Simple Summary
Propylaea quatuordecimpunctata (L.), an important natural enemy of crop pests, shows different levels of survival, reproduction, and predation at different high temperatures (32, 35, and 38 °C). Therefore, it is important to understand the molecular basis for this variation in the specie’s thermotolerance. In this study, high-throughput sequencing analysis confirmed that P. quatuordecimpunctata has more genes which are mobilized under high temperature (38 °C). Meanwhile, we found that the cytochrome P450 gene and the heat shock protein Hsp70 gene were significantly overexpressed after high temperature stress (35 and 38 °C). The reliability of transcriptome sequencing was confirmed by qRT-PCR, indicating that these differentially expressed genes (DEGs) play an important role in the resistance of P. quatuordecimpunctata to high temperature stress. Our study provides a molecular basis for further analysis of the thermotolerance mechanism of P. quatuordecimpunctata.
Abstract
In cotton-growing regions of northern Xinjiang, Propylaea quatuordecimpunctata (L.) (Coleoptera: Coccinellidae) is an important natural enemy that provides significant control of some pest hemipterans. Previous studies have shown that the survival and reproduction of P. quatuordecimpunctata differs under different high temperatures. However, its molecular mechanism for thermotolerance is poorly understood. In this study, transcriptome sequencing analysis was performed on P. quatuordecimpunctata, after its exposure to different temperatures (32–38 °C) for 24 h, using high-throughput sequencing technology. The results showed that the 35 vs. 38 °C groups had the most DEGs (1425), indicating that P. quatuordecimpunctata has more genes that can be mobilized under high temperature (38 °C). The results of functional analysis showed that the DEGs were mainly involved in “Oxidation–reduction process”, “Oxidoreductase activity”, “Metabolic pathways”, and “Small molecule metabolic processing” groups. We randomly selected DEGs (eleven P450 genes and one Hsp70 gene) of interest for qRT-PCR validation. The qRT-PCR results were consistent with the transcriptome data, indicating that the transcriptome data were reliable. In summary, these genes involved in these pathways play an important role in the resistance of P. quatuordecimpunctata to high temperature stress. Our study enriched our understanding of the molecular mechanism for thermotolerance in P. quatuordecimpunctata.
1. Introduction
As poikilotherms, insects are very sensitive to ambient environmental conditions. When the ambient temperature is too high, their growth, development, survival, and reproduction will be negatively affected [1,2]. For example, the aphids Rhopalosiphum padi (L.), Sitobion avenae (Fabricius), and Metopolophium dirhodum (Walker) present in corn fields in the northeastern part of the Iberian Peninsula were affected by high summer temperatures, causing their populations to decline from late June to mid-August [3]. Similarly, Laodelphax striatellus (Fallén) (Hemiptera: Delphacidae) showed a lower survival rate and slower development under heat stress (33 °C), which was consistent with the sharp decline of its population during the high temperatures of summer [4]. Temperature is also an important factor affecting species distribution. Oliveira et al. [5] found that temperature can directly affect the geographic distribution of two important natural enemies of mealybugs in South America: Tenuisvalvae notata (Mulsant) and Cryptolaemus montrouzieri (Mulsant). Two forest pests in Central Europe, namely Lymantria dispar (L.) and Lymantria monacha (L.), have been found to move northward in both northern and southern boundaries under climate warming scenarios [6].
With the decreasing cost of next-generation sequencing technology, high-throughput sequencing has been widely used to reveal the molecular mechanism of insect response to heat stress [7,8,9], at the levels of ecology, physiology, and biochemistry [10,11,12]. High temperature stress can change the overall gene expression pattern of insects [7,13]. Through high-throughput sequencing, we can simultaneously measure the expression levels of tens of thousands of genes and identify differentially expressed genes under different experimental treatments within the whole transcriptome [14]. Transcriptome sequencing technology has been used to study the molecular mechanism for thermotolerance in a large number of insects, such as Bombyx mori (L.) [15], Locusta migratoria tibetensis (Chen) [16], Dastarcus helophoroides (Fairmaire) [17], Frankliniella occidentalis (Pergande) [18], and Spodoptera frugiperda (Smith) [19].
Given that natural enemy insects contribute to natural biological control [20,21], it is crucial to investigate the molecular mechanism for thermotolerance of natural enemy insects. Propylea quatuordecimpunctata (L.) (Coleoptera: Coccinellidae) is an important natural enemy in cotton fields in northern Xinjiang, China. It contributes to the regulation of multiple crop pests such as aphids, mites, and thrips [22,23,24]. Based on its favorable intrinsic characteristics, such as its population’s rate of increase, doubling time, and control capabilities, it is considered promising for broad application in biological control programs [25,26,27]. Previous studies have mainly focused on the effects of high temperature on the growth, development, predation rate, and antioxidant capacity of this ladybug. For example, Papanikolaou et al. [28] found that the development time of immature-stage ladybugs was shortened with an increase in temperature from 17 to 32.5 °C, and the eggs could not hatch at 35 °C. In our previous study, we also found that the survival rate, reproduction, predation ability, and total antioxidant capacity of P. quatuordecimpunctata adults decreased gradually with an increase in temperature (32, 35, and 38 °C) [29]. However, the molecular response mechanisms to high temperature stress were not reported.
In this study, high-throughput sequencing technology was used to reveal the transcriptome changes of P. quatuordecimpunctata adults under different high temperatures (32, 35, and 38 °C). The DEGs were enriched and analyzed by GO (Gene Ontology) and KEGG (Kyoto Encyclopedia of Genes and Genomes) to further understand the biological functions of the genes. At the same time, quantitative real-time PCR (qRT-PCR) technology was used to verify the screened genes related to the high temperature tolerance of P. quatuordecimpunctata, to confirm the accuracy of the transcriptome data. The experimental data obtained fill in the blanks for the themolecular mechanism for the thermotolerance of P. quatuordecimpunctata.
