Genome-Wide Identification and Expression Analysis of WRKY Genes during Anthocyanin Biosynthesis in the Mango (Mangifera indica L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification and Annotation of Mango WRKY Genes
2.2. Phylogenetic Tree Construction and Sequence Analysis
2.3. Chromosomal Distribution and Syntenic Analysis
2.4. Promoter Cis-Acting Element Analysis
2.5. Plant Materials and Treatments
2.6. Acquisition and Analysis of RNA-Seq Data
2.7. Measurement of Anthocyanin Content of Mango Peel
2.8. RNA Extraction and Gene Expression Analysis
2.9. Statistical Analysis
3. Results
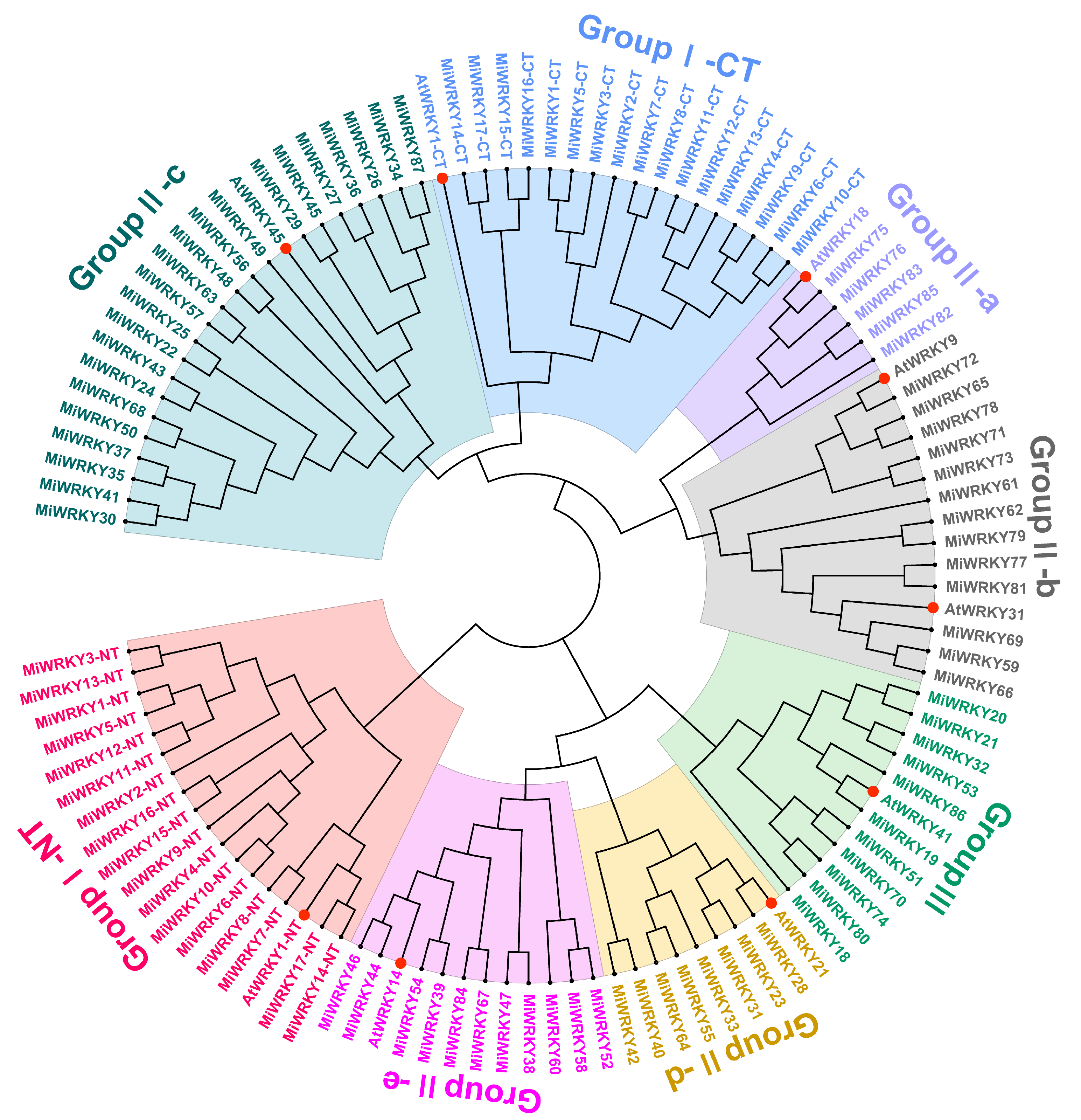
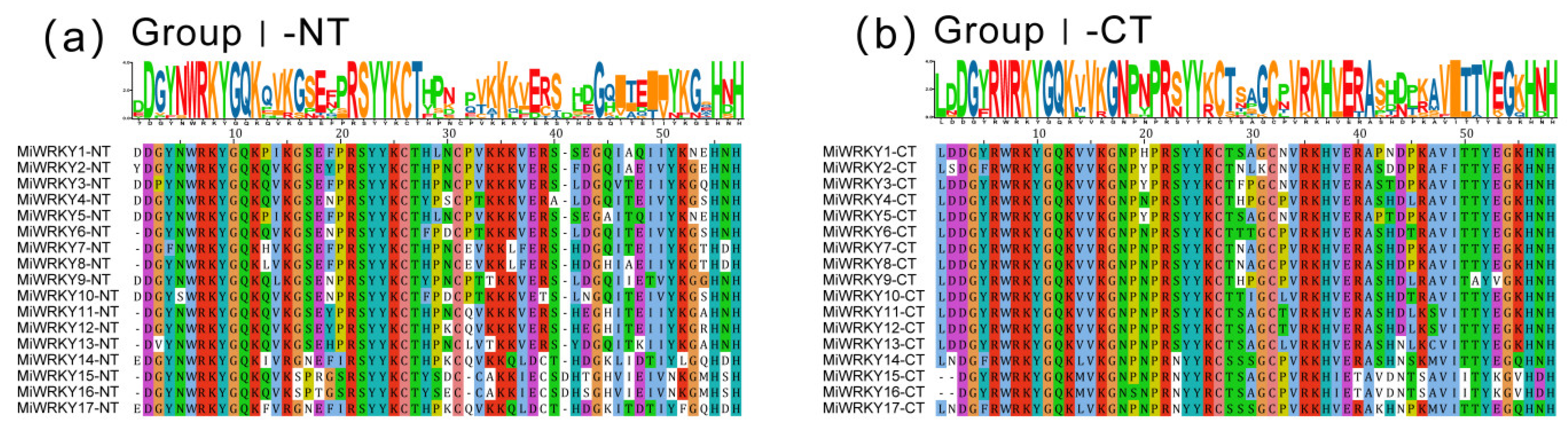
3.1. Identification, Multiple Sequence Alignment and Phylogenetic Analysis of MiWRKY Proteins
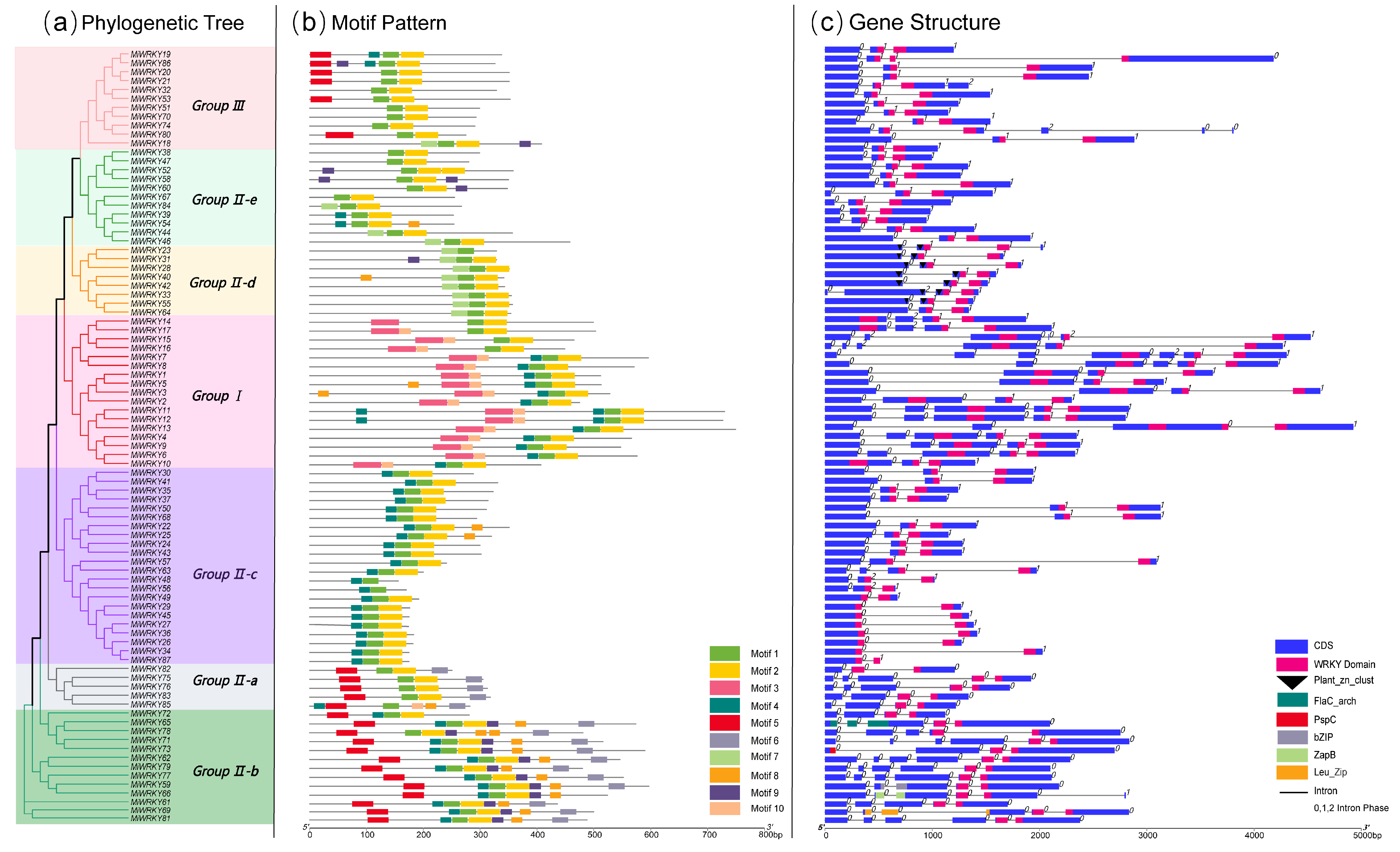
3.2. Gene Structure and Conserved Motif Analysis of MiWRKY Proteins
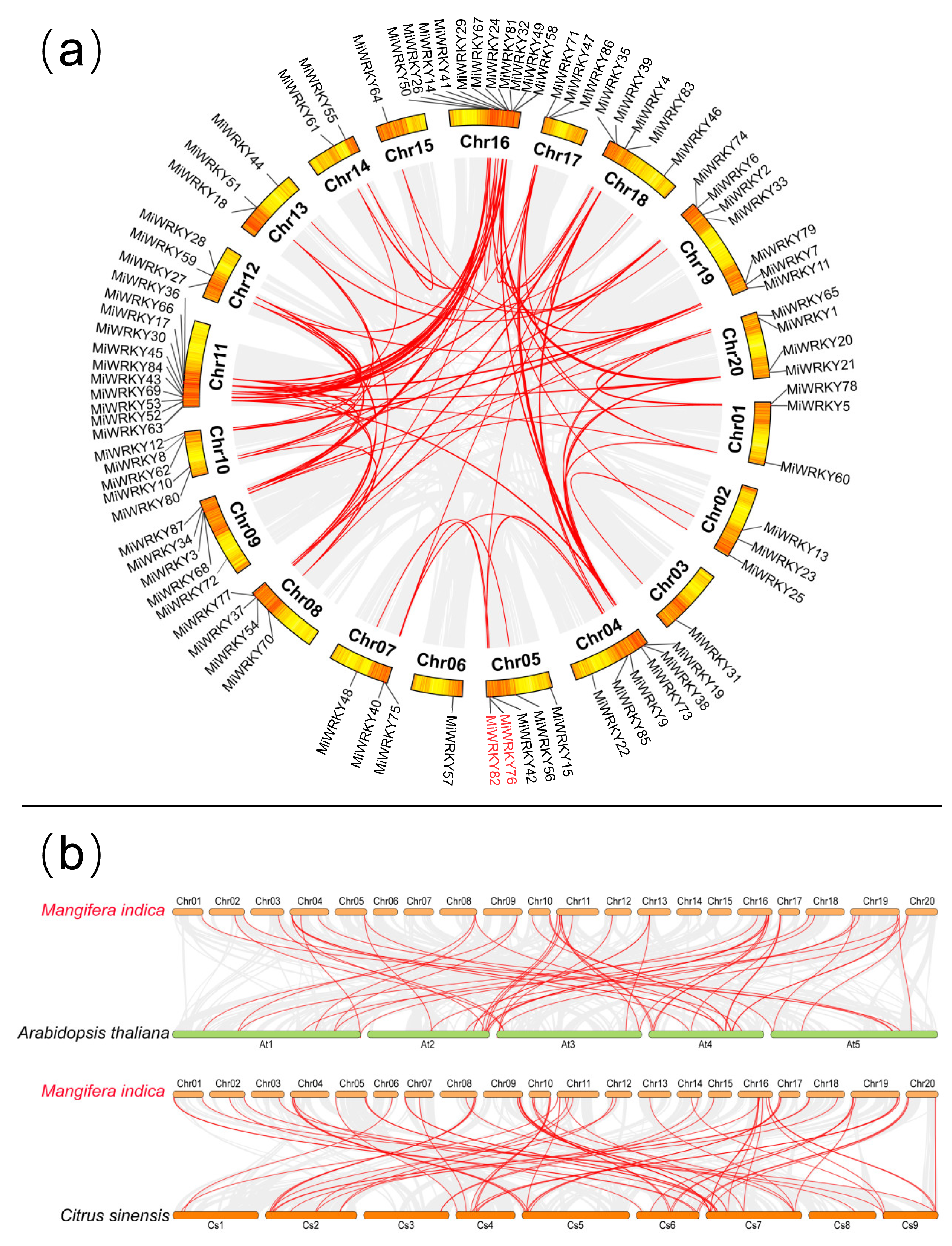
3.3. Chromosomal Distribution and Syntenic Analysis of MiWRKY Genes
3.4. Cis-Acting Element Analysis of MiWRKY Genes
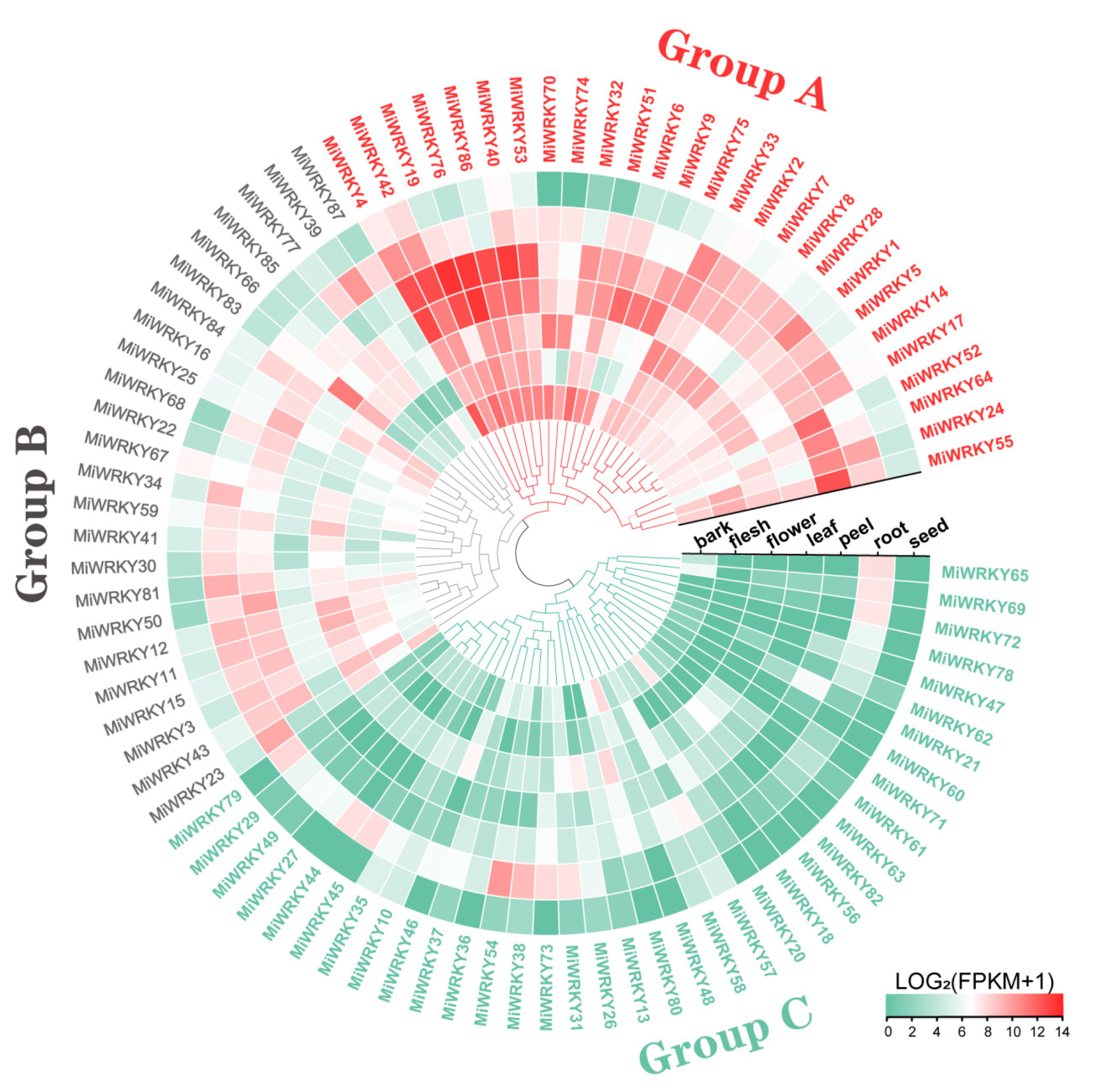
3.5. Tissue Specific Expression Profiling of MiWRKY Genes
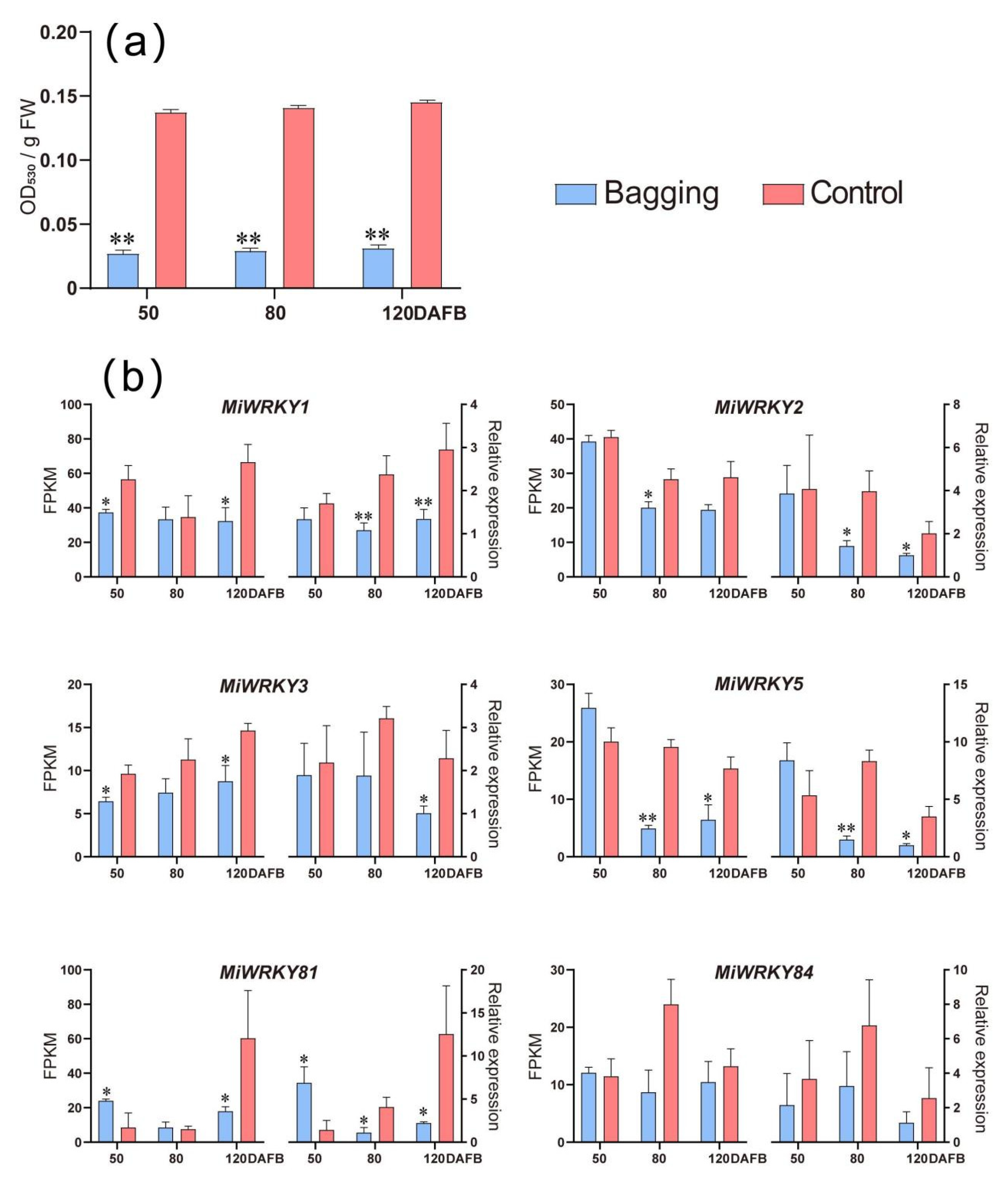
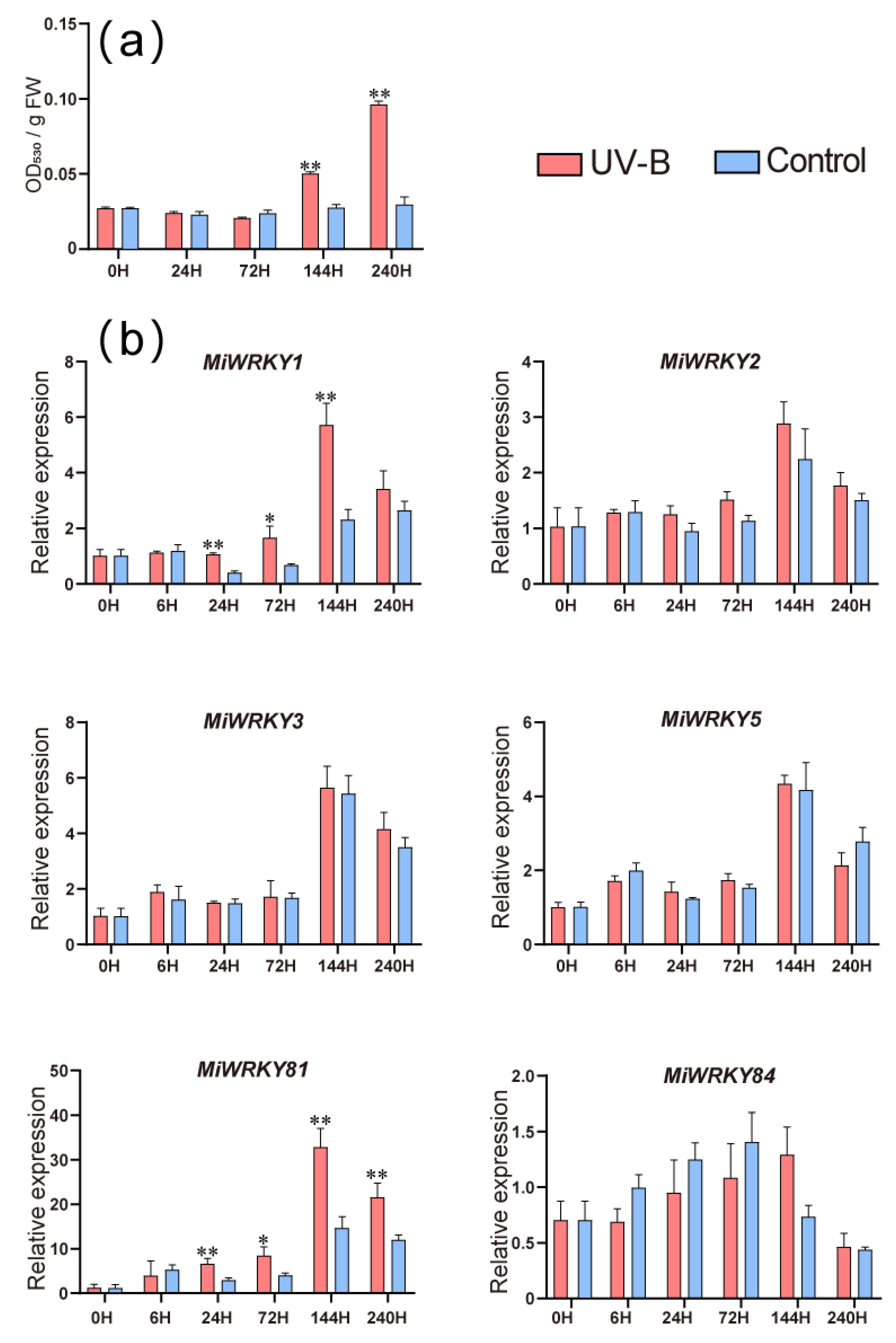
3.6. Expression Patterns of MiWRKY Genes during Anthocyanin Accumulation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Wu, K.L.; Guo, Z.J.; Wang, H.H.; Li, J. The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res. 2005, 12, 9–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, T.; Chen, C.; Li, C.; Liu, J.; Liu, C.; He, Y. Genome-wide investigation of WRKY gene family in pineapple: Evolution and expression profiles during development and stress. BMC Genomics 2018, 19, 490. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Vannozzi, A.; Wang, G.; Liang, Y.H.; Tornielli, G.B.; Zenoni, S.; Cavallini, E.; Pezzotti, M.; Cheng, Z.M. Genome and transcriptome analysis of the grapevine (Vitis vinifera L.) WRKY gene family. Hort. Res. 2014, 1, 16. [Google Scholar] [CrossRef] [Green Version]
