Genome Identification and Characterization of WRKY Transcription Factor Gene Family in Mandarin (Citrus reticulata)
Abstract
1. Introduction
2. Materials and Methods
2.1. Database Search, Classification, and Retrieval of Sequences
2.2. Analyzing the Structure of Genes
2.3. Phylogenetic Analysis
2.4. Cis-Elements and Conserved Motifs Analysis
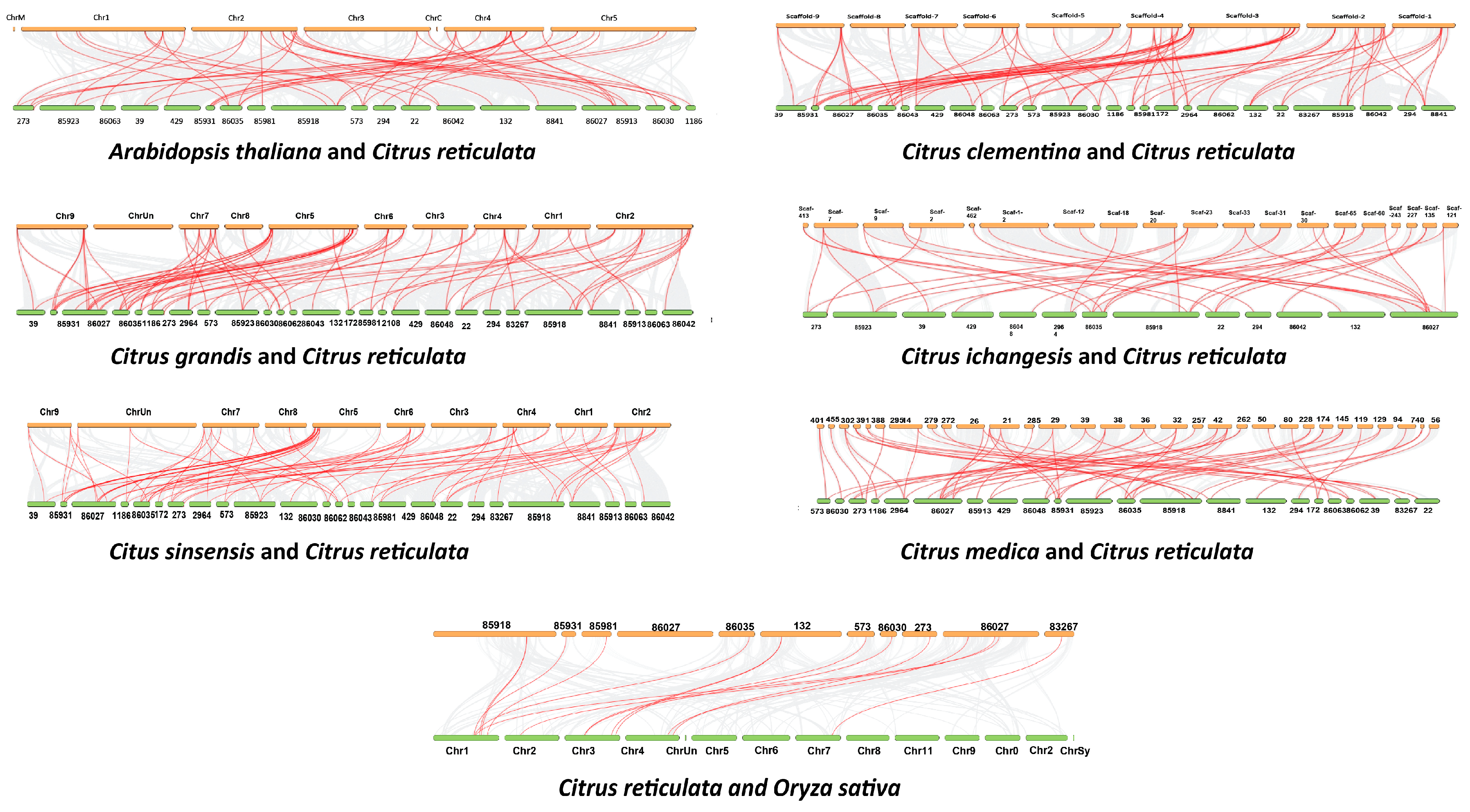
2.5. Analysis of Synteny and Duplication
2.6. Expression Analysis of CrWRKY in Different Organs
2.7. Comprehensive Analysis of microRNAs
3. Results
3.1. Identification of WRKY TFs in Mandarin (C. reticulata)
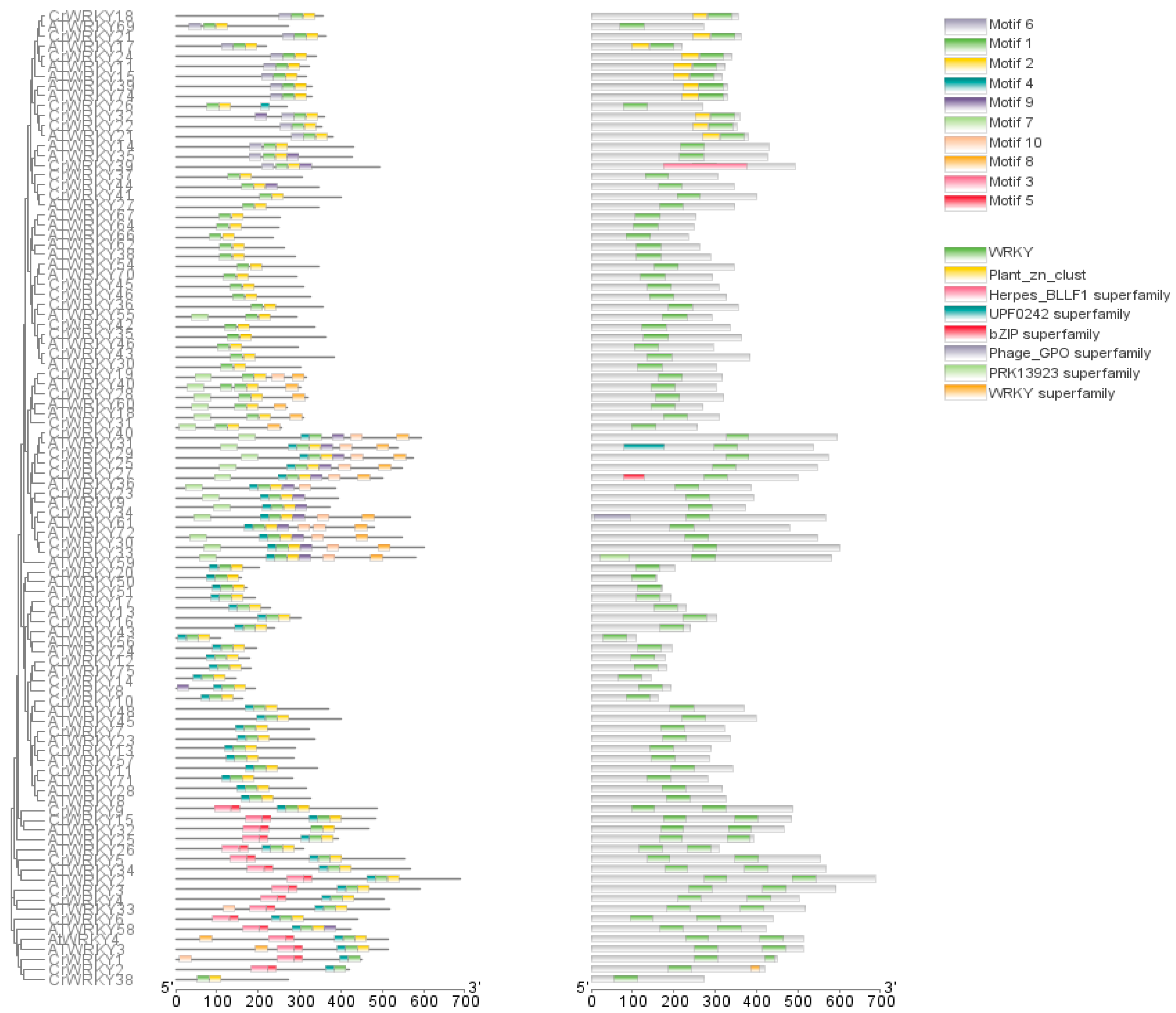
3.2. Gene Structure, Conserved Motif, and Domain
3.3. Comparative Phylogenetic Relationship of C. reticulata WRKY Gene Family with Arabidopsis
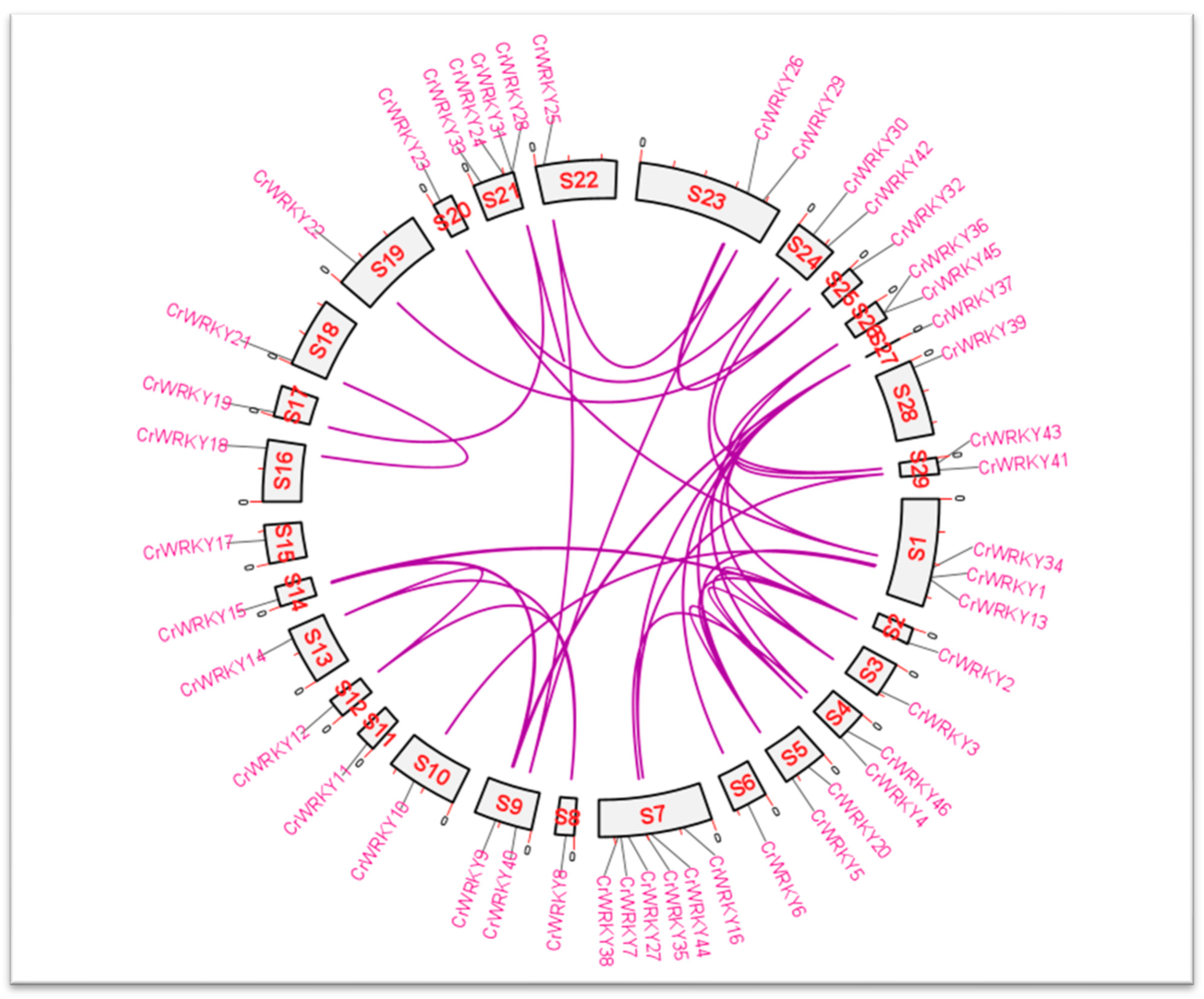
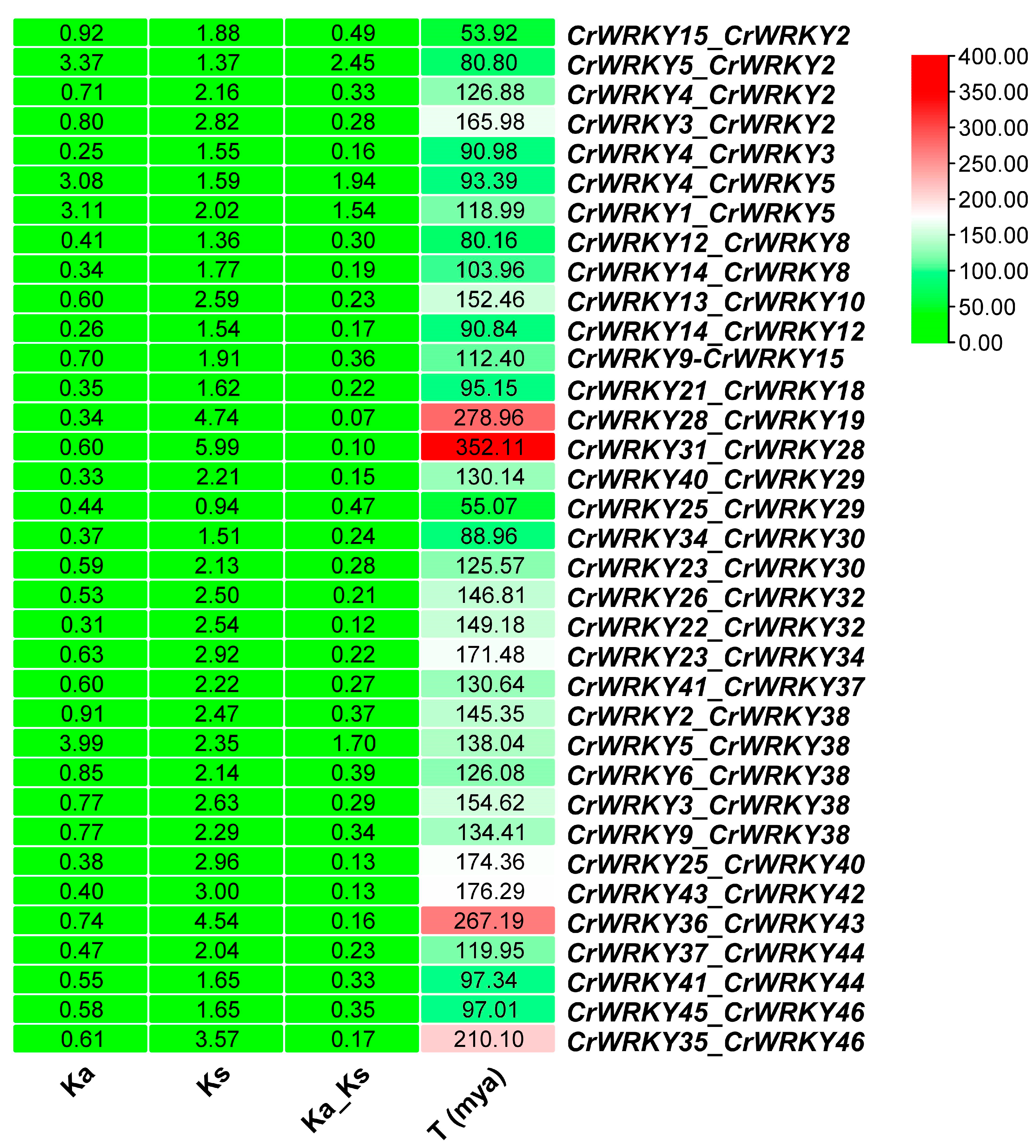
3.4. Gene Location and Duplication
Orthologous Gene Pairs between C. reticulata and Other Plant Species
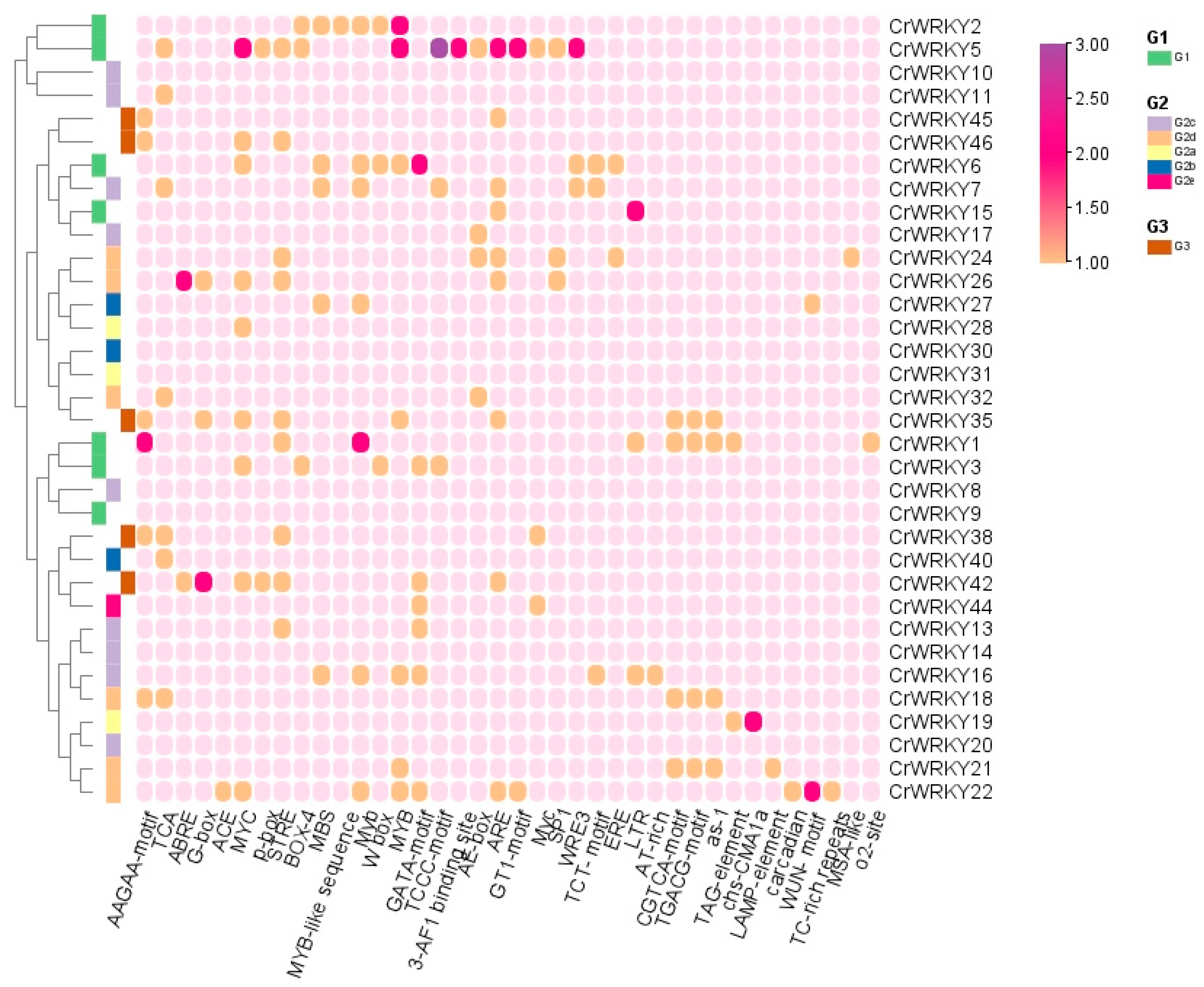
3.5. Cis-Regulatory Elements Analysis
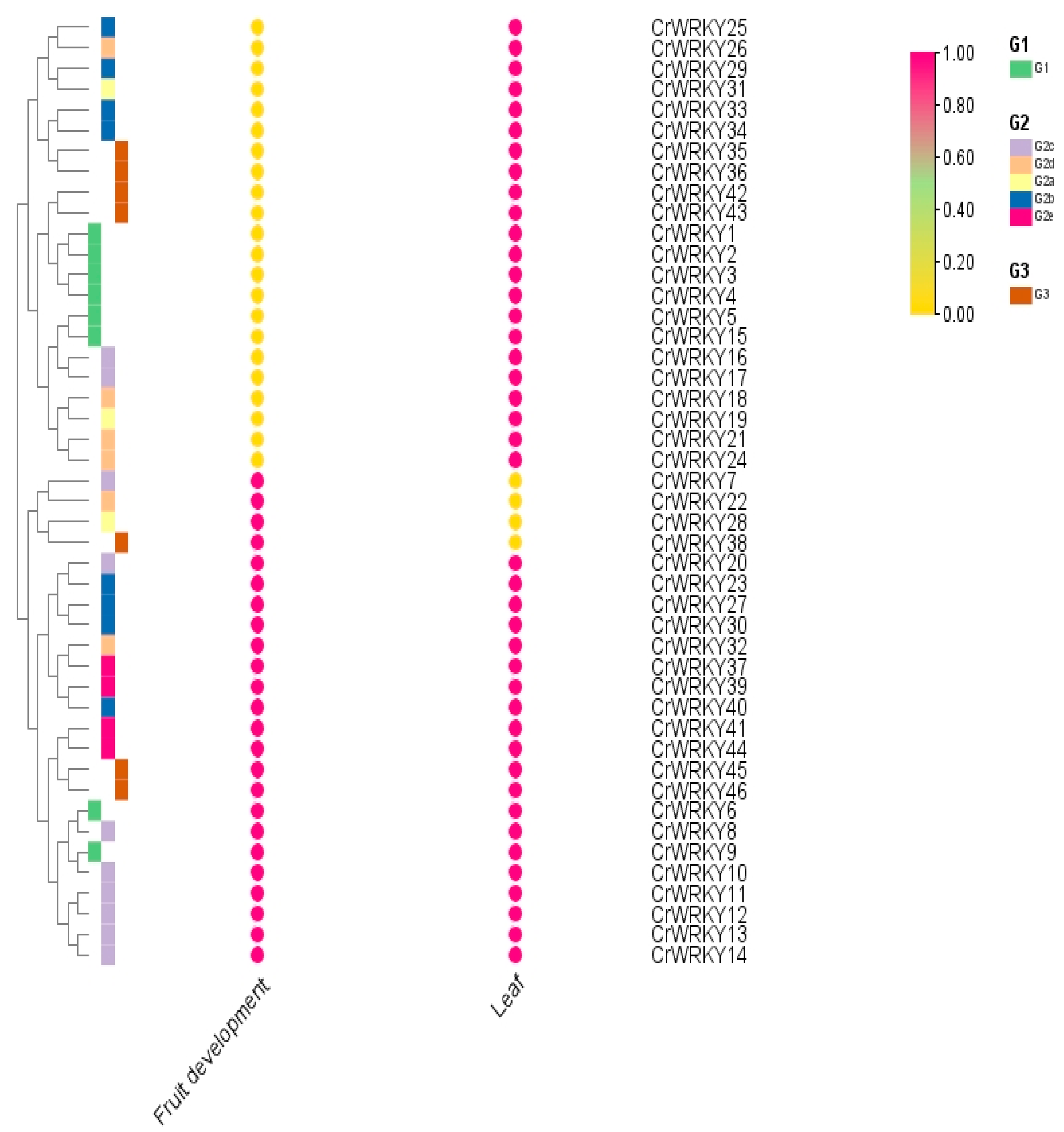
3.6. Expression Analysis of the CrWRKY Gene in Different Organs
3.7. Expression Analysis of CrWRKY Genes during Nucellarembryony Initiation
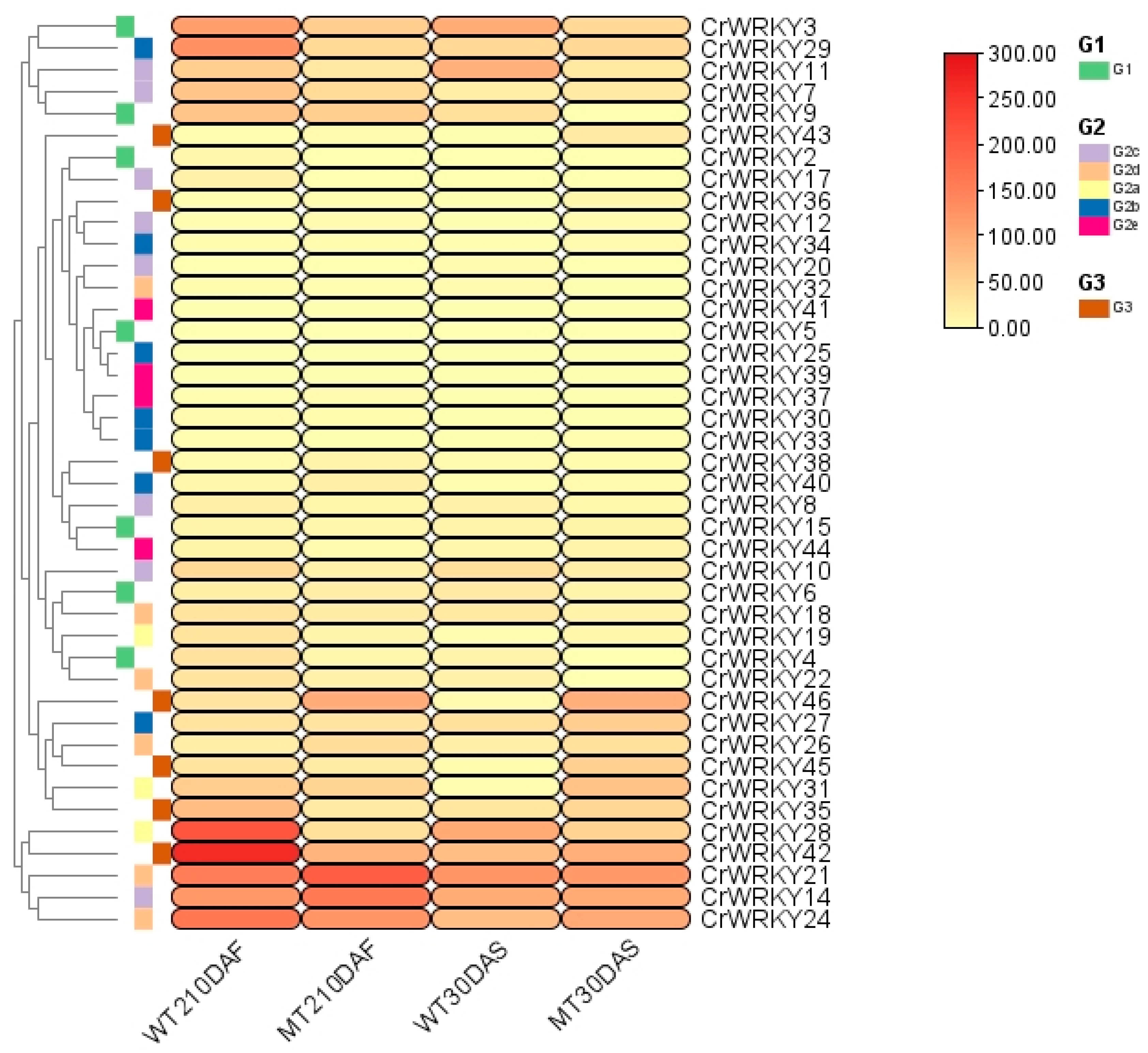
3.8. Fruit Color Expressed in Wild- and Mutant-Type of Stay-Green Citrus
3.9. CrWRKY Gene Expression Profiles in HBL Infection
3.10. miRNA Targets