2. Materials and Methods
2.1. Insect Rearing
Propylaea quatuordecimpunctata and Aphis gossypii Glover were collected from un-sprayed cotton plots at the experimental field station of Shihezi University (44.32° N, 85.92° E) (Shihezi, Xinjiang Uygur Autonomous Region, China) in 2019. Diagnostic keys were used to confirm ladybug and aphid species’ identity [30,31]. Next, field-caught individuals were transferred to the Langfang experimental station, Chinese Academy of Agricultural Sciences (CAAS; 39.53°N, 116.70°E) in Langfang, Hebei Province. Ladybugs were reared in plastic containers (diameter: 8 cm; height: 11.5 cm) within a controlled climate chamber (RXZ500D, Ningbo Jiangnan Instrument Factory, Ningbo, China) at 32 ± 1 °C, 70 ± 5% RH, and 16:8 h (L:D) photoperiod. Cotton aphids (A. gossypii) were reared on Cucurbita pepo L. (Xinzaoqing seed, Tianjin City Ji Nong Seed Co., Ltd., Tianjin, China) and maintained within screened cages (55 × 35 × 50 cm) in a greenhouse at 28–30 °C, 50 ± 5% RH, and 16:8 h (L:D) photoperiod. Ladybugs were fed daily ad libitum with cotton aphids (500 aphids per ladybug) and used for experiments after the ladybug population had stabilized.
2.2. Heat Stress Treatments
In the early stage, we combined the actual temperatures in Xinjiang and found that P. quatuordecimpunctata adults have different survival and reproduction at different temperatures (32, 35, and 38 °C) [29]. Therefore, we still choose these three temperatures to study the thermotolerance molecular mechanism of P. quatuordecimpunctata.
Newly emerged one-day-old P. quatuordecimpunctata adults (<12 h) were held for 24 h at 32 °C (as the control), or 35 °C (medium–high temperature), or 38 °C (high temperature) in climatic chambers (70 ± 5% RH and 16:8 h L:D). There were three replicates of each temperature, and each replicate consisted of two adult P. quatuordecimpunctata. Upon completion, these P. quatuordecimpunctata were immediately frozen in liquid nitrogen and stored at −80 °C before RNA extraction.
2.3. RNA Extraction and Transcriptome Sequencing
The whole bodies of these P. quatuordecimpunctata adults were used for RNA extraction. Total RNA was extracted using TRIzol reagent (TransGen Biotech, Beijing, China), and the integrity, purity, and concentration of the RNA were verified by Agilent 2100 (Agilent Technologies, Santa Clara, CA, USA) and Nanodrop 2000 (IMPLEN, Munich, Germany) [32].
After the RNA had been completely extracted, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. Then, the enriched mRNA was fragmented into short fragments using a fragmentation buffer. Single-stranded cDNA was synthesized by reverse transcription with random hexamers, and then double-stranded cDNA was synthesized by adding buffer, dNTPs, and DNA polymerase Ⅰ. The double-stranded cDNA fragments were purified using AMPure XP Beads. The purified double-stranded cDNA was repaired and then added to poly(A) and adapters. AMPure XP Beads were used to select 250 bp fragment size for double-stranded cDNA, and then PCR amplification was performed to construct cDNA libraries. Finally, the mRNA library was sequenced on the Illumina sequencing platform by Allwegene technologies Co., Ltd. (Beijing, China).
2.4. Quality Control, Analysis, and Functional Annotation
Raw data (raw reads) of fastq format were first processed through in-house perl scripts. In this step, clean data (clean reads) were obtained by removing reads containing adapter, reads containing ploy-N (content higher than 5%), and low-quality reads (quality below 15) from the raw data. At the same time, the Q20, Q30, GC content, and the level of sequence duplication level of the clean data were calculated. All the downstream analyses were based on clean data with high quality.
The adaptor sequences and low-quality sequence reads were removed from the data sets. Raw sequences were transformed into clean reads after data processing. Assembly of these clean reads was performed using Trinity [33]. Finally, Tgicl [34] was used to cluster the transcripts of each sample twice for redundancy removal, and the final unigenes for subsequent analysis.
Unigene function was annotated based on the following databases: Nr (NCBI non-redundant protein sequences); Nt (NCBI non-redundant nucleotide sequences); Pfam (Protein family); KOG/COG (Clusters of Orthologous Groups of proteins); Swiss-Prot (A manually annotated and reviewed protein sequence database); KO (KEGG Ortholog database); GO (Gene Ontology).
2.5. Differentially Expressed Genes (DEGs) and Functional Enrichment Analysis
Differential expression analysis of two conditions/groups was performed using the DESeq R package (1.10.1) [35]. DESeq provides statistical routines for determining differential expression in digital gene expression data using a model based on the negative binomial distribution. The resulting p values were adjusted using Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with an adjusted p-value < 0.05 found by DESeq were assigned as differentially expressed. Then, the differential genes were functionally analyzed by GO enrichment (GOseq software, http://bioinf.wehi.edu.au/software/goseq/, accessed on 29 May 2020) [36] and KEGG pathway enrichment (online database: http://www.genome.jp/kegg/, accessed on 29 May 2020) [37].
2.6. Quantitative Real-Time PCR (qRT-PCR)
We selected DEGs of interest (eleven P450 and one Hsp70) for qRT-PCR detection to verify the reliability of the transcriptomic data. Twelve different genes were selected from the detection results and verified by qRT-PCR. EF1ɑ was selected as the internal reference gene [38]. Using Primer3Plus (http://www.primer3plus.com, accessed on 1 June 2021), we designed qRT-PCR primers (amplification efficiency and specificity of primer were measured) (Table 1). The sequencing RNA (1 μg) was used to synthesize the first-strand cDNA (template) using a reverse transcription kit (Tiangen Biotechnology (Beijing) Co., Ltd., Beijing, China) and then mixed into a 20 μL reaction system by using a fluorescence quantitative kit (Tiangen Biotechnology (Beijing) Co., Ltd., Beijing, China). Relative changes in gene expression were assessed using the 2−ΔΔCt method [39]. Three biological replicates were performed for each treatment, and three technical replicates were performed for each sample.