- Chi, Y.; Yang, Y.; Zhou, Y.; Zhou, J.; Fan, B.; Yu, J.Q.; Chen, Z. Protein–Protein interactions in the regulation of WRKY transcription factors. Mol. Plant 2013, 6, 287–300. [Google Scholar] [CrossRef] [Green Version]
- Ishiguro, S.; Nakamura, K. Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and β-amylase from sweet potato. Mol. Gen. Genet. 1994, 244, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Ross, C.A.; Liu, Y.; Shen, Q.J. The WRKY gene family in rice (Oryza sativa). J. Integr. Plant Biol. 2007, 49, 827–842. [Google Scholar] [CrossRef]
- Cai, C.; Niu, E.; Du, H.; Zhao, L.; Feng, Y.; Guo, W. Genome-wide analysis of the WRKY transcription factor gene family in Gossypium raimondii and the expression of orthologs in cultivated tetraploid cotton. Crop J. 2014, 2, 87–101. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Li, K.; Xu, X.; Yao, Z.; Jin, C.; Zhang, S. Genome-wide analysis of WRKY transcription factors in white pear (Pyrus bretschneideri) reveals evolution and patterns under drought stress. BMC Genomics 2015, 16, 1104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, D.; Li, Y.; Bai, Y.; Li, M.; Cheng, L. Genome-wide identification and characterization of WRKY transcriptional factor family in apple and analysis of their responses to waterlogging and drought stress. Plant Physiol. Biochem. 2016, 103, 71–83. [Google Scholar] [CrossRef]
