfiglio, S.; Ginja, C.; De Gaetano, A.; Achilli, A.; Olivieri, A.; Colli, L.; Tesfaye, K.; Agha, S.H.; Gama, L.T.; Cattonaro, F.; et al. Origin and spread of Bos taurus: New clues from mitochondrial genomes belonging to haplogroup T1. PLoS ONE 2012, 7, e38601. [Google Scholar] [CrossRef]
- Scheu, A.; Powell, A.; Bollongino, R.; Vigne, J.-D.; Tresset, A.; Çakırlar, C.; Benecke, N.; Burger, J. The genetic prehistory of domesticated cattle from their origin to the spread across Europe. BMC Genet. 2015, 16, 54. [Google Scholar] [CrossRef]
- Decker, J.E.; McKay, S.D.; Rolf, M.M.; Kim, J.; Alcalá, A.M.; Sonstegard, T.S.; Hanotte, O.; Götherström, A.; Seabury, C.M.; Praharani, L.; et al. Worldwide patterns of ancestry, divergence, and admixture in domesticated cattle. PLoS Genet. 2014, 10, e1004254. [Google Scholar] [CrossRef]
- Achilli, A.; Bonfiglio, S.; Olivieri, A.; Malusà, A.; Pala, M. The multifaceted origin of taurine cattle reflected by the mitochondrial genome. PLoS ONE 2009, 4, e5753. [Google Scholar] [CrossRef]
- Gurke, M.; Vidal-Gorosquieta, A.; Pajimans, J.L.A.; Wȩcek, K.; Barlow, A.; González-Fortes, G.; Hartmann, S.; Grandal-d’Anglade, A.; Hofreiter, M. Insight into the introduction of domestic cattle and the process of Neolithization to the Spanish region Galicia by genetic evidence. PLoS ONE 2021, 16, e0249537. [Google Scholar] [CrossRef] [PubMed]
- Cubric-Curik, V.; Novosel, D.; Brajkovic, V.; Stabelli, O.R.; Krebs, S.; Sölkner, J.; Salamon, D.; Ristov, S.; Berger, B.; Trivizaki, S.; et al. Large-scale mitogenome sequencing reveals consecutive expansions of domestic taurine cattle and supports sporadic aurochs introgression. Evol. Appl. 2022, 15, 663–678. [Google Scholar] [CrossRef]
- Bollongino, R.; Edwards, C.J.; Alt, K.W.; Burger, J.; Bradley, D.G. Early history of European domestic cattle as revealed by ancient DNA. Biol. Lett. 2006, 2, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Janák, V.; Novák, K.; Kyselý, R. Late History of Cattle Breeds in Central Europe in Light of Genetic and Archaeogenetic Sources—Overview, Thoughts, and Perspectives. Animals 2024, 14, 645. [Google Scholar] [CrossRef]
- Kukučková, V.; Kasarda, R.; Žitný, J.; Moravčíková, N. Genetic markers and biostatistical methods as appropriate tools to preserve genetic resources. AGROFOR Intern. J. 2018, 3, 41–48. [Google Scholar] [CrossRef]
- Stock, F.; Edwards, C.J.; Bollongino, R.; Finlay, E.K.; Burger, J.; Bradley, D.G. Cytochrome b sequences of ancient cattle and wild ox support phylogenetic complexity in the ancient and modern bovine populations. Anim. Genet. 2009, 40, 694–700. [Google Scholar] [CrossRef]
- Necula, D.; Ciupe, S.; Tamas-Krumpe, O.; Todoran, D.; Ognean, L. Analysis of the Current Opportunities for Valorization and Conservation of the Main Autochthonous Cattle Breeds in the Conditions of the Carpathian Mountain Areas. Vet. Med. 2024, 81, 1–9. [Google Scholar] [CrossRef]
- Popa, R.; Popa, D.; Maftei, M.; Dronca, D.; Băcilă, V. Animal biodiversity conservation, a key of sustainable agriculture. Case study: The Romanian Pinzgau breed in Transylvania region. Sci. Pap. Anim. Sci. Ser. D 2012, LV, 25–29. [Google Scholar]
- Maciuc, V.; Pânzaru, C.; Ciocan-Alupii, M.; Radu-Rusu, C.-G.; Radu-Rusu, R.-M. Comparative Assessment of the Nutritional and Sanogenic Features of Certain Cheese Sorts Originating in Conventional Dairy Farms and in “Mountainous” Quality System Farms. Agriculture 2024, 14, 172. [Google Scholar] [CrossRef]
- Cărătuș, N.; Vidu, L.; Mărginean, G.E. Study on the exploitation of cattle in Transylvania. Ann. “Valahia” Univ. Targoviste 2019, 13, 57–62. [Google Scholar] [CrossRef]
- Necula, D.; Cătună-Boca, C.; Tamas-Krumpe, O.M.; Ognean, L. Evolution of the Transylvanian Pinzgau cattle breed in its natural habitat in the Carpathian mountain areas (A short review). ABAH Bioflux 2022, 14, 96–101. [Google Scholar]
- PCR Master Mix—Promega Corporation. PCR Master Mix. Available online: https://www.promega.ro/products/pcr/taq-polymerase/master-mix-pcr/?catNum=M7502 (accessed on 29 March 2024).