Table 1.
Primers used in qRT-PCR.
2.7. Data Analysis
Five dilution gradients were formed by diluting the template cDNA according to five-fold gradients. The qRT-PCR reaction was performed on the diluted template cDNA, and the standard curve was produced according to the Ct value. The amplification efficiency (E) was calculated by the formula E = [(5−1/slope of standard curve) − 1] × 100%. The amplification efficiency should be 90%~105%, and the correlation coefficient (R2) of the standard curve should be greater than 98%.
One-way analysis of variance (ANOVA) was used to analyze the expression of candidate genes of P. quatuordecimpunctata at different temperatures, and Tukey’s test was used to identify significant differences between specific temperatures in the expression of candidate genes in P. quatuordecimpunctata (p < 0.05). All statistical analyses were conducted using SPSS 25.0 (IBM Co., Ltd., Armonk, NS, USA) software and Microsoft Excel 2010 (Microsoft Co., Ltd., Redmond, WA, USA), while charts were generated using OriginPro 9.0 (OriginLab Co., Ltd., Northampton, MA, USA) and GraphPad Prism 8.0 (GraphPad Software Co., Ltd., La Jolla, CA, USA).
3. Results
3.1. Transcriptome Sequencing Quality Assessment and Functional Annotation
The results of sequencing analysis of experimental insects under different temperature treatments are shown in Table 2. In all tested samples, the base error rate was 0.02%; the Q20 value was in the range of 97.83~98.17%; the Q30 value was in the range of 93.91~94.72%; and the GC content was in the range of 39.42~40.31%. This indicates that the construction quality of the sequencing library is good, and the data obtained by sequencing are accurate and reliable.

Table 2.
Quality control result of sequencing data of P. quatuordecimpunctata under different temperature stress.
The sequences obtained above were assembled and analyzed to obtain 73,966 unigene sequences. The obtained unigene sequences were annotated with seven databases (NR, NT, KEGG, SwissProt, Pfam, GO, and KOG) for gene function. The highest number of unigenes was found in the NR database (24,304 unigenes), accounting for 32.86% of the total. A total of 741 unigenes sequences were annotated for all data, accounting for 1% of the total (Table 3).

Table 3.
Numbers of all annotated unigenes of P. quatuordecimpunctata in seven databases.
To understand the similarity of gene sequences between P. quatuordecimpunctata and other related species, unigene sequences of P. quatuordecimpunctata were compared and annotated with the NR database (Figure 1). We found that 44.56% of the gene sequences of P. quatuordecimpunctata were the same as the genome of Tribolium castaneum (Herbst), proving that it and T. castaneum have high similarity and share a close genetic relationship.
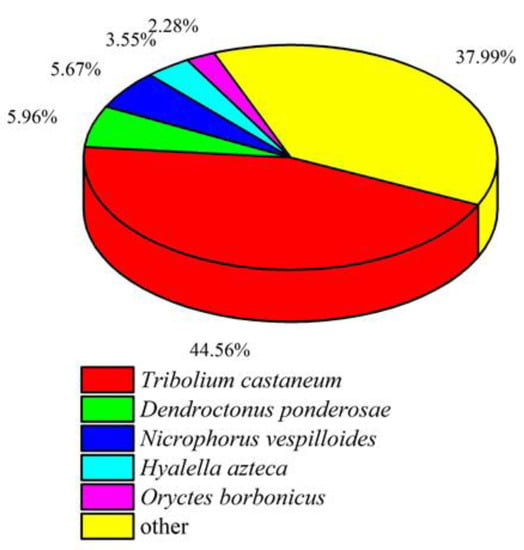
Figure 1.
All unigene sequences for P. quatuordecimpunctata that had blast annotations to the NR database were analyzed for species distribution.
3.2. Differentially Expressed Genes (DEGs)
In this study, the significant DEGs among different treatments were screened by their expression levels. The standard for DEGs screening is q-value < 0.05 (after correction, FDR value). We conducted statistical analysis of the DEGs, and we found an overlapping relationship of DEGs in P. quatuordecimpunctata under three temperature treatments (see Venn diagram in Figure 2). We found that the 38 vs. 35 °C groups had the largest number of DEGs (1425), including 762 upregulated genes and 663 downregulated genes. The 35 vs. 32 °C group had the lowest number of DEGs (781), including 524 upregulated genes and 257 downregulated genes. This indicates that P. quatuordecimpunctata have more genes that can be mobilized under high temperature (38 °C).
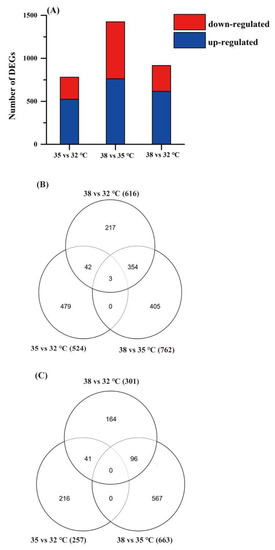
Figure 2.
Differentially expressed genes (DEGs) in P. quatuordecimpunctata under different temperature stress. (A) Total numbers of individual transcripts that were significantly up- or downregulated in different temperature groups. (B) Venn diagram illustrating the number of upregulated genes in the different temperature groups. (C) Venn diagram illustrating the number of downregulated genes in the different temperature groups.