- Zhang, C.Q.; Xu, Y.; Lu, Y.; Yu, H.X.; Gu, M.H.; Liu, Q.Q. The WRKY transcription factor OsWRKY78 regulates stem elongation and seed development in rice. Planta 2011, 234, 541–554. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Hu, R.; Wang, H.; Cao, Y.; He, G.; Fu, C.; Zhou, G. MiWRKY12, a novel Miscanthus transcription factor, participates in pith secondary cell wall formation and promotes flowering. Plant Sci. 2013, 212, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Robatzek, S.; Somssich, I.E. A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence- and defence-related processes. Plant J. 2001, 28, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Ramamoorthy, R.; Jiang, S.Y.; Kumar, N.; Venkatesh, P.N.; Ramachandran, S. A comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatments. Plant Cell Physiol. 2008, 49, 865–879. [Google Scholar] [CrossRef]
- Chen, L.; Song, Y.; Li, S.; Zhang, L.; Zou, C.; Yu, D. The role of WRKY transcription factors in plant abiotic stresses. BBA Gene Regul. Mech. 2012, 1819, 120–128. [Google Scholar] [CrossRef]
- Banerjee, A.; Roychoudhury, A. WRKY Proteins: Signaling and regulation of expression during abiotic stress responses. Sci. World J. 2015, 2015, 807560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petroni, K.; Tonelli, C. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 2011, 181, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Holton, T.A.; Cornish, E.C. Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 1995, 7, 1071–1083. [Google Scholar] [CrossRef]
- Allan, A.C.; Hellens, R.P.; Laing, W.A. MYB transcription factors that colour our fruit. Trends Plant Sci. 2008, 13, 99–102. [Google Scholar] [CrossRef]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB–bHLH–WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef]
- Jaakola, L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef] [Green Version]