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Sayed, E.; Habbasha, E.; Abouziena, H. Agriculture Biotechnology for Management of Multiple Biotic and Abiotic Environmental Stress in Crops Sustainable Management of Adverse Impacts on Farming and Soil Ecosystem Associated with Long Term Use of Low Quality Irrigation Water View Project Sustainable Management of Adverse Impacts on Farming and Soil Ecosystem Associated with Long Term Use of Low Quality Irrigation Water View Project. 2015. Available online: www.jocpr.com (accessed on 28 August 2022).
- Huang, S.; Gao, Y.; Liu, J.; Peng, X.; Niu, X.; Fei, Z.; Cao, S.; Liu, Y. Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol. Genet. Genom. 2012, 287, 495–513. [Google Scholar] [CrossRef]
- Baillo, E.H.; Kimotho, R.N.; Zhang, Z.; Xu, P. Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes 2019, 10, 771. [Google Scholar] [CrossRef]
- Ishiguro, S.; Nakamura, K. Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and β-amylase from sweet potato. Molec. Gen. Genet. 1994, 244, 563–571. [Google Scholar] [CrossRef]
- Agarwal, P.; Reddy, M.P.; Chikara, J. WRKY: Its structure, evolutionary relationship, DNA-binding selectivity, role in stress tolerance and development of plants. Mol. Biol. Rep. 2011, 38, 3883–3896. [Google Scholar] [CrossRef]
- Hsin, K.T.; Hsieh, M.C.; Lee, Y.H.; Lin, K.C.; Cheng, Y.S. Insight into the Phylogeny and Binding Ability of WRKY Transcription Factors. Int. J. Mol. Sci. 2022, 23, 2895. [Google Scholar] [CrossRef]