3.3. GO Enrichment Analysis of DEGs
A large number of DEGs were identified from P. quatuordecimpunctata after different temperature treatments. According to the enrichment significance (p-value), a histogram was drawn for the first 30 GO terms of P. quatuordecimpunctata (Figure 3). In all groups, all DEGs were found to be mainly enriched in two categories: biological processes and molecular functions. In the 35 vs. 32 °C groups, the DEGs were mainly enriched in the “carbohydrate metabolic” pathway (7 upregulated genes and 4 downregulated genes) of biological processes and the “catalytic activity” (74 upregulated genes and 17 downregulated genes) and “hydrolase activity” pathway (36 upregulated genes and 6 downregulated genes) of molecular functions (Figure 3A, Supplemental Table S1). However, in the 38 vs. 35 °C groups, the DEGs were mainly enriched in the area of “small molecule metabolic process” (33 upregulated genes and 9 downregulated genes) and “oxidation−reduction” pathways (31 upregulated genes and 5 downregulated genes) of biological processes, as well as the “catalytic activity” (124 upregulated genes and 61 downregulated genes) and “oxidoreductase activity” pathways (49 upregulated genes and 9 downregulated genes) of molecular functions (Figure 3B, Supplemental Table S1). These results suggest possible reasons for the differences in the response of P. quatuordecimpunctata to different high temperatures (35 and 38 °C).
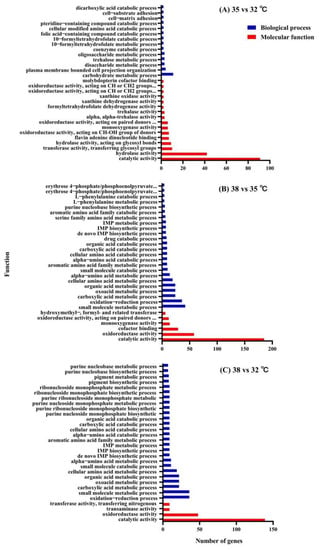
Figure 3.
GO enrichment analysis of adult P. quatuordecimpunctata under different temperature stress.
3.4. KEGG Enrichment Analysis of DEGs
The enrichment analysis based on KEGG was used to further resolve the mechanism of thermotolerance in P. quatuordecimpunctata. The DEGs of the 35 vs. 32 °C groups, the 38 vs. 35 °C groups, and the 38 vs. 32 °C groups were enriched to 183, 180, and 164 KEGG metabolic pathways, respectively. Here, we showed the top 20 most significant “metabolic pathways” that were enriched (Figure 4). “ABC transporters”, “Ascorbate and aldarate metabolism”, “Chemical carcinogenesis”, “Drug metabolism−other enzymes”, “Metabolic pathways”, “Pentose and glucuronate interconversions”, “Retinol metabolism”, “Starch and sucrose metabolism”, and “Steroid hormone biosynthesis” were the co-enriched pathways in the 35 vs. 32 °C and 38 vs. 35 °C groups (Figure 4, Supplemental Table S2). The DEGs involved in “Metabolic pathways” were further analyzed. Most of the DEGs (35 genes) of the metabolic pathways were obviously upregulated in 35 vs. 32 °C, and only a few DEGs (7 genes) were obviously downregulated in 35 vs. 32 °C. Similarly, most of the DEGs (87 genes) of the metabolic pathways were obviously upregulated in 35 vs. 32 °C, and only a few DEGs (26 genes) were obviously downregulated in 35 vs. 32 °C (Supplemental Table S2).
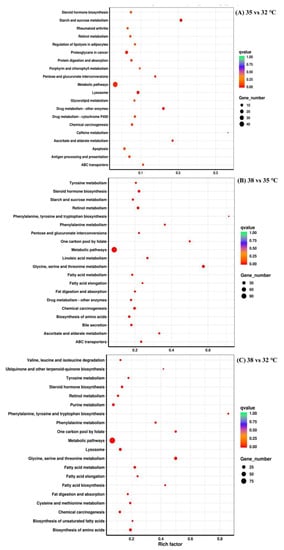
Figure 4.
KEGG enrichment analysis of adult P. quatuordecimpunctata under different temperature stress.
3.5. Real-Time Fluorescence Quantitative PCR Validation
Through functional enrichment analysis, a large number of DEGs related to heat resistance were found (Supplemental Tables S1 and S2). We selected twelve DEGs (eleven P450 and one Hsp70) of interest for qRT-PCR verification. The FPKM values of DEGs obtained by sequencing are shown in Table 4.

Table 4.
Statistics of FPKM values of DEGs at different temperatures of P. quatuordecimpunctata.
qRT-PCR was performed to verify the genes of the cytochrome P450 family and the heat shock protein Hsp70 family genes of P. quatuordecimpunctata. Twelve genes were selected: eleven P450 genes and one Hsp70 gene. First, we detected the primers and found that the amplification efficiency of all the primers was between 90% and 105% (Table 2). In addition, the dissolution curves were all single peaks, indicating that these primers could be used for qRT-PCR reaction. The results were consistent with those of transcriptome sequencing (RNA-Seq) (Table 4, Figure 5), indicating that the transcriptome sequencing results were reliable. After different high temperature stresses, the expression of genes such as cytochrome P450 and heat-shock protein Hsp70 were significantly upregulated (p < 0.001, Figure 5).
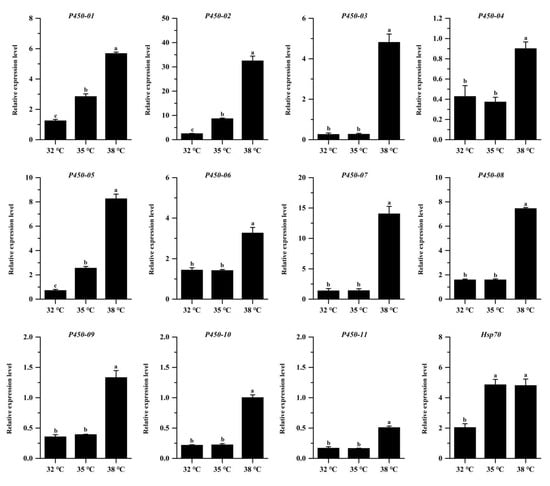
Figure 5.
