Identification of Thermo-Sensitive Chloroplast Development Gene TSCD5 Required for Rice Chloroplast Development under High Temperature
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Measurement of Chlorophyll Content
2.3. Measurement of Photosynthetic Rate
2.4. Transmission Electron Microscopy (TEM)
2.5. TUNEL Assay
2.6. Nitro Blue Tetrazolium (NBT) and 3, 3′-Diaminobenzidine (DAB) Staining
2.7. Measurement of Physiological Indices Related to ROS
2.8. Genetic Analysis and Fine Mapping
2.9. Sequence and Phylogenetic Analysis
2.10. Plasmid Construction and Rice Transformation
2.11. Subcellular Localization of TSCD5 Protein
2.12. GUS Assay
2.13. RNA Extraction and Quantitative Real-Time PCR (qRT-PCR) Analysis
2.14. Statistical Analysis
3. Results
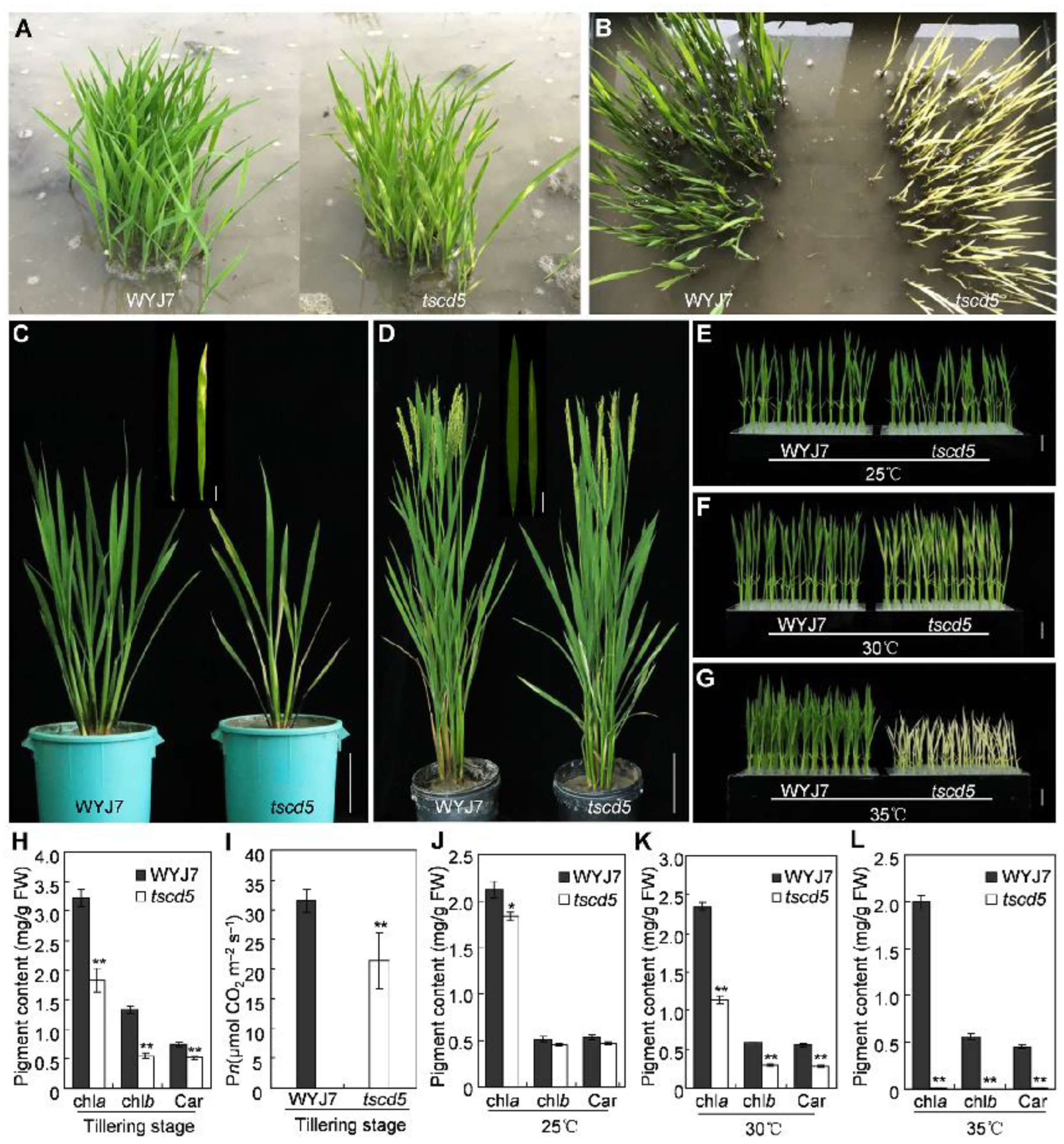
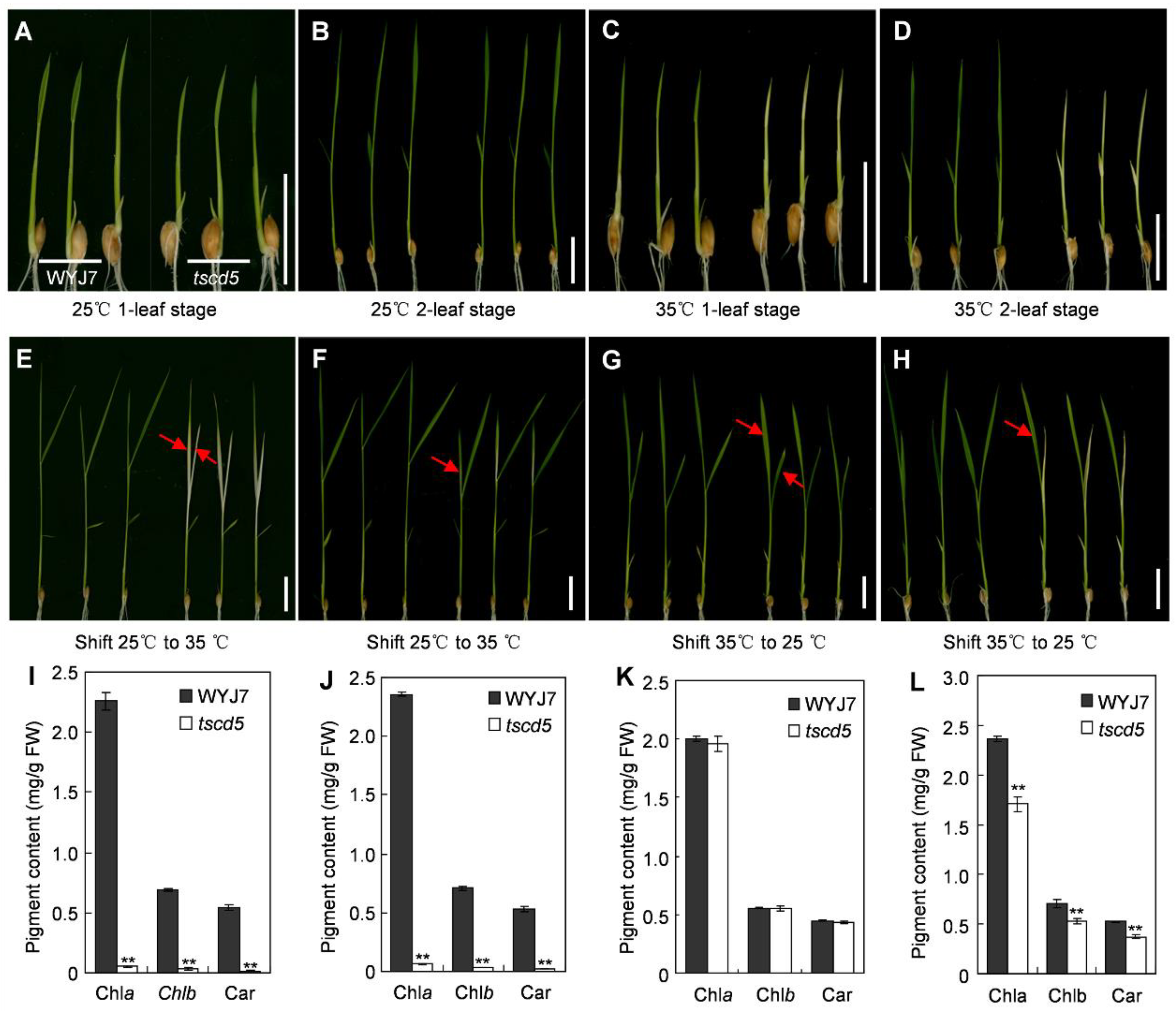
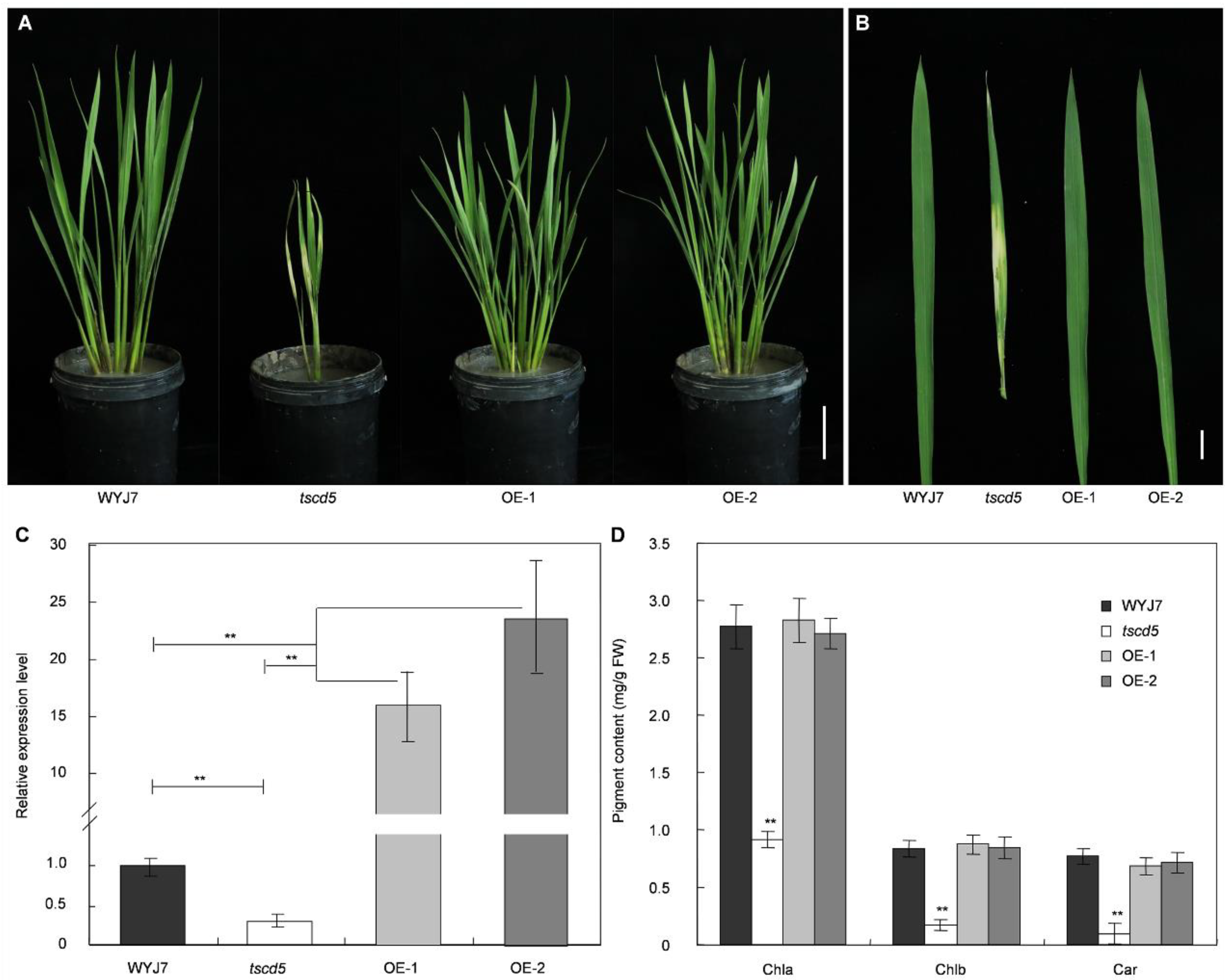
3.1. The Mutant Phenotype of tscd5 Is Sensitive to High Temperature
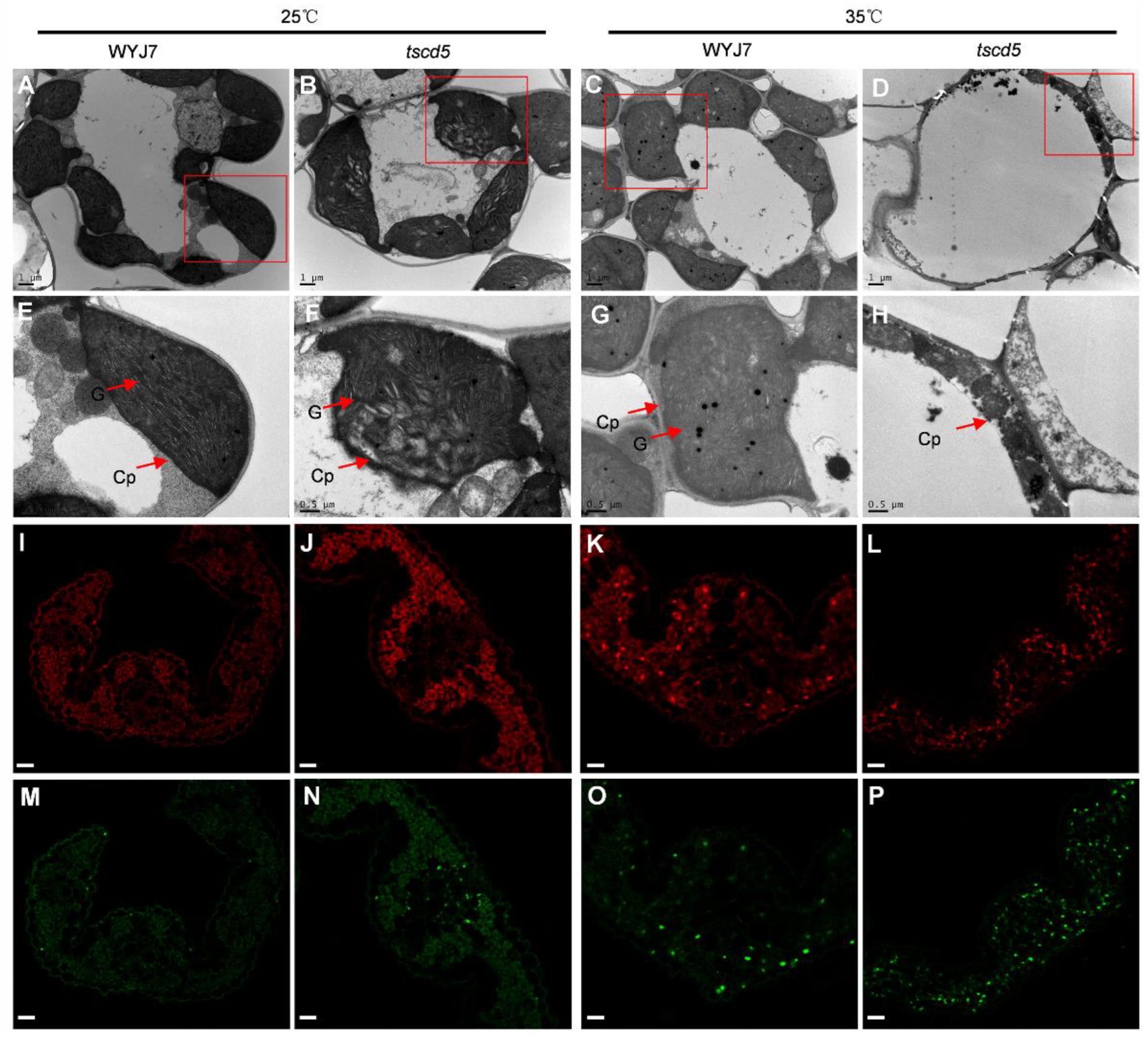
3.2. Chloroplast Development Is Hampered and Cell Death Is Exacerbated in tscd5 at High Temperature
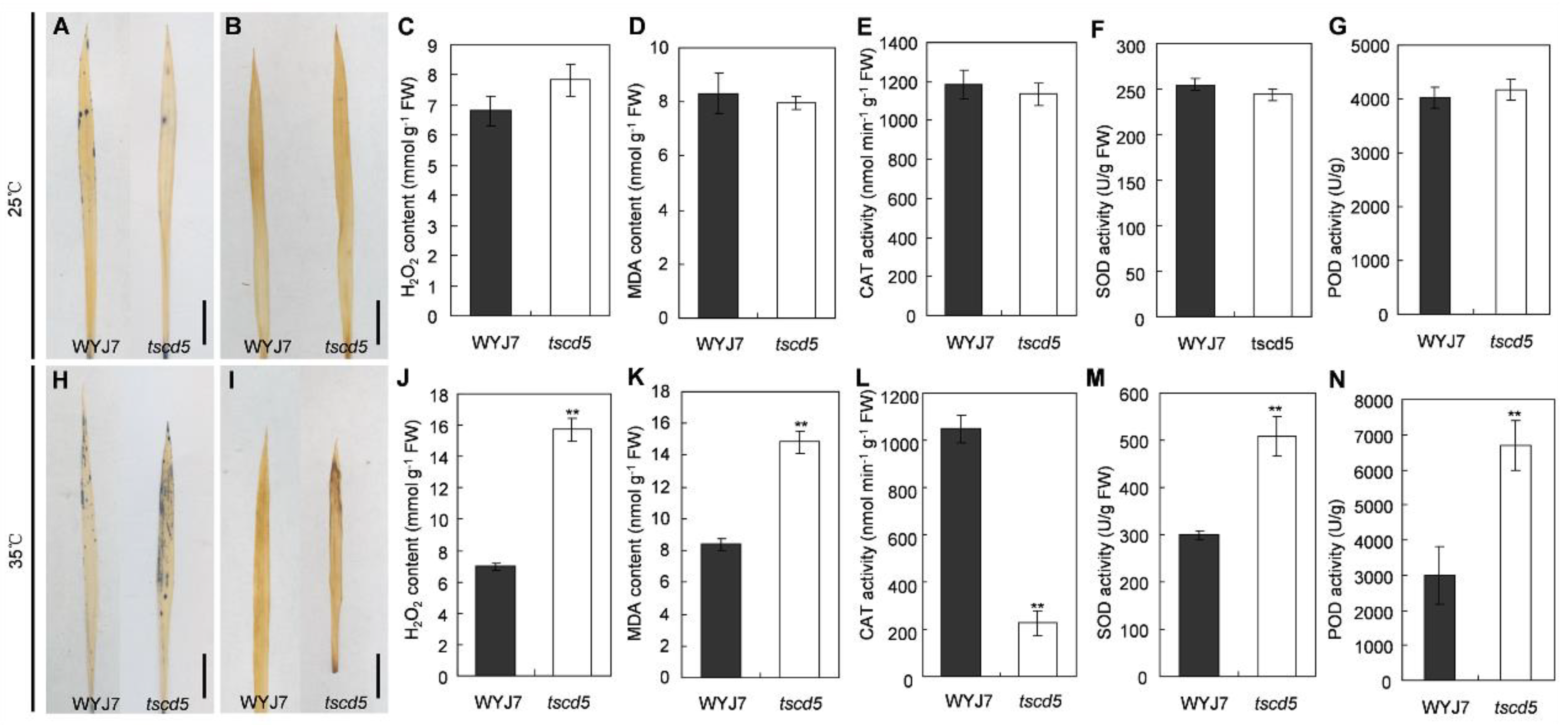
3.3. Excess ROS Accumulation in tscd5 at High Temperature
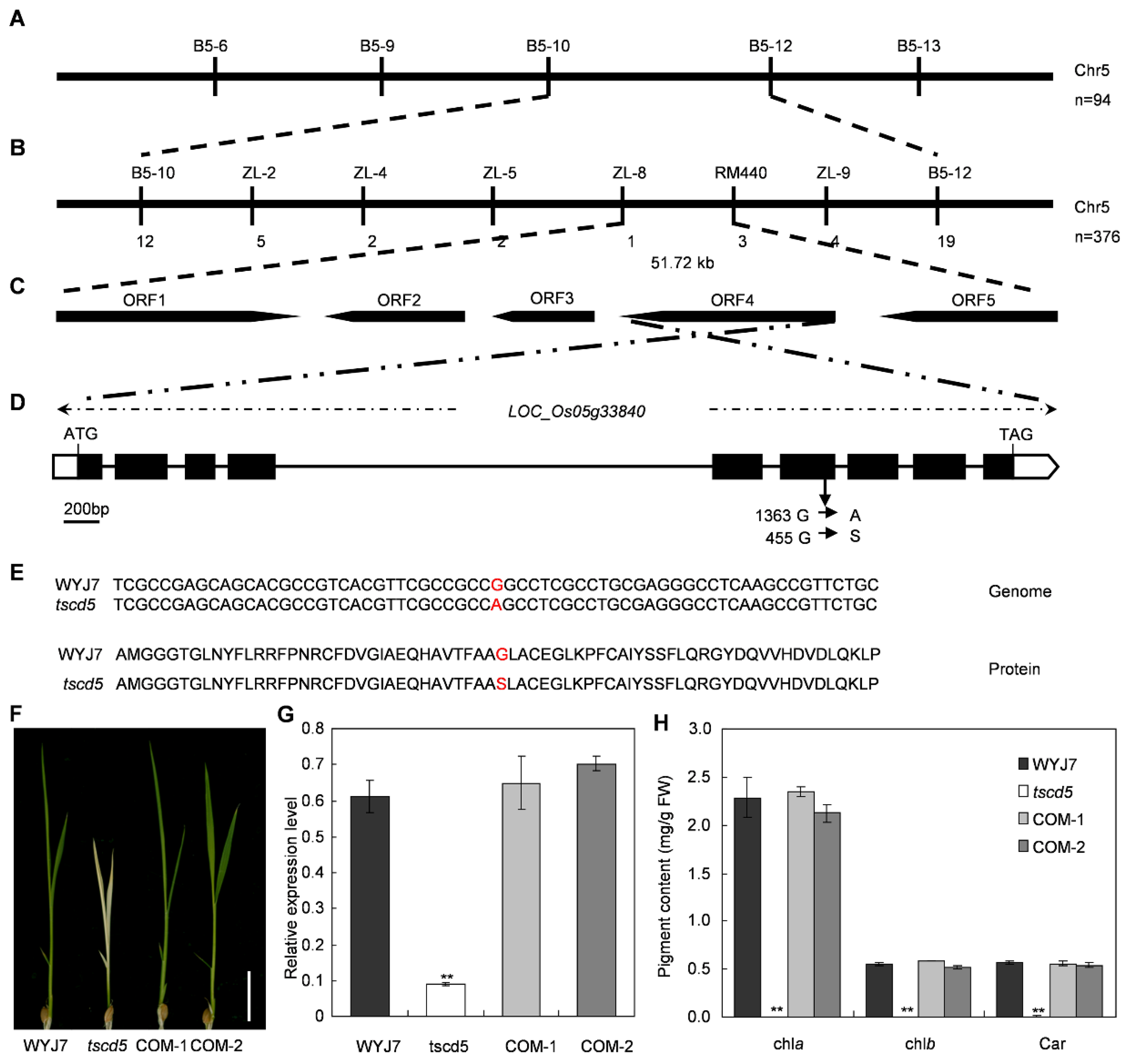
3.4. Map-Based Cloning of TSCD5
3.5. TSCD5 Proteins Are Highly Conserved in Plants
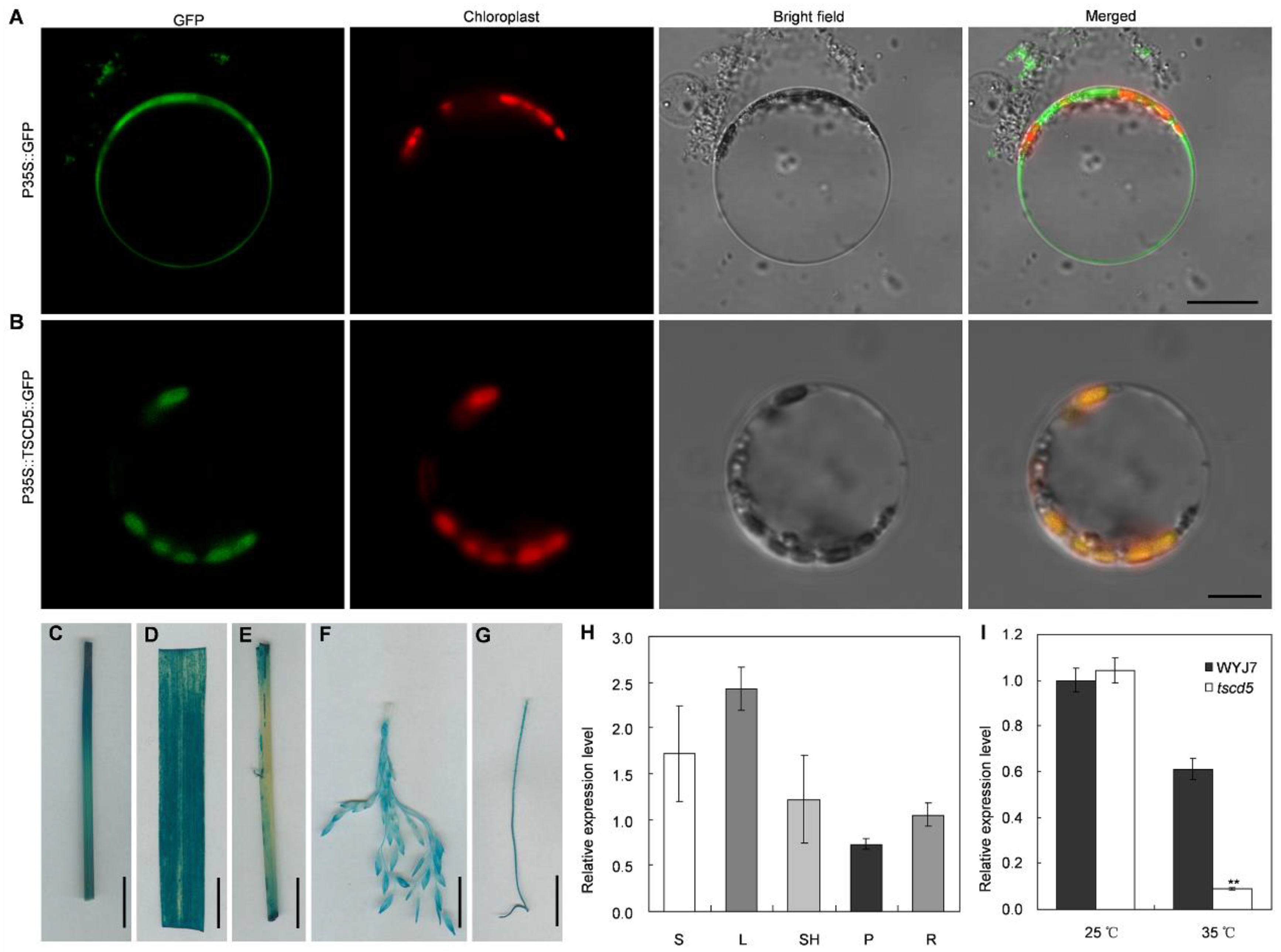
3.6. TSCD5 Protein Localized in Chloroplasts in Rice
3.7. TSCD5 Is Ubiquitously Expressed in Rice Tissues and Significantly Lower in tscd5 Leaves Than That in WYJ7 Leaves under HIGH Temperature
3.8. Expression of Genes Related to Chlorophyll Synthesis and Degradation, Chloroplast Development and Photosynthesis Are Affected Dramatically in tscd5 under High Temperature
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ye, W.; Hu, S.; Wu, L.; Ge, C.; Cui, Y.; Chen, P.; Wang, X.; Xu, J.; Ren, D.; Dong, G.; et al. White stripe leaf 12 (WSL12), encoding a nucleoside diphosphate kinase 2 (OsNDPK2), regulates chloroplast development and abiotic stress response in rice (Oryza sativa L.). Mol Breed. 2016, 36, 57. [Google Scholar] [CrossRef]
- Zhao, C.; Xu, J.; Chen, Y.; Mao, C.; Zhang, S.; Bai, Y.; Jiang, D.; Wu, P. Molecular cloning and characterization of OsCHR4, a rice chromatin-remodeling factor required for early chloroplast development in adaxial mesophyll. Planta 2012, 236, 1165–1176. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, Z.; Gu, H.; You, J.; Hu, M.; Zhang, Y.; Zhu, Z.; Wang, Y.; Liu, S.; Chen, L.; et al. Identification and Phenotypic Characterization of ZEBRA LEAF16 Encoding a β-Hydroxyacyl-ACP Dehydratase in Rice. Front. Plant Sci. 2018, 9, 782. [Google Scholar] [CrossRef] [PubMed]