- An, J.P.; Qu, F.J.; Yao, J.F.; Wang, X.N.; You, C.X.; Wang, X.F.; Hao, Y.J. The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Hort. Res. 2017, 4, 17023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, H.; Wang, K.L.; Wang, H.; Gu, C.; Dare, A.P.; Espley, R.V.; He, H.; Allan, A.C.; Han, Y. Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J. 2015, 82, 105–121. [Google Scholar] [CrossRef]
- Gou, J.Y.; Felippes, F.F.; Liu, C.J.; Weigel, D.; Wang, J.W. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 2011, 23, 1512–1522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, M.; Ni, J.; Niu, Q.; Bai, S.; Bao, L.; Li, J.; Sun, Y.; Zhang, D.; Teng, Y. Response of miR156-SPL Module during the red peel coloration of bagging-treated Chinese sand pear (Pyrus pyrifolia Nakai). Front. Physiol. 2017, 8, 550. [Google Scholar] [CrossRef] [Green Version]
- Ni, J.; Bai, S.; Zhao, Y.; Qian, M.; Tao, R.; Yin, L.; Gao, L.; Teng, Y. Ethylene response factors Pp4ERF24 and Pp12ERF96 regulate blue light-induced anthocyanin biosynthesis in ‘Red Zaosu’ pear fruits by interacting with MYB114. Plant Mol. Biol. 2019, 99, 67–78. [Google Scholar] [CrossRef]
- Ni, J.; Premathilake, A.T.; Gao, Y.; Yu, W.; Tao, R.; Teng, Y.; Bai, S. Ethylene-activated PpERF105 induces the expression of the repressor-type R2R3-MYB gene PpMYB140 to inhibit anthocyanin biosynthesis in red pear fruit. Plant J. 2021, 105, 167–181. [Google Scholar] [CrossRef]
- Bai, S.; Tao, R.; Yin, L.; Ni, J.; Yang, Q.; Yan, X.; Yang, F.; Guo, X.; Li, H.; Teng, Y. Two B-box proteins, PpBBX18 and PpBBX21, antagonistically regulate anthocyanin biosynthesis via competitive association with Pyrus pyrifolia ELONGATED HYPOCOTYL 5 in the peel of pear fruit. Plant J. 2019, 100, 1028–1223. [Google Scholar] [CrossRef]