Real-time PCR validation of adult P. quatuordecimpunctata DEGs under different temperature stress. For each genes, different letters indicate statistically significant differences (ANOVA; Tukey’s post hoc test; p < 0.05).
4. Discussion
Temperature is one of the important factors affecting insect growth and development, physiology, behavior, and geographical distribution [40,41,42]. By evaluating the effects of high temperature stress on P. quatuordecimpunctata, we were able to describe the adaptive capabilities of P. quatuordecimpunctata beetles to different high temperature environments [29]. Transcriptome sequencing is an important and effective method to explore the effects that heat stress may have in terms of tissue molecular composition, cellular transcriptional regulation, and functional genomic elements [43,44]. In this study, we found that the most DEGs (1425: 762 upregulated and 663 downregulated) were detected in the 38 vs. 35 °C group, and there were fewer overlapping genes between the 35 vs. 32 °C groups. Previous investigations have also shown that P. quatuordecimpunctata adults have different survival and reproduction at different temperatures (32, 35, and 38 °C) [29], indicating that the types of genes regulated in the comparison of the 38 vs. 35 °C groups was different from other groups and may be more complex. The differences in gene expression observed in this study between different temperature groups indicated that P. quatuordecimpunctata has different gene regulatory mechanisms for high temperatures (35 and 38 °C), which should be further explored in future studies.
In addition, we analyzed a large number of DEGs for GO and KEGG enrichment, and GO enrichment analysis showed that the first 30 pathways that showed significance enrichment in all temperature groups belonged to two categories, either genes for biological processes or those for molecular functions. In the medium–high temperature group (35 °C) vs. the 32 °C control, the DEGs were significantly enriched in the pathways for carbohydrate metabolic processes and catalytic activity, and trehalase was upregulated (Table 1). Trehalase hydrolyzes trehalose to provide energy for insects to maintain their life activities [45]. We speculated that this increase in trehalase gene expression level at 35 °C enhanced the energy supply process during high temperature stress. Comparing the highest temperature group (38 °C) to the moderate–high temperature (35 °C) group, even more metabolic pathways related to biological processes were enhanced. Specifically, there were a large number of DEGs that stimulated increases in the oxidation–reduction process and oxidoreductase activity pathway. Most of the DEGs involved in these pathways were oxidoreductases, and most of them were upregulated. Liu et al. [46] also found that some anti-oxidation-related genes in the crambid moth Glyphodes pyloalis (Walker) were significantly upregulated under heat stress (exposure to 40 °C for 4h compared with 25 °C). Meanwhile, previous studies also found that the activities of the anti-oxidative enzymes SOD and CAT in P. quatuordecimpunctata increased under high temperature stress [30], implying that these genes play important roles in the high temperature resistance process. “Monooxygenase activity” and “Oxidoreductase activity, acting on paired donors ...” were both upregulated in DEGs at 35 vs. 32 °C, while downregulated genes were found at 38 vs. 35 °C (Supplemental Table S1). Therefore, we inferred that the medium–high temperature (35 °C) may stimulate gene expression in this pathway, while the high temperature (38 °C) might inhibit expression.
High temperature stress affects the metabolic process of insects by affecting their enzymatic activity and enzymatic reactions. Many gene families are involved in the process of heat stress in insects, of which the cytochrome family is the most typical [47,48]. High temperature stress causes a large amount of reactive oxygen species (ROS) to be deposited in insect cells, resulting in the production of toxic substances [49]. As a typical detoxification enzyme system, the cytochrome P450 will assist the body to metabolize toxic substances. In our study, GO and KEGG enrichment analysis found that a large number of cytochrome P450 genes were enriched and significantly upregulated in all three temperature groups of P. quatuordecimpunctata, implying that these genes also play an important role in the process of thermotolerance of P. quatuordecimpunctata. The expression levels of the CYP450 genes (LtCYP4g1, LtCYP4g15, and LtCYP301A1) of Liriomyza trifolii (Burgess) were significantly higher than in the controls (25 °C) after 1 h treatment at different temperatures (35, 37.5, and 40 °C) [50]. Meanwhile, we also found that the expression level of the heat shock protein Hsp70 gene was significantly increased and continued to increase with the increase of temperature. Studies have shown that adverse conditions (e.g., high temperature) can increase the expression of heat shock protein genes in organisms or cells, protecting cells from damage or reducing damage [51]. A high expression level of the Hsp70 gene was also found in a transcriptome study of high temperature stress in the crambid moth Cnaphalocrocis medialis (Guenée) [52], the delphacid planthopper Sogatella furcifera (Horvath) [53], and the whitefly Bemisia tabaci (Gennadius) [54]. At the same time, RNA interference (RNAi) technology confirmed that a heat shock protein gene plays a key role in the high temperature tolerance process of the silkmoth Antheraea pernyi (Guérin-Méneville) [46], A. hygrophila [55], and the honeybee Apis cerana cerana (Fabricius) [56]. KEGG enrichment analysis also found that the DEGs related to the glucose metabolic pathway were continuously highly expressed at 35 vs. 32 °C and 38 vs. 35 °C, such as Glucose dehydrogenase and UDP-glucuronosyltransferase. However, the specific functions of these genes in the thermotolerance process of P. quatuordecimpunctata require further validation and confirmation.
Therefore, through the transcriptome sequencing analysis and DEG analysis of P. quatuordecimpunctata under different temperatures (32, 35, and 38 °C), we found that the cytochrome P450 family gene and the heat shock protein Hsp70 gene were both upregulated in the process of increasing the insect’s heat tolerance. These genes likely play an important role in the thermotolerance of P. quatuordecimpunctata, but their specific functions remain unclear. We will further verify the specific functions of these genes in the process of thermotolerance in P. quatuordecimpunctata through reliable methods, such as RNAi [57], gene knockout [58], and yeast two-hybrid [59].