- Minhas-Khan, A.; Ghafar-Zadeh, M.; Shaffaf, T.; Forouhi, S.; Scime, A.; Magierowski, S.; Ghafar-Zadeh, E. UV-Vis Spectrophotometric Analysis of DNA Retrieval for DNA Storage Applications. Actuators 2021, 10, 246. [Google Scholar] [CrossRef]
- Koetsier, G.; Cantor, E. A Practical Guide to Analyzing Nucleic Acid Concentration and Purity with Microvolume Spectrophotometers. In Technical Note; New England Biolabs Inc.: Ipswich, MA, USA, 2019; pp. 1–8. [Google Scholar]
- GenBank Overview-NCBI. Bos taurus Complete Mitochondrial Genome. Available online: https://www.ncbi.nlm.nih.gov/nuccore/V00654 (accessed on 9 December 2023).
- Seroussi, E.; Yakobson, E. Bovine mtDNA D-loop haplotypes exceed mutations in number despite reduced recombination: An effective alternative for identity control. Animal 2010, 4, 1818–1822. [Google Scholar] [CrossRef]
- Arbizu, C.I.; Ferro-Mauricio, R.D.; Chávez-Galarza, J.C.; Vásquez, H.V.; Maicelo, J.L.; Poemape, C.; Gonzales, J.; Quilcate, C.; Corredor, F.-A. The Complete Mitochondrial Genome of a Neglected Breed, the Peruvian Creole Cattle (Bos taurus), and Its Phylogenetic Analysis. Data 2022, 7, 76. [Google Scholar] [CrossRef]
- Hiendleder, S.; Lewalski, H.; Janke, A. Complete Mitochondrial Genomes of Bos taurus and Bos indicus Provide New Insights into Intra-Species Variation, Taxonomy and Domestication. Cytogenet. Genome Res. 2008, 120, 150–156. [Google Scholar] [CrossRef]
- UCSC Genome Browser. Available online: https://genome.ucsc.edu/ (accessed on 9 September 2023).
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Furutani, S.; Furutani, N.; Kawai, Y.; Nakayama, A.; Nagai, H. Rapid DNA Sequencing Technology Based on the Sanger Method for Bacterial Identification. Sensors 2022, 22, 2130. [Google Scholar] [CrossRef]
- Li, M.; Nordborg, M.; Li, L.M. Adjust quality scores from alignment and improve sequencing accuracy. Nucleic Acids Res. 2004, 32, 5183–5191. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Chen, W.; Kenney, T.; Bielawski, J.; Gu, H. Testing adequacy for DNA substitution models. BMC Bioinform. 2019, 20, 329. [Google Scholar] [CrossRef] [PubMed]
- Sinding, M.H.S.; Gilbert, M.T.P. The Draft Genome of Extinct European Aurochs and its Implications for De-Extinction. Open Quat. 2016, 2, 7. [Google Scholar] [CrossRef]
- Gouy, M.; Tannier, E.; Comte, N.; Parsons, D.P. Seaview version 5: A multiplatform software for multiple sequence alignment, molecular phylogenetic analyses, and tree reconciliation. Mult. Seq. Alignment Methods Protoc. 2021, 2231, 241–260. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Sánchez-DelBarrio, J.C.; Messeguer, X.; Rozas, R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 2003, 19, 2496–2497. [Google Scholar] [CrossRef] [PubMed]
- Tarekegn, G.M.; Ji, X.Y.; Bai, X.; Liu, B.; Zhang, W.; Birungi, J.; Djikeng, A.; Tesfaye, K. Variations in mitochondrial cytochrome b region among Ethiopian indigenous cattle populations assert Bos taurus maternal origin and historical dynamics. Asian-Australas. J. Anim. Sci. 2018, 31, 1393–1400. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.L.; Lin, R.Y.; Xu, L.X.; Cheng, L. Analysis of polymorphisms of mitochondrial DNA D-loop and Mc1R gene in Chinese Wuchuan Black cattle. J. Appl. Anim. Res. 2014, 42, 487–491. [Google Scholar] [CrossRef]