- Xu, Y.-P.; Xu, H.; Wang, B.; Su, X.D. Crystal structures of N-terminal WRKY transcription factors and DNA complexes. Protein Cell 2020, 11, 208–213. [Google Scholar] [CrossRef]
- Villano, C.; Esposito, S.; D’Amelia, V.; Garramone, R.; Alioto, D.; Zoina, A.; Aversano, R.; Carputo, D. WRKY genes family study reveals tissue-specific and stress-responsive TFs in wild potato species. Sci. Rep. 2020, 10, 7196. [Google Scholar] [CrossRef]
- Liu, Q.N.; Liu, Y.; Xin, Z.Z.; Zhang, D.Z.; Ge, B.M.; Yang, R.P.; Wang, Z.F.; Yang, L.; Tang, B.P.; Zhou, C.L. Genome-wide identification and characterization of the WRKY gene family in potato (Solanum tuberosum). Biochem. Syst. Ecol. 2017, 71, 212–218. [Google Scholar] [CrossRef]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, J.; Zhou, X.; Liu, S.; Zhuang, Y. Identification of WRKY gene family and characterization of cold stress-responsive WRKY genes in eggplant. PeerJ 2020, 8, e8777. [Google Scholar] [CrossRef]
- Wani, S.H.; Anand, S.; Singh, B.; Bohra, A.; Joshi, R. WRKY transcription factors and plant defense responses: Latest discoveries and future prospects. Plant Cell Rep. 2021, 40, 1071–1085. [Google Scholar] [CrossRef]
- Li, Y.; Li, X.; Wei, J.; Cai, K.; Zhang, H.; Ge, L.; Ren, Z.; Zhao, C.; Zhao, X. Genome-Wide Identification and Analysis of the WRKY Gene Family and Cold Stress Response in Acer truncatum. Genes 2021, 12, 1867. [Google Scholar] [CrossRef]