5. Conclusions
In this study, we compared the transcriptome of P. quatuordecimpunctata under different high temperature (32, 35, and 38 °C) stress treatments using high-throughput sequencing, which increased our knowledge of the genomic resources of P. quatuordecimpunctata and provided an improved basis for further research. There were unique DEGs in both temperature groups (32 vs. 35 °C and 35 vs. 38 °C), indicating that the different genes were mobilized in P. quatuordecimpunctata under different high temperatures. In this study, through GO and KEGG enrichment analysis, we found that the DEGs were mainly involved in “Oxidation–reduction process”, “Oxidoreductase activity”, ”Metabolic pathways”, and “Small molecule metabolic processing”. Furthermore, the qRT-PCR results confirmed the authenticity of transcriptome sequencing, demonstrating that genes involved in these pathways play an important role in the high temperature tolerance of P. quatuordecimpunctata. This study filled in the gaps in the research on the molecular mechanism for thermotolerance of P. quatuordecimpunctata, and the key genes screened in this paper will be functionally verified in the further exploration of the thermotolerance molecular mechanism in P. quatuordecimpunctata.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/agriculture12081088/s1. Table S1: GO enrichment analysis (the DEGs number in per pathway) of adults of P. quatuordecimpunctata under different temperature stress; Table S2: KEGG enrichment analysis (the DEGs number in per pathway) of adults of P. quatuordecimpunctata under different temperature stress.
Author Contributions
Conceptualization, Y.L. and Y.Y.; data curation, Q.Y.; formal analysis, Q.Y. and J.L.; funding acquisition, Y.L.; investigation, Q.Y.; methodology, Y.L.; project administration, Y.L.; resources, Y.L.; software, Y.L.; supervision, Y.L. and Y.Y.; validation, Y.L.; visualization, Y.L.; writing—original draft, Q.Y. and J.L.; writing—review and editing, Y.L. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the National Natural Science Funds of China (No. U2003212) and the China Agriculture Research System (CARS-15-21).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the first author at yangqingzzr@126.com.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Michel, D.K.; Fiaboe, K.K.M.; Kekeunou, S.; Nanga, S.N.; Kuate, A.F.; Tonnang, H.E.Z.; Gnanvossou, D.; Hanna, R. Temperature-based phenology model to predict the development, survival, and reproduction of the Oriental fruit fly Bactrocera dorsalis. J. Therm. Biol. 2021, 97, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Keena, M.A.; Moore, P.M.; Bradford, G. Effects of temperature on Anoplophora chinensis (Coleoptera: Cerambycidae) adult survival, reproduction, and egg hatch. Forests 2021, 12, 432. [Google Scholar] [CrossRef]
- Luis, A.; Xavier, P. Effect of high temperature on the growth and reproduction of corn aphids (Homoptera: Aphididae) and implications for their population dynamics on the Northeastern Iberian Peninsula. Environ. Entomol. 2001, 6, 1127–1134. [Google Scholar]
- Liu, X.D.; Zhang, A.M. High temperature determines the ups and downs of small brown planthopper Laodelphax striatellus population. Insect Sci. 2013, 20, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, C.M.D.; Silvatorres, C.S.A.D.; Torres, J.B.; Silva, G.D.S. Estimation of population growth for two species of lady beetles (Coleoptera: Coccinellidae) under different temperatures. Biocontrol Sci. Technol. 2022, 32, 74–89. [Google Scholar] [CrossRef]
- Vanhanen, H.; Veleli, T.O.; Paivinen, S.; Kellomaki, S.; Niemela, P. Climate change and range shifts in two insect defoliators: Gypsy moth and moth-a model study. Silva Fenni. 2007, 41, 621–638. [Google Scholar] [CrossRef] [Green Version]
- Jiang, F.Z.; Zheng, L.Y.; Guo, J.X.; Zhang, G.R. Effects of temperature stress on insect fertility and its physiological and biochemical mechanisms. J. Environ. Entomol. 2015, 37, 653–663. [Google Scholar]
- Zhang, Q.L.; Yuan, M.L. Progress in insect transcriptomics based on the next-generation sequencing technique. Acta Entomol. Sin. 2013, 56, 1489–1508. [Google Scholar]
- Liu, Y.C.; Su, H.; Li, R.Q.; Li, X.T.; Xu, Y.S.; Dai, X.P.; Zhou, Y.Y.; Wang, H.B. Comparative transcriptome analysis of Glyphodes pyloalis Walker (Lepidoptera: Pyralidae) reveals novel insights into heat stress tolerance in insects. BMC Genom. 2017, 18, 974. [Google Scholar] [CrossRef]
- Neven, L.G. Physiological responses of insects to heat. Postharvest Biol. Technol. 2000, 21, 103–111. [Google Scholar] [CrossRef]
- Ma, C.S.; Ma, G.; Pincebourde, S. Survive a warming climate: Insect Responses to Extreme High Temperatures. Annu. Rev. Entomol. 2021, 66, 163–184. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.H.; Cai, T.W.; Ren, Z.J.; Liu, Y.; Yuan, M.J.; Cai, Y.F.; Yu, C.; Shu, R.H.; He, S.; Li, J.H.; et al. Decline in symbiont-dependent host detoxification metabolism contributes to increased insecticide susceptibility of insects under high temperature. ISME J. 2021, 15, 3693–3703. [Google Scholar] [CrossRef] [PubMed]