- Bai, S.; Tao, R.; Tang, Y.; Yin, L.; Ma, Y.; Ni, J.; Yan, X.; Yang, Q.; Wu, Z.; Zeng, Y. BBX16, a B-box protein, positively regulates light-induced anthocyanin accumulation by activating MYB10 in red pear. Plant Biotechnol. J. 2019, 17, 1985–1997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaakola, L.; Poole, M.; Jones, M.O.; Kämäräinen-Karppinen, T.; Koskimäki, J.J.; Hohtola, A.; Häggman, H.; Fraser, P.D.; Manning, K.; King, G.J. A SQUAMOSA MADS box gene involved in the regulation of anthocyanin accumulation in bilberry fruits. Plant Physiol. 2010, 153, 1619–1629. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Wu, J.; Hu, K.D.; Wei, S.W.; Sun, H.Y.; Hu, L.Y.; Han, Z.; Yao, G.F.; Zhang, H. PyWRKY26 and PybHLH3 cotargeted the PyMYB114 promoter to regulate anthocyanin biosynthesis and transport in red-skinned pears. Hort. Res. 2020, 7, 37. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Xu, H.; Yang, G.; Wang, N.; Zhang, J.; Wang, Y.; Jiang, S.; Fang, H.; Zhang, Z.; Chen, X. Molecular mechanism of MYB111 and WRKY40 involved in anthocyanin biosynthesis in red-fleshed apple callus. Plant Cell Tiss. Org. 2019, 139, 467–478. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, Z.; Zhao, Y.; Guo, D.; Zhao, X.; Gao, W.; Zhang, J.; Song, B. StWRKY13 promotes anthocyanin biosynthesis in potato (Solanum tuberosum) tubers. Funct. Plant Biol. 2022, 49, 102–114. [Google Scholar] [CrossRef]
- Mao, Z.; Jiang, H.; Wang, S.; Wang, Y.; Yu, L.; Zou, Q.; Liu, W.; Jiang, S.; Wang, N.; Zhang, Z. The MdHY5-MdWRKY41-MdMYB transcription factor cascade regulates the anthocyanin and proanthocyanidin biosynthesis in red-fleshed apple. Plant Sci. 2021, 306, 110848. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Yang, T.; Li, Y.; Zhang, J.; Wu, T.; Song, T.; Yao, Y.; Tian, J. The long noncoding RNA MdLNC499 bridges MdWRKY1 and MdERF109 function to regulate early-stage light-induced anthocyanin accumulation in apple fruit. Plant Cell 2021, 33, 3309–3330. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Fang, H.; Wang, J.; Yue, X.; Su, M.; Mao, Z.; Zou, Q.; Jiang, H.; Guo, Z.; Yu, L.; et al. Ultraviolet B-induced MdWRKY72 expression promotes anthocyanin synthesis in apple. Plant Sci. 2020, 292, 110377. [Google Scholar] [CrossRef]