- Wang, N.; Song, G.; Zhang, F.; Shu, X.; Cheng, G.; Zhuang, W.; Wang, T.; Li, Y.; Wang, Z. Characterization of the WRKY Gene Family Related to Anthocyanin Biosynthesis and the Regulation Mechanism under Drought Stress and Methyl Jasmonate Treatment in Lycoris radiata. Int. J. Mol. Sci. 2023, 24, 2423. [Google Scholar] [CrossRef]
- Ning, P.; Liu, C.; Kang, J.; Lv, J. Genome-wide analysis of WRKY transcription factors in wheat (Triticumaestivum L.) and differential expression under water deficit condition. PeerJ 2017, 5, e3232. [Google Scholar] [CrossRef]
- Ye, H.; Qiao, L.; Guo, H.; Guo, L.; Ren, F.; Bai, J.; Wang, Y. Genome-Wide Identification of Wheat WRKY Gene Family Reveals That TaWRKY75-A Is Referred to Drought and Salt Resistances. Front. Plant Sci. 2021, 12, 2677. [Google Scholar] [CrossRef]
- Zhang, Y.; Can Feng, J. Identification and Characterization of the Grape WRKY Family. BioMed Res. Int. 2014, 2014, 787680. [Google Scholar] [CrossRef]
- Sahebi, M.; Hanafi, M.M.; Rafii, M.Y.; Mahmud TM, M.; Azizi, P.; Osman, M.; Abiri, R.; Taheri, S.; Kalhori, N.; Shabanimofrad, M.; et al. Improvement of Drought Tolerance in Rice (Oryza sativa L.): Genetics, Genomic Tools, and the WRKY Gene Family. BioMed Res. Int. 2018, 2018, 3158474. [Google Scholar] [CrossRef]
- Wu, K.-L.; Guo, Z.-J.; Wang, H.-H.; Li, J. The WRKY Family of Transcription Factors in Rice and Arabidopsis and Their Origins. DNA Res. 2005, 12, 9–26. Available online: http://www.ncbi.nlm.nih.gov (accessed on 26 August 2022). [CrossRef]