- Du, R.; Ma, C.S.; Zhao, Q.H.; Ma, G.; Yang, H.P. Effects of heat stress on physiological and biochemical mechanisms of insects: A literature review. Acta Ecol. Sin. 2007, 4, 1565–1572. [Google Scholar]
- Mutz, K.O.; Heilkenbrinker, A.; Lönne, M.; Walter, J.G.; Stahl, F. Transcriptome analysis using next-generation sequencing. Curr. Opin. Biotechnol. 2013, 24, 22–30. [Google Scholar] [CrossRef]
- Guo, H.Z.; Huang, C.L.; Jiang, L.; Cheng, T.C.; Feng, T.S.; Xia, Q.Y. Transcriptome analysis of the response of silkworm to drastic changes in ambient temperature. Appl. Microbiol. Biot. 2018, 102, 10161–10170. [Google Scholar] [CrossRef] [PubMed]
- Ran, L.Y.; Jiang, C.X.; Yang, Q.F.; Wang, H.J.; Bai Ma, T.Z.; Chen, L.; Kuang, J.K.; Huang, T.T.; Li, Q. Comparative transcriptome analysis of Locusta migratoria tibetensis Chen (Orthoptera: Oedipodidae) under high- and low-temperature stress. J. Appl. Entomol. 2020, 144, 181–190. [Google Scholar] [CrossRef]
- Shen, H.Y.; He, H.; Lu, C.D.; Liang, Y.; Wu, H.M.; Zheng, L.Z.; Wang, X.Y.; Liang, G.H. Comparative transcriptome analysis of two populations of Dastarcus helophoroides (Fairmaire) under high temperature stress. Forests 2022, 13, 13. [Google Scholar] [CrossRef]
- Yuan, J.W.; Qin, J.; Wei, R.; Xie, H.F.; Du, Y.Z. Transcriptome analysis of responses of western flower thrips Frankliniella occidentalis to high and low temperature stresses. J. Plant Protec. 2021, 48, 1400–1410. [Google Scholar]
- Vatanparast, M.; Park, Y. Differential transcriptome analysis reveals genes related to low- and high-temperature stress in the Fall Armyworm, Spodoptera frugiperda. Front. Physiol. 2022, 12, 827077. [Google Scholar] [CrossRef]
- Wyckhuys, K.A.G.; Lu, Y.H.; Morales, H.; Vazquez, L.L.; Legaspi, J.C.; Eliopoulos, P.A.; Hernandez, L.M. Current status and potential of conservation biological control for agriculture in the developing world. Biol. Control 2013, 65, 152–167. [Google Scholar] [CrossRef]
- Wyckhuys, K.A.G.; Lu, Y.H.; Zhou, W.W.; Cock, M.J.W.; Naranjo, S.E.; Fereti, A.; Williams, F.E.; Furlong, M.J. Ecological pest control fortifies agricultural growth in Asia-Pacific economies. Nat. Ecol. Evol. 2020, 4, 1522–1530. [Google Scholar] [CrossRef] [PubMed]
- Obrycki, J.J.; Orr, D.B.; Orr, C.J.; Wallendorf, M.; Flanders, R.V. Comparative developmental and reproductive biology of three populations of Propylea quatuordecimpunctata (Coleoptera: Coccinellidae). Biol. Control 1993, 3, 27–33. [Google Scholar] [CrossRef]
- Kalushkov, P.; Hodek, I. The effect of six species of aphids on some life history parameters of the ladybird Propylea quatuordecimpunctata (Coleoptera: Coccinellidae). Eur. J. Entomol. 2005, 102, 449–452. [Google Scholar] [CrossRef] [Green Version]
- Papanikolaou, N.E.; Martinou, A.F.; Kontodimas, D.C.; Matsinos, Y.G.; Milonas, P.G. Functional responses of immature stages of Propylea quatuordecimpunctata (Coleoptera: Coccinellidae) to Aphis fabae (Hemiptera: Aphididae). Eur. J. Entomol. 2011, 108, 391–395. [Google Scholar] [CrossRef] [Green Version]
- Kontodimas, D.C.; Milonas, P.G.; Stathas, G.J.; Papanikolaou, N.E.; Skourti, A.; Matsinos, Y.G. Life table parameters of the aphid predators Coccinella septempunctata, Ceratomegilla undecimnotata and Propylea quatuordecimpunctata (Coleoptera: Coccinellidae). Eur. J. Entomol. 2008, 105, 427–430. [Google Scholar] [CrossRef] [Green Version]
- Pervez, A.; Omkar. Ecology of aphidophagous ladybird Propylea species: A review. J. Asia-Pac. Entomol. 2011, 14, 357–365. [Google Scholar] [CrossRef]
- Lu, Y.H.; Wu, K.M.; Jiang, Y.Y.; Guo, Y.Y.; Desneux, N. Widespread adoption of Bt cotton and insecticide decrease promotes biocontrol services. Nature 2012, 487, 362–365. [Google Scholar] [CrossRef]
- Papanikolaou, N.E.; Milonas, P.G.; Kontodimas, D.C.; Demiris, N.; Matsinos, Y.G. Temperature-dependent development, survival, longevity, and fecundity of Propylea quatuordecimpunctata (Coleoptera: Coccinellidae). Ann. Entomol. Soc. Am. 2013, 106, 228–234. [Google Scholar] [CrossRef] [Green Version]
- Yang, Q.; Liu, J.P.; Wyckhuys, K.A.G.; Yang, Y.Z.; Lu, Y.H. Impact of heat stress on the predatory ladybugs Hippodamia variegata and Propylaea quatuordecimpunctata. Insects 2022, 13, 306. [Google Scholar] [CrossRef]
- Ren, S.X.; Wang, X.M.; Pang, H.; Peng, Z.Q.; Zeng, T. Colored Pictorial Handbook of Ladybird Beetles in China; Science Press: Beijing, China, 2009. [Google Scholar]
- Li, G.Y. Cotton Diseases and Pests in Xinjiang, 1st ed.; China Agricultural Press: Beijing, China, 2017; Volume 4, p. 168. [Google Scholar]