- Kukučková, V.; Moravčíková, N.; Ferenčaković, M.; Simčič, M.; Mészáros, G.; Sölkner, J.; Trakovická, A.; Kadlečík, O.; Curik, I.; Kasarda, R. Genomic characterization of Pinzgau cattle: Genetic conservation and breeding perspectives. Conserv. Genet. 2017, 18, 893–910. [Google Scholar] [CrossRef]
- Singhla, T.; Boonyayatra, S.; Chulakasian, S.; Lukkana, M.; Alvarez, J.; Sreevatsan, S.; Wells, S.J. Determination of the sensitivity and specificity of bovine tuberculosis screening tests in dairy herds in Thailand using a Bayesian approach. BMC Vet. Res. 2019, 15, 149. [Google Scholar] [CrossRef]
- Kasarda, R.; Moravčí-ková, N.; Trakovická, A.; Mészáros, G.; Kadlečí-k, O. Genome-wide selection signatures in Pinzgau cattle. Potravin. Slovak J. Food Sci. 2015, 9, 268–274. [Google Scholar] [CrossRef]
- Miluchová, M.; Gábor, M.; Gašper, J. Analysis of the Genetic Structure of Slovak Holstein Cattle Using Seven Candidate Genes Related to Milk Quality. Diversity 2022, 14, 989. [Google Scholar] [CrossRef]
- Smitz, N.; Berthouly, C.; Cornélis, D.; Heller, R.; Van Hooft, P.; Chardonnet, P.; Caron, A.; Prins, H.; van Vuuren, B.J.; De Iongh, H.; et al. Pan-African genetic structure in the African buffalo (Syncerus caffer): Investigating intraspecific divergence. PLoS ONE 2013, 8, e56235. [Google Scholar] [CrossRef]
- Li, M.H.; Zerabruk, M.; Vangen, O.; Olsaker, I.; Kantanen, J. Reduced genetic structure of north Ethiopian cattle revealed by Y-chromosome analysis. Heredity 2007, 98, 214–221. [Google Scholar] [CrossRef]
- Felius, M.; Koolmees, P.A.; Theunissen, B.; European Cattle Genetic Diversity Consortium; Lenstra, J.A. On the Breeds of Cattle—Historic and Current Classifications. Diversity 2011, 3, 660–692. [Google Scholar] [CrossRef]
- Dorji, J.; Vander-Jagt, C.; Chamberlain, A.; Cocks, B.; MacLeod, I.; Daetwyler, H. Cattle Maternal Diversity Inferred from 1,883 Taurine and Indicine Mitogenomes-Preprint. Res. Sq. 2021, 1–35. [Google Scholar] [CrossRef]
- Dorji, J.; Vander Jagt, C.J.; Chamberlain, A.J.; Cocks, B.G.; MacLeod, I.M.; Daetwyler, H.D. Recovery of mitogenomes from whole genome sequences to infer maternal diversity in 1883 modern taurine and indicine cattle. Sci. Rep. 2022, 12, 5582. [Google Scholar] [CrossRef]
- Liu, H.; Zhai, J.; Wu, H.; Wang, J.; Zhang, S.; Li, J.; Niu, Z.; Shen, C.; Zhang, K.; Liu, Z.; et al. Diversity of Mitochondrial DNA Haplogroups and Their Association with Bovine Antral Follicle Count. Animals 2022, 12, 2350. [Google Scholar] [CrossRef] [PubMed]
- Baumung, R.; Sölkner, J. Analysis of pedigrees of Tux-Zillertal, Carinthian Blond and Original Pinzgau cattle population in Austria. J. Anim. Breed. Genet. 2002, 119, 175–181. [Google Scholar] [CrossRef]
- Matiuti, M.; Bogdan, A.T.; Crainiceanu, E. A case study of Transylvanian Pinzgau in Banat areal. Sci. Pap. Anim. Sci. Biotechnol. 2009, 42, 169–174. [Google Scholar]
- Mannen, H.; Kohno, M.; Nagata, Y.; Tsuji, S.; Bradley, D.G.; Yeo, J.S.; Nyamsamba, D.; Zagdsuren, Y.; Yokohama, M.; Nomura, K.; et al. Independent mitochondrial origin and historical genetic differentiation in North Eastern Asian cattle. Mol. Phylogenet Evol. 2004, 32, 539–544. [Google Scholar] [CrossRef]