- Hu, W.; Ren, Q.; Chen, Y.; Xu, G.; Qian, Y. Genome-wide identification and analysis of WRKY gene family in maize provide insights into regulatory network in response to abiotic stresses. BMC Plant Biol. 2021, 21, 427. [Google Scholar] [CrossRef]
- Chen, C.; Chen, X.; Han, J.; Lu, W.; Ren, Z. Genome-wide analysis of the WRKY gene family in the cucumber genome and transcriptome-wide identification of WRKY transcription factors that respond to biotic and abiotic stresses. BMC Plant Biol. 2020, 20, 443. [Google Scholar] [CrossRef]
- Xie, T.; Chen, C.; Li, C.; Liu, J.; Liu, C.; He, Y. Genome-wide investigation of WRKY gene family in pineapple: Evolution and expression profiles during development and stress. BMC Genom. 2018, 19, 490. [Google Scholar] [CrossRef]
- Zheng, J.; Zhang, Z.; Tong, T.; Fang, Y.; Zhang, X.; Niu, C.; Li, J.; Wu, Y.; Xue, D.; Zhang, X.; et al. Genome-Wide Identification of WRKY Gene Family and Expression Analysis under Abiotic Stress in Barley. Agronomy 2021, 11, 521. [Google Scholar] [CrossRef]
- Goyal, P.; Manzoor, M.M.; Vishwakarma, R.A.; Sharma, D.; Dhar, M.K.; Gupta, S. A comprehensive transcriptome-Wide Identification and Screening of WRKY Gene Family Engaged in Abiotic Stress in Glycyrrhizaglabra. Sci. Rep. 2020, 10, 373. [Google Scholar] [CrossRef]
- Pandey, S.P.; Somssich, I.E. Update on WRKY Transcription Factors in Plant Defense The Role of WRKY Transcription Factors in Plant Immunity. Am. Soc. Plant Biol. 2009, 150, 1648–1655. [Google Scholar] [CrossRef]