- Koussevitzky, S.; Nott, A.; Mockler, T.C.; Hong, F.; Sachetto-Martins, G.; Surpin, M.; Lim, J.; Mittler, R.; Chory, J. Signals from Chloroplasts Converge to Regulate Nuclear Gene Expression. Science 2007, 316, 715–719. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Feng, P.; Li, Y.; Yu, P.; Yu, G.; Sang, X.; Ling, Y.; Zeng, X.; Li, Y.; Huang, J.; et al. VIRESCENT-ALBINO LEAF 1 regulates leaf colour development and cell division in rice. J Exp Bot. 2018, 69, 4791–4804. [Google Scholar] [CrossRef] [PubMed]
- Zheng, K.; Zhao, J.; Lin, D.; Chen, J.; Xu, J.; Zhou, H.; Teng, S. The rice TCM5 gene encoding a novel Deg protease protein is essential for chloroplast development under high temperatures. Rice 2016, 9, 13. [Google Scholar] [CrossRef]
- Lv, Y.; Shao, G.; Qiu, J.; Jiao, G.; Sheng, Z.; Xie, L.; Wu, Y.; Tang, S.; Wei, X.; Hu, P. White Leaf and Panicle 2, encoding a PEP-associated protein, is required for chloroplast biogenesis under heat stress in rice. J. Exp. Bot. 2017, 68, 5147–5160. [Google Scholar] [CrossRef]
- Qiu, Z.; Kang, S.; He, L.; Zhao, J.; Zhang, S.; Hu, J.; Zeng, D.; Zhang, G.; Dong, G.; Gao, Z.; et al. The newly identified, heat-stress sensitive albino 1, gene affects chloroplast development in rice. Plant Sci. 2017, 267, 168–179. [Google Scholar] [CrossRef]
- Fang, G.; Yang, S.; Ruan, B.; Liu, C.; Zhang, A.; Jiang, H.; Ding, S.; Tian, B.; Zhang, Y.; Jaha, N.; et al. Isolation of TSCD11 Gene for Early Chloroplast Development under High Temperature in Rice. Rice 2020, 13, 49. [Google Scholar] [CrossRef]
- Kusumi, K.; Mizutani, A.; Nishimura, M.; Iba, K. A virescent gene V1 determines the expression timing of plastid genes for transcription/translation apparatus during early leaf development in rice. Plant J. 1997, 12, 1241–1250. [Google Scholar] [CrossRef]
- Sugimoto, H.; Kusumi, K.; Noguchi, K.; Yano, M.; Yoshimura, A.; Iba, K. The rice nuclear gene, VIRESCENT 2, is essential for chloroplast development and encodes a novel type of guanylate kinase targeted to plastids and mitochondria. Plant J. 2007, 52, 512–527. [Google Scholar] [CrossRef]
- Yoo, S.C.; Cho, S.H.; Sugimoto, H.; Li, J.; Kusumi, K.; Koh, H.J.; Iba, K.; Paek, N.C. Rice Virescent3 and Stripe1 encoding the large and small subunits of ribonucleotide reductase are required for chloroplast biogenesis during early leaf development. Plant Physiol. 2009, 150, 388–401. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Mei, J.; Gong, X.D.; Xu, J.L.; Zhang, J.H.; Teng, S. IImportance of the rice TCD9 encoding α subunit of chaperonin protein 60 (Cpn60α) for the chloroplast development during the early leaf stage. Plant Sci. 2014, 215–216, 172–179. [Google Scholar] [CrossRef]