- An, J.P.; Zhang, X.W.; You, C.X.; Bi, S.Q.; Wang, X.F.; Hao, Y.J. MdWRKY40 promotes wounding-induced anthocyanin biosynthesis in association with MdMYB1 and undergoes MdBT2-mediated degradation. New Phytol. 2019, 224, 380–395. [Google Scholar] [CrossRef]
- Kour, R.; Singh, M.; Gill, P.P.S.; Jawandha, S.K. Ripening quality of Dusehri mango in relation to harvest time. J. Food Sci. Technol. 2018, 55, 2395–2400. [Google Scholar] [CrossRef]
- Shi, B.; Wu, H.; Zheng, B.; Qian, M.; Gao, A.; Zhou, K. Analysis of light-independent anthocyanin accumulation in mango (Mangifera indica L.). Horticulturae 2021, 7, 423. [Google Scholar] [CrossRef]
- Kanzaki, S.; Ichihi, A.; Tanaka, Y.; Fujishige, S.; Koeda, S.; Shimizu, K. The R2R3-MYB transcription factor MiMYB1 regulates light dependent red coloration of ‘Irwin’ mango fruit skin. Sci. Hortic. 2020, 272, 109567. [Google Scholar] [CrossRef]
- Kanzaki, S.; Kamikawa, S.; Ichihi, A.; Tanaka, Y.; Shimizu, K.; Koeda, S. Isolation of UDP: Flavonoid 3-O-glycosyltransferase (UFGT)-like genes and expression analysis of genes associated with anthocyanin accumulation in mango ‘Irwin’ skin. Hort. J. 2019, 88, 435–443. [Google Scholar] [CrossRef] [Green Version]
- Karanjalker, G.R.; Ravishankar, K.V.; Shivashankara, K.S.; Dinesh, M.R.; Roy, T.K.; Sudhakar Rao, D.V. A study on the expression of genes involved in carotenoids and anthocyanins during ripening in fruit peel of green, yellow, and red colored mango cultivars. Appl. Biochem. Biotechnol. 2018, 184, 140–154. [Google Scholar] [CrossRef]