- Moore, R.C.; Purugganan, M.D. The evolutionary dynamics of plant duplicate genes. Curr. Opin. Plant Biol. 2005, 8, 122–128. [Google Scholar] [CrossRef]
- Hasan, N.; Kamruzzaman, M.; Islam, S.; Hoque, H.; HossainBhuiyan, F.; Prodhan, S.H. Development of partial abiotic stress tolerant Citrus reticulata Blanco and Citrus sinensis (L.) Osbeck through Agrobacterium-mediated transformation method. J. Genet. Eng. Biotechnol. 2019, 17, 14. [Google Scholar] [CrossRef]
- Ayadi, M.; Hanana, M.; Kharrat, N.; Merchaoui, H.; ben Marzoug, R.; Lauvergeat, V.; Rebaï, A.; Mzid, R. The WRKY Transcription Factor Family in Citrus: Valuable and Useful Candidate Genes for Citrus Breeding. Appl. Biochem. Biotechnol. 2010, 180, 516–543. [Google Scholar] [CrossRef]
- Humann, J.L.; Piaskowski, J.; Jung, S. Resources in the Citrus Genome Database that Enable Basic, Translational, and Applied Research. In Proceedings of the 5th International Research Conference on Huanglongbing, Orlando, FL, USA, 14–17 March 2017; Available online: https://ashs.confex.com/ashs/2017/meetingapp.cgi/Paper/26424 (accessed on 26 August 2022).
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Analysis Tools on the ExPASy Server 571 571 from: The Proteomics Protocols Handbook Protein Identification and Analysis Tools on the ExPASy Server 2005. Available online: http://www.expasy.org/tools/ (accessed on 26 August 2022).