- Wu, L.; Wu, J.; Liu, Y.; Gong, X.; Xu, J.; Lin, D.; Dong, Y. The Rice Pentatricopeptide Repeat Gene TCD10 Is Needed for Chloroplast Development Under Cold Stress. Rice 2016, 9, 67. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, J.; Shi, X.; Peng, Y.; Li, P.; Lin, D.; Dong, Y.; Teng, S. Temperature-sensitive albino gene TCD5, encoding a monooxygenase, affects chloroplast development at low temperatures. J. Exp. Bot. 2016, 67, 5187–5202. [Google Scholar] [CrossRef]
- Lin, D.; Zhang, L.; Mei, J.; Chen, J.; Piao, Z.; Lee, G.; Dong, Y. Mutation of the rice TCM12 gene encoding 2,3-bisphosphoglycerate-independent phosphoglycerate mutase affects chlorophyll synthesis, photosynthesis and chloroplast development at seedling stage at low temperatures. Plant Biol. 2019, 21, 585–594. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Wang, Y.; Wu, J.; Han, X.; Gu, X.; Lu, T.; Zhang, Z. The RNA editing factor DUA1 is crucial to chloroplast development at low temperature in rice. New Phytol. 2019, 221, 834–849. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z.; Zhu, L.; He, L.; Chen, D.; Zeng, D.; Chen, G.; Hu, J.; Zhang, G.; Ren, D.; Dong, G.; et al. DNA damage and reactive oxygen species cause cell death in the rice local lesions 1 mutant under high light and high temperature. New Phytol. 2019, 222, 349–365. [Google Scholar] [CrossRef]
- Li, X.; Cai, C.; Wang, Z.; Fan, B.; Zhu, C.; Chen, Z. Plastid translation elongation factor Tu is Prone to Heat-Induced Aggregation Despite Its Critical Role in Plant Heat Tolerance. Plant Physiol. 2018, 176, 3027–3045. [Google Scholar] [CrossRef]
- Morita, S.; Siratsuchi, H.; Takanashi, J.; Fujita, K. Effect of high temperature on ripening in rice plant: Analysis of the effect of high night and high day temperature applied to the panicle in other parts of the plant. Jpn. J. Crop. 2004, 73, 77–83. [Google Scholar] [CrossRef]
- Wahid, A.; Gelani, S.; Ashraf, M.; Foolad, M.R. Heat tolerance in plants: An overview. Environ. Exp. Bot. 2007, 61, 199–223. [Google Scholar] [CrossRef]
- Ambavaram, M.M.R.; Basu, S.; Krishnan, A.; Ramegowda, V.; Batlang, U.; Rahman, L.; Baisakh, N.; Pereira, A. Coordinated regulation of photosynthesis in rice increases yield and tolerance to environmental stress. Nat. Commun. 2014, 5, 5302. [Google Scholar] [CrossRef] [PubMed]