- Karanjalker, G.R.; Ravishankar, K.V.; Shivashankara, K.S.; Dinesh, M.R. Influence of bagging on color, anthocyanin and anthocyanin biosynthetic genes in peel of red colored mango Cv. ‘Lily’. Erwerbs Obstbau. 2018, 60, 281–287. [Google Scholar] [CrossRef]
- Wang, P.; Luo, Y.; Huang, J.; Gao, S.; Zhu, G.; Dang, Z.; Gai, J.; Yang, M.; Zhu, M.; Zhang, H.; et al. The genome evolution and domestication of tropical fruit mango. Genome Biol. 2020, 21, 60. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [Green Version]
- Holub, E.B. The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2001, 2, 516–527. [Google Scholar] [CrossRef] [PubMed]
- Ling, J.; Jiang, W.; Zhang, Y.; Yu, H.; Mao, Z.; Gu, X.; Huang, S.; Xie, B. Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC Genomics 2011, 12, 471. [Google Scholar] [CrossRef] [Green Version]
- Wei, K.F.; Chen, J.; Chen, Y.F.; Wu, L.J.; Xie, D.X. Molecular phylogenetic and expression analysis of the complete WRKY transcription factor family in maize. DNA Res. 2012, 19, 153–164. [Google Scholar] [CrossRef] [Green Version]
- Wei, Y.; Shi, H.; Xia, Z.; Tie, W.; Ding, Z.; Yan, Y.; Wang, W.; Hu, W.; Li, K. Genome-wide identification and expression analysis of the WRKY gene family in cassava. Front. Plant Sci. 2016, 7, 25. [Google Scholar] [CrossRef] [Green Version]
- Xie, Z.; Zhang, Z.L.; Zou, X.; Huang, J.; Ruas, P.; Thompson, D.; Shen, Q.J. Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol. 2005, 137, 176–189. [Google Scholar] [CrossRef] [Green Version]
- van Verk, M.C.; Pappaioannou, D.; Neeleman, L.; Bol, J.F.; Linthorst, H.J.M. A novel WRKY transcription factor is required for induction of PR-1a gene expression by salicylic acid and bacterial elicitors. Plant Physiol. 2008, 146, 1983–1995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Q.Y.; Tian, A.G.; Zou, H.F.; Xie, Z.M.; Lei, G.; Huang, J.; Wang, C.M.; Wang, H.W.; Zhang, J.S.; Chen, S.Y. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 2008, 6, 486–503. [Google Scholar] [CrossRef]