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Rombauts, S.; Déhais, P.; van Montagu, M.; Rouzé. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 1999, 27, 295–296. Available online: https://academic.oup.com/nar/article/27/1/295/1238334 (accessed on 26 August 2022). [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Wang, Y.; Li, J.; Paterson, A.H. MCScanX-transposed: Detecting transposed gene duplications based on multiple colinearity scans. Bioinformatics 2013, 29, 1458–1460. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Zhu, F.; Luo, T.; Liu, C.; Wang, Y.; Zheng, L.; Xiao, X.; Zhang, M.; Yang, H.; Yang, W.; Xu, R.; et al. A NAC transcription factor and its interaction protein hinder abscisic acid biosynthesis by synergistically repressing NCED5 in Citrus reticulata. J. Exp. Bot. 2020, 71, 3613–3625. [Google Scholar] [CrossRef]
- Samad AF, A.; Sajad, M.; Nazaruddin, N.; Fauzi, I.A.; Murad AM, A.; Zainal, Z.; Ismail, I. MicroRNA and transcription factor: Key players in plant regulatory network. Front. Plant Sci. 2017, 8, 565. [Google Scholar] [CrossRef]
- Koralewski, T.E.; Krutovsky, K.V. Evolution of Exon-Intron Structure and Alternative Splicing. PLoS ONE 2011, 6, e18055. [Google Scholar] [CrossRef]
- Dong, J.; Chen, C.; Chen, Z. Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol. Biol. 2003, 51, 21–37. Available online: http://rana.stanford.edu/clustering/ (accessed on 26 August 2022). [CrossRef]
- Rozière, J.; Guichard, C.; Brunaud, V.; Martin, M.-L.; Coursol, S. A comprehensive map of preferentially located motifs reveals distinct proximal cis-regulatory elements in plants. Front. Plant Sci. 2022, 13, 976371. [Google Scholar] [CrossRef]
- Jones, D.M.; Vandepoele, K. Identification and evolution of gene regulatory networks: Insights from comparative studies in plants. Curr. Opin. Plant Biol. 2020, 54, 42–48. [Google Scholar] [CrossRef]
- Long, J.-M.; Liu, Z.; Wu, X.-M.; Fang, Y.-N.; Jia, H.-H.; Xie, Z.-Z.; Deng, X.-X.; Guo, W.-W. Genome-scale mRNA and small RNA transcriptomic insights into initiation of citrus apomixis. J. Exp. Bot. 2016, 67, 5743–5756. [Google Scholar] [CrossRef]
- Zhao, H.; Sun, R.; Albrecht, U.; Padmanabhan, C.; Wang, A.; Coffey, M.; Girke, T.; Wang, Z.; Close, T.; Roose, M.; et al. Small RNA profiling reveals phosphorus deficiency as a contributing factor in symptom expression for citrus huanglongbing disease. Mol. Plant 2013, 6, 301–310. [Google Scholar] [CrossRef]
- Bakshi, M.; Oelmüller, R. WRKY transcription factors. Plant Signal. Behav. 2014, 9, e27700. [Google Scholar] [CrossRef]
- Khan, M.K.; Zill-E-Huma; Dangles, O. A comprehensive review on flavanones, the major citrus polyphenols. J. Food Compos. Anal. 2014, 33, 85–104. [Google Scholar] [CrossRef]
- He, H.; Dong, Q.; Shao, Y.; Jiang, H.; Zhu, S.; Cheng, B.; Xiang, Y. Genome-wide survey and characterization of the WRKY gene family in Populustrichocarpa. Plant Cell Rep. 2012, 31, 1199–1217. [Google Scholar] [CrossRef]
- Xu, Y.-H.; Sun, P.-W.; Tang, X.-L.; Gao, Z.-H.; Zhang, Z.; Wei, J.-H. Genome-wide analysis of WRKY transcription factors in Aquilaria sinensis (Lour.) Gilg. Sci. Rep. 2020, 10, 3018. [Google Scholar] [CrossRef]
- Chen, X.; Li, C.; Wang, H.; Guo, Z. WRKY transcription factors: Evolution, binding, and action. Phytopathol. Res. 2019, 1, 13. [Google Scholar] [CrossRef]
- Bao, F.; Ding, A.; Cheng, T.; Wang, J.; Zhang, Q. Genome-wide analysis of members of the WRKY gene family and their cold stress response in prunusmume. Genes 2019, 10, 911. [Google Scholar] [CrossRef]
- Rushton, D.L.; Tripathi, P.; Rabara, R.C.; Lin, J.; Ringler, P.; Boken, A.K.; Langum, T.J.; Smidt, L.; Boomsma, D.D.; Emme, N.J.; et al. WRKY transcription factors: Key components in abscisic acid signaling. Plant Biotechnol. J. 2012, 10, 2–11. [Google Scholar] [CrossRef]
- Spanudakis, E.; Jackson, S. The role of microRNAs in the control of flowering time. J. Exp. Bot. 2014, 65, 365–380. [Google Scholar] [CrossRef]
- Li, Y.; Wang, L.F.; Bhutto, S.H.; He, X.R.; Yang, X.M.; Zhou, X.H.; Lin, X.Y.; Rajput, A.A.; Li, G.B.; Zhao, J.H.; et al. Blocking miR530 Improves Rice Resistance, Yield, and Maturity. Front. Plant Sci. 2021, 12, 729560. [Google Scholar] [CrossRef]
- Liu, Y.; Ke, L.; Wu, G.; Xu, Y.; Wu, X.; Xia, R.; Deng, X.; Xu, Q. miR3954 is a trigger of phasiRNAs that affects flowering time in citrus. Plant J. 2017, 92, 263–275. [Google Scholar] [CrossRef]
- Chen, J.-G.; Wu, G.; Wang, H.; Wang, Y.; Gao, R.; Gruber, M.Y.; Hannoufa, A. miR156/SPL10 Modulates Lateral Root Development, Branching and Leaf Morphology in Arabidopsis by Silencing AGAMOUS-LIKE 79. Front. Plant Sci. 2018, 1, 2226. [Google Scholar] [CrossRef]
- Millar, A.A.; Lohe, A.; Wong, G. Plants Biology and Function of miR159 in Plants. Plants 2019, 8, 255. [Google Scholar] [CrossRef]