- Howarth, C.J. Genetic improvements of tolerance to high temperature. In Abiotic Stresses: Plant Resistance through Breeding and Molecular Approaches; Ashraf, M., Harris, P.J.C., Eds.; Haworth Press Inc.: New York, NY, USA, 2005; pp. 277–300. [Google Scholar] [CrossRef]
- Suzuki, N.; Koussevitzky, S.; Mittler, R.; Miller, G. ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 2012, 35, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Saed-Moucheshi, A.; Shekoofa, A.; Pessarakli, M. Reactive oxygen species (ROS) generation and detoxifying in plants. J. Plant Nutr. 2014, 37, 1573–1585. [Google Scholar] [CrossRef]
- Mittler, R.; Vanderauwera, S.; Suzuki, N.; Miller, G.; Tognetti, V.B.; Vandepoele, K.; Gollery, M.; Shulaev, V.; Van, B.F. ROS signaling: The new wave? Trends Plant Sci. 2011, 16, 300–309. [Google Scholar] [CrossRef]
- Joyard, J.; Ferro, M.; Masselon, C.; Seigneurin-Berny, D.; Salvi, D.; Garin, J.; Rolland, N. Chloroplast Proteomics and the Compartmentation of Plastidial Isoprenoid Biosynthetic Pathways. Mol Plant. 2009, 2, 1154–1180. [Google Scholar] [CrossRef]
- You, M.K.; Lee, Y.J.; Kim, J.K.; Baek, S.A.; Jeon, Y.A.; Lim, S.H.; Ha, S.H. The organ-specific differential roles of rice DXS and DXR, the first two enzymes of the MEP pathway, in carotenoid metabolism in Oryza sativa leaves and seeds. BMC Plant Biol. 2020, 20, 167. [Google Scholar] [CrossRef]
- Lange, B.M.; Ghassemian, M. Genome organization in Arabidopsis thaliana: A survey for genes involved in isoprenoid and chlorophyll metabolism. Plant Mol Biol. 2003, 51, 925–948. [Google Scholar] [CrossRef]
- Pulido, P.; Toledo-Ortiz, G.; Phillips, M.A.; Wright, L.P.; Rodríguez-Concepción, M. Arabidopsis J-Protein J20 Delivers the First Enzyme of the Plastidial Isoprenoid Pathway to Protein Quality Control. Plant Cell. 2013, 25, 4183–4194. [Google Scholar] [CrossRef]
- Mandel, M.A.; Feldmann, K.A.; Herrera-Estrella, L.; Rocha-Sosa, M.; León, P. CLA1, a novel gene required for chloroplast development, is highly conserved in evolution. Plant J. 1996, 9, 649–658. [Google Scholar] [CrossRef]
- Estévez, J.M.; Cantero, A.; Reindl, A.; Reichler, S.; León, P. 1-Deoxy-D-xylulose-5-phosphate synthase, a limiting enzyme for plastidic isoprenoid biosynthesis in plants. J. Biol. Chem. 2001, 276, 22901–22909. [Google Scholar] [CrossRef] [PubMed]
- Sprenger, G.A.; Schorken, U.; Wiegert, T. Identification of a thiamin-dependent synthase in Escherichia coli required for the formation of the 1-deoxy-D-xylulose 5-phosphate precursor to isoprenoids, thiamin, and pyridoxol. Proc. Natl. Acad. Sci. USA 1997, 94, 12857–12862. [Google Scholar] [CrossRef] [PubMed]