- Pandey, B.; Grover, A.; Sharma, P. Molecular dynamics simulations revealed structural differences among WRKY domain-DNA interaction in barley (Hordeum vulgare). BMC Genomics 2018, 19, 132. [Google Scholar] [CrossRef] [Green Version]
- Mackay, J.P.; Crossley, M. Zinc fingers are sticking together. Trends Biochem. Sci. 1998, 23, 1–4. [Google Scholar] [CrossRef]
- Sun, C.; Palmqvist, S.; Olsson, H.; Borén, M.; Ahlandsberg, S.; Jansson, C. A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. Plant Cell 2003, 15, 2076–2092. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, B.; Wu, H.; Zhu, W.; Zheng, B.; Wang, S.; Zhou, K.; Qian, M. Genome-Wide Identification and Expression Analysis of WRKY Genes during Anthocyanin Biosynthesis in the Mango (Mangifera indica L.). Agriculture 2022, 12, 821. https://doi.org/10.3390/agriculture12060821
Shi B, Wu H, Zhu W, Zheng B, Wang S, Zhou K, Qian M. Genome-Wide Identification and Expression Analysis of WRKY Genes during Anthocyanin Biosynthesis in the Mango (Mangifera indica L.). Agriculture. 2022; 12(6):821. https://doi.org/10.3390/agriculture12060821
Chicago/Turabian StyleShi, Bin, Hongxia Wu, Wencan Zhu, Bin Zheng, Songbiao Wang, Kaibing Zhou, and Minjie Qian. 2022. "Genome-Wide Identification and Expression Analysis of WRKY Genes during Anthocyanin Biosynthesis in the Mango (Mangifera indica L.)" Agriculture 12, no. 6: 821. https://doi.org/10.3390/agriculture12060821
APA StyleShi, B., Wu, H., Zhu, W., Zheng, B., Wang, S., Zhou, K., & Qian, M. (2022). Genome-Wide Identification and Expression Analysis of WRKY Genes during Anthocyanin Biosynthesis in the Mango (Mangifera indica L.). Agriculture, 12(6), 821. https://doi.org/10.3390/agriculture12060821