- Kasarda, R.; Moravčíková, N.; Lehocká, K.; Olšanská, B.; Prišťák, J.; Trakovická, A.; Kadlečík, O. Haplotype block structure in the genome of Slovak Pinzgau cattle. In Proceedings of the X International Scientific Agricultural Symposium Agrosym, Jahorina, Bosnia and Herzegovina, 3–6 October 2019; pp. 1440–1445. [Google Scholar]
- Šidlová, V.; Kasarda, R.; Moravčíková, N.; Trakovická, A.; Kadlečík, O. Microsatellite analysis of population structure in Slovak Pinzgau cattle. Acta Agrar. Kaposváriensis 2014, 18, 24–29. [Google Scholar]
- Mang, N. Genetic Structure of Quantitative Characters in Pinzgau of Transilvania Breed from Hateg Region. Sci. Pap. Anim. Sci. Biotechnol. 2011, 44, 432–434. [Google Scholar]
- Brenig, B. Endangered Pinzgauer cattle subtype Jochberger Hummel are genetically distinct. Anim. Genet. 2020, 51, 590–594. [Google Scholar] [CrossRef] [PubMed]
- Kadlečík, O.; Kasarda, R.; Pavlík, I.; Hazuchová, E. Pedigree Analysis of Slovak Pinzgau Breed. Agric. Conspec. Sci. 2011, 76, 165–168. [Google Scholar]
- Kasarda, R.; Kadlecik, O.; Mészáros, G. Trends of endangered population of Pinzgau Cattle in Slovakia. Arch. Zootech. 2008, 11, 82–87. [Google Scholar]
- Šidlová, V.; Moravčíková, N.; Trakovická, A.; Ferenčaković, M.; Curik, I.; Kasarda, R. Production type of Slovak Pinzgau cattle in respect of related breeds. Acta Fytotechn. Zootechn. 2015, 18, 25–29. [Google Scholar] [CrossRef][Green Version]
- Kukučková, V.; Moravčíková, N.; Trakovická, A.; Kadlečík, O.; Kasarda, R. Genetic differentiation of Slovak Pinzgau, Simmental, Charolais and Holstein cattle based on the linkage disequilibrium, persistence of phase and effective population size. Acta Argic. Slov. 2016, 5, 37–40. [Google Scholar] [CrossRef]
- Melus, V.; Kasarda, R.; Kadlecik, O.; Trakovicka, A. Breeding potential of the Slovak Pinzgau cattle: Seeking biochemical and molecular biologic traits. Arch. Zootech. 2009, 12, 34–37. [Google Scholar]
- Mészáros, G.; Fuerst, C.; Fuerst-Waltl, B.; Kadlečík, O.; Kasarda, R.; Sölkner, J. Genetic evaluation for length of productive life in Slovak Pinzgau cattle. Arch. Anim. Breed. 2008, 51, 438–448. [Google Scholar] [CrossRef]
- Taberlet, P.; Coissac, E.; Pansu, J.; Pompanon, F. Conservation genetics of cattle, sheep, and goats. Comptes Rendus Biol. 2011, 334, 247–254. [Google Scholar] [CrossRef]
- Pavlík, I.; Sölkner, J.; Kadlečík, O.; Kasarda, R.; Mészáros, G.; Fuerst, C.; Fuerst-Waltl, B. Joint genealogical analysis as a tool for diversity evaluation in Pinzgau cattle populations. Arch. Anim. Breed. 2014, 57, 14. [Google Scholar] [CrossRef][Green Version]
- Magee, D.A.; MacHugh, D.E.; Edwards, C.J. Skip Nav Destination Interrogation of modern and ancient genomes reveals the complex domestic history of cattle. Anim. Front. 2014, 4, 7–22. [Google Scholar] [CrossRef]
- Zhang, Y.; Wei, Z.; Zhang, M.; Wang, S.; Gao, T.; Huang, H.; Zhang, T.; Cai, H.; Liu, X.; Fu, T.; et al. Population Structure and Selection Signal Analysis of Nanyang Cattle Based on Whole-Genome Sequencing Data. Genes 2024, 15, 351. [Google Scholar] [CrossRef]
- Verdugo, M.P.; Mullin, V.E.; Scheu, A.; Mattiangeli, V.; Daly, K.G.; Delser, P.M.; Hare, A.J.; Burger, J.; Collins, M.J.; Kehati, R.; et al. Ancient cattle genomics, origins, and rapid turnover in the Fertile Crescent. Science 2019, 365, 173–176. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).