- Lange, B.M.; Wildung, M.R.; Mccaskill, D.; Croteau, R. A family of transketolases that directs isoprenoid biosynthesis via a mevalonate-independent pathway. Proc. Natl. Acad. Sci. USA 1998, 95, 2100–2104. [Google Scholar] [CrossRef] [PubMed]
- Estévez, J.M. Analysis of the Expression of CLA1, a Gene That Encodes the 1-Deoxyxylulose 5-Phosphate Synthase of the 2-C-Methyl-D-Erythritol-4-Phosphate Pathway in Arabidopsis. Plant Physiol. 2000, 124, 95–104. [Google Scholar] [CrossRef]
- Hahn, F.M.; Eubanks, L.M.; Testa, C.A.; Blagg, B.S.; Baker, J.A.; Poulter, C.D. 1-Deoxy-d-Xylulose 5-Phosphate Synthase, the Gene Product of Open Reading Frame (ORF) 2816 and ORF 2895 in Rhodobacter capsulatus. J. Bacteriol. 2001, 183, 1–11. [Google Scholar] [CrossRef]
- Walter, M.H.; Hans, J.; Strack, D. Two distantly related genes encoding 1-deoxy-d-xylulose 5-phosphate synthases: Differential regulation in shoots and apocarotenoid-accumulating mycorrhizal roots. Plant J. 2002, 31, 243–254. [Google Scholar] [CrossRef]
- Kim, B.R.; Kim, S.U.; Chang, Y.J. Differential expression of three 1-deoxy-D-xylulose-5-phosphate synthase genes in rice. Biotechnol. Lett. 2005, 27, 997–1001. [Google Scholar] [CrossRef]
- Cordoba, E.; Porta, H.; Arroyo, A.; San Roman, C.; Medina, L.; Rodriguez-Concepcion, M.; Leon, P. Functional characterization of the three genes encoding 1-deoxy-D-xylulose 5-phosphate synthase in maize. J. Exp. Bot. 2011, 62, 2023–2038. [Google Scholar] [CrossRef]
- Okada, A.; Shimizu, T.; Okada, K.; Kuzuyama, T.; Koga, J.; Shibuya, N.; Nojiri, H.; Yamane, H. Elicitor induced activation of the methylerythritol phosphate pathway toward phytoalexins biosynthesis in rice. Plant Mol. Biol. 2007, 65, 177–187. [Google Scholar] [CrossRef]
- Floss, D.S.; Hause, B.; Lange, P.R.; Kuster, H.; Strack, D.; Walter, M.H. Knock-down of the MEP pathway isogene 1-deoxy-D-xylulose 5-phosphate synthase 2 inhibits formation of arbuscular mycorrhiza-induced apocarotenoids, and abolishes normal expression of mycorrhiza-specific plant marker genes. Plant J. 2008, 56, 86–100. [Google Scholar] [CrossRef]
- Carretero-Paulet, L.; Cairo, A.; Talavera, D.; Saura, A.; Imperial, S.; Rodriguez-Concepcion, M.; Campos, N.; Boronat, A. Functional and evolutionary analysis of DXL1, a non-essential gene encoding a 1-deoxy-D-xylulose 5-phosphate synthase like protein in Arabidopsis thaliana. Gene 2013, 524, 40–53. [Google Scholar] [CrossRef]