- Li, X.; Xie, X.; Li, J.; Cui, Y.; Hou, Y.; Zhai, L.; Wang, X.; Fu, Y.; Liu, R.; Bian, S. Conservation and diversification of the miR166 family in soybean and potential roles of newly identified miR166s. BMC Plant Biol. 2017, 17, 32. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Tan, C.; Cheng, X.; Zhao, X.; Li, T.; Jiang, J. miR168 targets Argonaute1A mediated miRNAs regulation pathways in response to potassium deficiency stress in tomato. BMC Plant Biol. 2020, 20, 477. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Chenid, X. Secrets of the MIR172 family in plant development and flowering unveiled. PLoS Biol. 2021, 19, e3001099. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liu, S.; Liu, L.; Li, R.; Guo, R.; Xia, X.; Wei, C. miR477 targets the phenylalanine ammonia-lyase gene and enhances the susceptibility of the tea plant (Camellia sinensis) to disease during Pseudopestalotiopsis species infection. Planta 2020, 251, 59. [Google Scholar] [CrossRef]
- Fang, Y.; Xie, K.; Xiong, L. Conserved miR164-targeted NAC genes negatively regulate drought resistance in rice. J. Exp. Bot. 2014, 65, 2119–2135. [Google Scholar] [CrossRef] [PubMed]
- Xia, R.; Chen, C.; Pokhrel, S.; Ma, W.; Huang, K.; Patel, P.; Wang, F.; Xu, J.; Liu, Z.; Li, J.; et al. 24-nt reproductive phasiRNAs are broadly present in angiosperms. Nat. Commun. 2019, 10, 627. [Google Scholar] [CrossRef]
- Tuteja, N.; Chai, L.; Li, M.; Zhang, W.; Xie, Y.; Xu, L.; Wang, Y.; Zhu, X.; Wang, R.; Zhang, Y.; et al. Identification of microRNAs and Their Target Genes Explores miRNA-Mediated Regulatory Network of Cytoplasmic Male Sterility Occurrence during Anther Development in Radish (Raphanus sativus L.). Front. Plant Sci. 2016, 1, 1054. [Google Scholar] [CrossRef]
- Li, M.J.; Yang, Y.H.; Chen, J.; Wang, Q.; Lin, W.X.; Yi, Y.J.; Zeng, L.; Yang, S.Y.; Zhang, Z.Y. Transcriptome/Degradome-Wide Identification of R. glutinosa miRNAs and Their Targets: The Role of miRNA Activity in the Replanting Disease. PLoS ONE 2013, 8, e68531. [Google Scholar] [CrossRef]
- Matthewman, C.A.; Kawashima, C.G.; Húska, D.; Csorba, T.; Dalmay, T.; Kopriva, S.; Schroeder, J. miR395 is a general component of the sulfate assimilation regulatory network in Arabidopsis. FEBS Lett. 2012, 586, 3242–3248. [Google Scholar] [CrossRef]
- Soto-Suárez, M.; Baldrich, P.; Weigel, D.; Rubio-Somoza, I.; Segundo, B.S. The Arabidopsis miR396 mediates pathogen-associated molecular pattern-triggered immune responses against fungal pathogens OPEN. Sci. Rep. 2017, 7, 44898. [Google Scholar] [CrossRef] [PubMed]
- Schranz, M.E.; Reyes, J.L.; Wang, S.; Budak, H.; Akpinar, B.A. Dissecting miRNAs in Wheat D Genome Progenitor, Aegilopstauschii. Front. Plant Sci. 2016, 7, 606. [Google Scholar] [CrossRef]
- Borges, F.; Parent, J.S.; van Ex, F.; Wolff, P.; Martínez, G.; Köhler, C.; Martienssen, R.A. Transposon-derived small RNAs triggered by miR845 mediate genome dosage response in Arabidopsis. Nat. Genet. 2018, 50, 186–192. [Google Scholar] [CrossRef]
- Wang, Y.; Xie, Y.; Li, X.; Lin, J.; Zhang, S.; Li, Z.; Huo, L.; Gong, R. MiR-876-5p acts as an inhibitor in hepatocellular carcinoma progression by targeting DNMT3A. Pathol. Res. Pract. 2018, 214, 1024–1030. [Google Scholar] [CrossRef] [PubMed]




| Plant Species | No. of WRKY Genes | References |
|---|---|---|
| Wheat (Triticumaestivum L.) | 171 | [15,16] |
| Grapes (Vitisvinifera) | 80 | [17] |
| Japonica (Oryza sativa subsp. Japonica) Indian Wild Rice (Oryza nivara) | 89 97 | [18] |
| Arabidopsis (A. thaliana) | 72 | [19] |
| Corn (Zea maize) | 140 | [20] |
| Cucumber (Cucumissativus) | 61 | [21] |
| Pineapple (Ananascomosus) | 54 | [22] |
| Barely (Hordeumvulgare) | 86 | [23] |
| Licorice (Glycyrrhizaglabra) | 87 | [24] |
| Papaya (Carica papaya) Sorghum (Sorghum bicolor) Poplar (Populus spp.) | 66 68 104 | [25] |
| Moss (Physcomitrella patens) | 38 | [25] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maheen, N.; Shafiq, M.; Sadiq, S.; Farooq, M.; Ali, Q.; Habib, U.; Shahid, M.A.; Ali, A.; Ali, F. Genome Identification and Characterization of WRKY Transcription Factor Gene Family in Mandarin (Citrus reticulata). Agriculture 2023, 13, 1182. https://doi.org/10.3390/agriculture13061182
Maheen N, Shafiq M, Sadiq S, Farooq M, Ali Q, Habib U, Shahid MA, Ali A, Ali F. Genome Identification and Characterization of WRKY Transcription Factor Gene Family in Mandarin (Citrus reticulata). Agriculture. 2023; 13(6):1182. https://doi.org/10.3390/agriculture13061182
Chicago/Turabian StyleMaheen, Nimra, Muhammad Shafiq, Saleha Sadiq, Muhammad Farooq, Qurban Ali, Umer Habib, Muhammad Adnan Shahid, Asjad Ali, and Fawad Ali. 2023. "Genome Identification and Characterization of WRKY Transcription Factor Gene Family in Mandarin (Citrus reticulata)" Agriculture 13, no. 6: 1182. https://doi.org/10.3390/agriculture13061182
APA StyleMaheen, N., Shafiq, M., Sadiq, S., Farooq, M., Ali, Q., Habib, U., Shahid, M. A., Ali, A., & Ali, F. (2023). Genome Identification and Characterization of WRKY Transcription Factor Gene Family in Mandarin (Citrus reticulata). Agriculture, 13(6), 1182. https://doi.org/10.3390/agriculture13061182