- Gambino, G.; Perrone, I.; Gribaudo, I. A rapid and effective method for RNA extraction from different tissues of grapevine and other woody plants. Phytochem. Anal. 2008, 19, 520–525. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pertea, G.; Huang, X.Q.; Liang, F.; Antonescu, V.; Sultana, R.; Karamycheva, S.; Lee, Y.; White, J.; Cheung, F.; Parvizi, B.; et al. TIGR Gene Indices clustering tools (TGICL): A software system for fast clustering of large EST datasets. Bioinformatics 2003, 19, 651–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.X.; Liu, T.H.; Khashaveh, A.; Yi, C.Q.; Liu, X.X.; Zhang, Y.J. Identification and evaluation of suitable reference genes for RT-qPCR analysis in Hippodamia variegata (Coleoptera: Coccinellidae) under different biotic and abiotic conditions. Front. Physiol. 2021, 12, 669510. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101. [Google Scholar] [CrossRef]
- Abram, P.K.; Boivin, G.; Moiroux, J.; Brodeur, J. Behavioural effects of temperature on ectothermic animals: Unifying thermal physiology and behavioural plasticity. Biol. Rev. 2017, 92, 1859–1876. [Google Scholar] [CrossRef]
- Hensen, M.C.; Hernandez, M.I.M.; da Silva, P.G.; Amore, V.; Lobo, J.M. Distribution of Canthon rutilans rutilans and Canthon rutilans cyanescens along spatio-temporal and temperature gradients. Insects 2018, 9, 124. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Huang, L.F.; Wang, W.L.; Chen, E.H.; Chen, H.S.; Jiang, J.J. Effects of temperature on growth and development of the brown planthopper, Nilaparvata lugens (Homoptera: Delphacidae). Environ. Entomol. 2021, 50, 1–11. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Quan, N.; Palfreyman, R.W.; Chan, L.C.L.; Steven, R.; Nielsen, L.K. Transcriptome sequencing of and microarray development for a Helicoverpa zea cell line to investigate in vitro insect cell–baculovirus interactions. PLoS ONE 2012, 7, e36324. [Google Scholar]
- Qin, J.M.; Luo, S.D.; He, S.Y.; Wu, J. Researching in characters and functions of trehalose and trehalase in insects. J. Environ. Entomol. 2015, 37, 163–169. [Google Scholar]
- Liu, Q.N.; Zhu, B.J.; Dai, L.S.; Fu, W.W.; Lin, K.Z.; Liu, C.L. Overexpression of small heat shock protein 21 protects the Chinese oak silkworm Antheraea pernyi against thermal stress. J. Insect Physiol. 2013, 59, 848–854. [Google Scholar] [CrossRef]
- Bühler, A.; Lanzrein, B.; Wille, H. Influence of temperature and carbon dioxide concentration on juvenile hormone titre and dependent parameters of adult worker honey bees (Apis mellifera L.). J. Insect Physiol. 1983, 29, 885–893. [Google Scholar] [CrossRef]
- Claudianos, C.; Ranson, H.; Johnson, R.M.; Biswas, S.; Schuler, M.A.; Berenbaum, M.R.; Feyereisen, R.; Oakeshott, J.G. A deficit of detoxification enzymes: Pesticide sensitivity and environmental response in the honeybee. Insect Mol. Biol. 2006, 15, 615–636. [Google Scholar] [CrossRef] [Green Version]
- Green, D.R.; Reed, J.C. Mitochondria and apoptosis. Science 1998, 281, 1309–1312. [Google Scholar] [CrossRef]
- Wang, Y.C.; Chang, Y.W.; Bai, J.; Zhang, X.X.; Iqbal, J.; Lu, M.X.; Hu, J.; Du, Y.Z. High temperature stress induces expression of CYP450 genes and contributes to insecticide tolerance in Liriomyza trifolii. Pestic. Biochem. Phys. 2021, 174, 104826. [Google Scholar] [CrossRef] [PubMed]
- Carranco, R.; Almoguera, C.; Jordano, J. A plant small heat shock protein gene expressed during zygotic embryogenesis but noninducible by heat stress. J. Biol. Chem. 1997, 272, 27470–27475. [Google Scholar] [CrossRef] [Green Version]
- Gu, L.L.; Li, M.Z.; Wang, G.R.; Liu, X.D. Multigenerational heat acclimation increases thermal tolerance and expression levels of Hsp70 and Hsp90 in the rice leaf folder larvae. J. Therm. Biol. 2019, 81, 103–109. [Google Scholar] [CrossRef]
- Zhou, C.; Yang, X.B.; Yang, H.; Long, G.Y.; Wang, Z.; Jin, D.C. Effects of abiotic stress on the expression of Hsp70 genes in Sogatella furcifera (Horvath). Cell Stress Chaperones 2020, 25, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Liu, X.N.; Lu, M.X.; Du, Y.Z. Transcriptional profiling of MED Bemisia tabaci exposed to thermal stress and verification of HSP70 expression. Entomol. Res. 2021, 51, 251–262. [Google Scholar] [CrossRef]
- Jin, J.S.; Zhao, M.; Wang, Y.; Zhou, Z.S. Induced thermotolerance and expression of three key Hsp genes (Hsp70, Hsp21, and sHsp21) and their roles in the high temperature tolerance of Agasicles hygrophila. Front. Physiol. 2020, 10, 1593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.Y.; Liu, Y.L.; Guo, X.L.; Li, Y.L.; Gao, H.R.; Guo, X.Q.; Xu, B.H. sHsp22.6, an intronless small heat shock protein gene, is involved in stress defense and development in Apis cerana cerana. Insect Biochem. Mol. 2014, 53, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.Y.; Palli, S.R. Mechanisms, applications, and challenges of insect RNA interference. Annu. Rev. Entomol. 2020, 65, 293–311. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.X.; Li, Y.; Zheng, X.F.; Guo, H.W. Modern Molecular Biology, 5th ed.; Higher Education Press: Beijing, China, 2019. [Google Scholar]
- Nelson, D.L.; Cox, M.M. Lehninger Principles of Biochemistry, 7th ed.; W. H. Freeman and Company: New York, NY, USA, 2017. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).