- Peng, G.; Wang, C.; Song, S.; Fu, X.; Azam, M.; Grierson, D.; Xu, C. The role of 1-deoxy-d-xylulose-5-phosphate synthase and phytoene synthase gene family in citrus carotenoid accumulation. Plant Physiol. Biochem. 2013, 71, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, Y.; Liu, L.; Liu, Q.; Bi, Z.; Yu, N.; Cheng, S.; Cao, L. Fine mapping of the lesion mimic and early senescence 1 (lmes1) in rice (Oryza sativa). Plant Physiol. Biochem. 2014, 80, 300–307. [Google Scholar] [CrossRef]
- Schwartzman, R.A.; Cidlowski, J.A. Apoptosis: The biochemistry and molecular biology of programmed cell death. Endocr. Rev. 1993, 14, 133–151. [Google Scholar] [CrossRef]
- Oberhammer, F.; Wilson, J.W.; Dive, C.; Morris, I.D.; Hickman, J.A.; Wakeling, A.E.; Walker, P.R.; Sikorska, M. Apoptotic death in epithelial cells: Cleavage of DNA to 300 and/or 50 kb fragments prior to or in the absence of internucleosomal fragmentation. EMBO J. 1993, 12, 3679–3684. [Google Scholar] [CrossRef]
- Huang, Y.J.; Lu, K.S. TUNEL staining and electron microscopy studies of apoptotic changes in the guinea pig vallate taste cells after unilateral glossopharyngeal denervation. Anat Embryol. 2001, 204, 493–501. [Google Scholar] [CrossRef] [PubMed]
- Ruan, B.; Hua, Z.; Zhao, J.; Zhang, B.; Ren, D.; Liu, C.; Yang, S.; Zhang, A.; Jiang, H.; Yu, H.; et al. OsACL-A2 negatively regulates cell death and disease resistance in rice. Plant. Biotechnol. J. 2019, 17, 1344–1356. [Google Scholar] [CrossRef]
- Ramel, F.; Sulmon, C.; Bogard, M.; Couée, I.; Gouesbet, G. Differential patterns of reactive oxygen species and antioxidative mechanisms during atrazine injury and sucrose-induced tolerance in Arabidopsis thalianaplantlets. BMC Plant Biol. 2009, 9, 28. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Li, D. Hydrogen peroxide is not involved in HrpN from Erwinia amylovora-induced hypersensitive cell death in maize leaves. Plant Cell Rep. 2011, 30, 1273–1279. [Google Scholar] [CrossRef]
- Rao, Y.; Yang, Y.; Xu, J.; Li, X.; Leng, Y.; Dai, L.; Huang, L.; Shao, G.; Ren, D.; Hu, J.; et al. EARLY SENESCENCE 1 encodes a SCAR-like protein2 that affects water loss in rice. Plant Physiol. 2015, 169, 1225–1239. [Google Scholar] [CrossRef]
- Ruan, B.; Shang, L.; Zhang, B.; Hu, J.; Wang, Y.; Lin, H.; Zhang, A.; Liu, C.; Peng, Y.; Zhu, L.; et al. Natural variation in the promoter of TGW2 determines grain width and weight in rice. New Phytol. 2020, 227, 629–640. [Google Scholar] [CrossRef]
- Pospíšil, P. Production of reactive oxygen species by photosystem II as a response to light and temperature stress. Front Plant Sci. 2016, 7, 1950. [Google Scholar] [CrossRef]
- Leister, D. Chloroplast research in the genomic age. Trends Genet. 2003, 19, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, Y.; Chai, J.; Wang, L.; Wang, C.; Long, W.; Wang, D.; Wang, Y.; Zheng, M.; Peng, C.; et al. Green-revertible Chlorina 1 (grc1), is required for the biosynthesis of chlorophyll and the early development of chloroplasts in rice. J. Plant Biol. 2013, 56, 326–335. [Google Scholar] [CrossRef]
- Kusumi, K.; Sakata, C.; Nakamura, T.; Kawasaki, S.; Yoshimura, A.; Iba, K. A plastid protein NUS1 is essential for build-up of the genetic system for early chloroplast development under cold stress conditions. Plant J. 2011, 68, 1039–1050. [Google Scholar] [CrossRef] [PubMed]
- Price, A.H.; Atherton, N.M.; Hendry, G.A. Plants under drought-stress generate activated oxygen. Free Radic. Res. Commun. 1989, 8, 61–66. [Google Scholar] [CrossRef]
- Chang, C.C.C.; Ball, L.; Fryer, M.J.; Baker, N.R.; Karpinski, S.; Mullineaux, P.M. Induction of ASCORBATE PEROXIDASE 2 expression in wounded Arabidopsis leaves does not involve known wound-signalling pathways but is associated with changes in photosynthesis. Plant J. 2004, 38, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.P.; Chao, D.Y.; Lin, H.X. Understanding abiotic stress tolerance mechanisms: Recent studies on stress response in rice. J. Integr. Plant Biol. 2007, 49, 742–750. [Google Scholar] [CrossRef]
- Wang, H.S.; Yu, C.; Tang, X.F.; Wang, L.Y.; Dong, X.C.; Meng, Q.W. Antisense-mediated depletion of tomato endoplasmic reticulum omega-3 fatty acid desaturase enhances thermal tolerance. J. Integr. Plant Biol. 2010, 52, 568–577. [Google Scholar] [CrossRef]
- Zhao, K.F.; Song, J.; Fan, H.; Zhou, S.; Zhao, M. Growth response to ionic and osmotic stress of NaCl in salt-tolerant and salt-sensitive maize. J. Integr. Plant Biol. 2010, 52, 468–475. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.; Su, Q.; Lin, D.; Jiang, Q.; Xu, J.; Zhang, J.; Teng, S.; Dong, Y. The rice OsV4 encoding a novel pentatricopeptide repeat protein is required for chloroplast development during the early leaf stage under cold stress. J. Integr. Plant Biol. 2014, 56, 400–410. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Wu, C.; He, L.; Qiu, Z.; Zhang, S.; Zhang, Y.; Guo, L.; Zeng, D.; Hu, J.; Ren, D.; et al. Knocking out the gene RLS1 induces hypersensitivity to oxidative stress and premature leaf senescence in rice. Int. J. Mol. Sci. 2018, 19, 2853. [Google Scholar] [CrossRef] [PubMed]
- Przybyla, D.; Göbel, C.; Imboden, A.; Hamberg, M.; Feussner, I.; Apel, K. Enzymatic, but not non-enzymatic, 1O2-mediated peroxidation of polyunsaturated fatty acids forms part of the EXECUTER1-dependent stress response program in the flu mutant of Arabidopsis thaliana. Plant J. 2008, 54, 236–248. [Google Scholar] [CrossRef] [PubMed]
- Triantaphylidès, C.; Krischke, M.; Hoeberichts, F.A.; Ksas, B.; Gresser, G.; Havaux, M.; Breusegem, F.V.; Mueller, M.J. Singlet Oxygen Is the Major Reactive Oxygen Species Involved in Photooxidative Damage to Plants. Plant Physiol. 2008, 148, 960–968. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhang, T.; Peng, H.; Luo, S.; Tan, J.; Jiang, K.; Heng, Y.; Zhang, X.; Guo, X.; Zheng, J.; et al. Rice Premature Leaf Senescence 2, encoding a glycosyltransferase (GT), is involved in leaf senescence. Front. Plant Sci. 2018, 9, 560. [Google Scholar] [CrossRef]
- Miller, G.; Suzuki, N.; Ciftci-Yilmaz, S.; Mittler, R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010, 33, 453–467. [Google Scholar] [CrossRef]
- Hamurcu, M.; Khan, M.K.; Pandey, A.; Ozdemir, C.; Avsaroglu, Z.Z.; Elbasan, F.; Omay, A.H.; Gezgin, S. Nitric oxide regulates watermelon (Citrullus lanatus) responses to drought stress. 3 Biotech. 2020, 10, 494. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, S.; Fang, G.; Ruan, B.; Zhang, A.; Zhou, Y.; Ye, G.; Su, W.; Guo, H.; Wang, J.; Gao, Z. Identification of Thermo-Sensitive Chloroplast Development Gene TSCD5 Required for Rice Chloroplast Development under High Temperature. Agriculture 2023, 13, 563. https://doi.org/10.3390/agriculture13030563
Yang S, Fang G, Ruan B, Zhang A, Zhou Y, Ye G, Su W, Guo H, Wang J, Gao Z. Identification of Thermo-Sensitive Chloroplast Development Gene TSCD5 Required for Rice Chloroplast Development under High Temperature. Agriculture. 2023; 13(3):563. https://doi.org/10.3390/agriculture13030563
Chicago/Turabian StyleYang, Shenglong, Guonan Fang, Banpu Ruan, Anpeng Zhang, Yun Zhou, Guangji Ye, Wang Su, Heng Guo, Jian Wang, and Zhenyu Gao. 2023. "Identification of Thermo-Sensitive Chloroplast Development Gene TSCD5 Required for Rice Chloroplast Development under High Temperature" Agriculture 13, no. 3: 563. https://doi.org/10.3390/agriculture13030563
APA StyleYang, S., Fang, G., Ruan, B., Zhang, A., Zhou, Y., Ye, G., Su, W., Guo, H., Wang, J., & Gao, Z. (2023). Identification of Thermo-Sensitive Chloroplast Development Gene TSCD5 Required for Rice Chloroplast Development under High Temperature. Agriculture, 13(3), 563. https://doi.org/10.3390/agriculture13030563






