Abstract
Salinity is one of the most destructive environmental challenges for citriculture worldwide, and all climate change scenarios are predicting an increased impact of salinity on citrus orchards. Citrus cultivars are grown as grafts on various rootstocks to provide specific adaptation to abiotic stress and tolerance to major diseases such as citrus tristeza virus. To understand rootstock–scion interactions with regard to salinity, transcriptome profiling of mRNA expression was analyzed for 12 candidate genes in leaves, shoots, and roots of five Hernandina clementine scions grafted on Rangpur lime (LR), Volkamer lemon (CV), Carrizo citrange (CC), sour orange (Big), and Cleopatra mandarin (MC) rootstocks in response to moderate and severe salinity. qRT-PCR analysis revealed differential gene expression that varied by rootstock, salinity level, and tissue. The majority of induced genes were those involved in ion transporter proteins (mainly NHX1 and HKT1 genes), Cl− homeostasis (CCC1 gene), biosynthesis and accumulation of compatible osmolytes, proline (P5CS gene) and glycine betaine (CMO gene), accumulation of proteins (LEA2 gene), and ROS scavenging antioxidant activity (mainly APX). We show that these expression patterns could explain the relative tolerance of the used rootstocks and report new insights on the main salt tolerance mechanisms activated by these rootstocks.
1. Introduction
Climate change is part of a larger challenge for sustainable development and human welfare globally. Among the prime factors, salinity is a major unfavorable abiotic stress factor causing land degradation [1], affecting almost all aspects of plant development, and drastically reducing agricultural productivity [2]. Yield losses due to irrigation with saline water largely depend on factors including climate, type of soil, evaporative demand, drainage, irrigation water quality, method and time of irrigation, and plant species tolerance [3,4].
Citrus is a major fruit crop with high economic importance around the world. The extensive use of rootstocks has enabled citrus to be grown under various environmental conditions worldwide. Rootstocks provide resistance to various diseases such as citrus tristeza virus (CTV) and abiotic stresses [5]. This is particularly important, as contamination by CTV has been reported in many previously virus-free areas, such as Tunisia [6]. Citrus rootstocks enable growers to avoid juvenility and ensure early fruiting, and they can be used to control plant vigor and tree size to allow high-density planting options [7,8,9]. It was shown that rootstocks affect the productivity and fruit quality of the scion [10,11,12].
Citrus is also a salt-sensitive crop [3,13]. To reduce the negative effects of salinity on citrus, important factors to consider include the use of appropriate rootstocks and proper agricultural and irrigation practices [14]. Evaluating salt-tolerant citrus rootstocks and studying the rootstock–scion interaction should enable us to determine the best selection and use of rootstocks that can improve resistance to this constraint [8].
Salinity is the concentration of soluble salts [15], mainly sodium (Na+) and chloride (Cl−), increased concentrations of which in soil solution cause undesirable morphological, physiological, and biochemical effects in different organs of citrus plants [16,17,18,19]. High salinity can affect different aspects of the physiological status of plants, such as oxidative stress, ion toxicity, membrane disorganization, nutritional disorders, alteration of metabolic processes, and reduction of cell division and expansion [2,20,21,22]. These effects can decrease plant growth, development, and survival. Certain concentrations of salinity can cause three important interrelated stresses in citrus: osmotic stress, which causes quick and temporary changes in plant–water relations, and salt-specific and oxidative stress, which occur later, depending on the species, and cause salt-induced injury [1,23]. Salinity causes stress by damaging ionic and osmotic balances in plants [19].
Salt-specific or ionic stress starts with the plant’s absorption of ions and is the result of the harmful effect of these ions inside plant cells. Salt absorbed by the roots goes through the transpiration stream to shoots, and finally accumulates in leaves [23]; this occurs very rapidly in sensitive genotypes that lack efficient salt exclusion or compartmentalization mechanisms, causing different physiological abnormalities [1]. Plants may face secondary stresses due to osmotic and ionic stresses, which can include oxidative stress and the accumulation of toxic compounds; severe damage to the cellular structure and inhibition of photosynthesis are caused by reactive oxygen species (ROS) [24,25,26], including H2O2 [27]. Oxidative damage to lipids, proteins, and DNA is caused by increased ROS levels, modifying fundamental membrane properties such as stability and enzymatic activity and decreasing overall homeostasis, which can lead to plant cell death [28].
At the cellular, molecular, physiological, and biochemical levels, plants have built different mechanisms to overcome all of these detrimental conditions [29,30,31,32]. Plants that are able to develop under high NaCl concentrations are also able to promote osmotic homeostasis and prevent uptake, transport, compartmentalization, extrusion, and mobilization of Na+ (and Cl−) ions [33]. In citrus, resistance to salinity is more dependent on ion avoidance mechanisms (Cl− and Na+ exclusion) than effective vacuolar compartmentalization of ions in leaf cells [34]. Additionally, studies have shown that Cl− homeostasis is the physiological basic for citrus resistance to salinity [35,36].
Lately, studies have been able to explain the molecular mechanisms behind plant salt stress responses and adaptation with the help of molecular genetics, genomics analysis, transcriptomic and gene expression regulation, and metabolomics [30,37,38,39,40,41]. Many candidate genes responsible for salt tolerance response were identified and proved to be valuable markers for the selection of rootstocks that provide salt tolerance in the grafted citrus cultivars [36,42,43].
Under salt stress, Na+ homeostasis is regulated mainly by the Salt Overly Sensitive (SOS) signal pathway, the vacuolar membrane Na+/H+ exchanger (NHX) gene, and the membrane transporter-class HKT1 gene, while chloride exclusion relies on cation Cl− cotransporter (CCC) genes, which regulate root Cl− uptake in the epidermis and/or long-distance chloride transport in the vascular cylinder [31,35]. Plants can survive oxidative stress due to many antioxidants and detoxifying enzymes that can eliminate ROS [2,23]. ROS scavenging enzymes include catalase (CAT), superoxide dismutase (SOD), and peroxidase (POD), while antioxidant enzymes are ascorbate peroxidase (APX) and glutathione reductase (GR) [44,45,46]. Many plants, including wheat, vines, and maize, show a correlation between antioxidant enzyme activation and tolerance to salinity [47,48,49].
Osmotic stress regulation is achieved through the activation of pathways to induce the biosynthesis and accumulation of compatible osmolytes, which are very important in reducing the cell osmotic potential and stabilizing proteins and cellular structures [50,51]. The types of osmolytes synthesized under salt stress are species- and tissue-specific. However, proline, glycine betaine, and sugars have been commonly shown to accumulate in roots and leaves [40]. Proline synthesis in the cytosol or chloroplasts is mostly catalyzed by Δ-1-pyrroline-5-carboxylate synthetase (P5CS). Plants overexpressing the P5CS gene showed enhanced accumulation of proline amino acid and improved salt tolerance [52], whereas P5CS1 gene silencing in Arabidopsis thaliana (L.) Heynh. impaired stress-induced proline synthesis, and plants were hypersensitive to salinity [53]. Choline mono-oxygenase (CMO) is involved in the biosynthesis of osmoticum glycine betaine [54]. It was shown that glycine betaine acts as an osmolyte to protect PSII under saline conditions [55].
Studies have implicated other proteins and metabolites in abiotic stress (mainly salinity) tolerance. These include late embryogenesis abundant (LEA) proteins [56,57], sucrose [58], and lipid transfer proteins (LTPs) [59,60]. In many plants, there is a relation between LEA protein expression and tolerance against salinity, freezing, and drought [61,62]. Duan and Cai [63] identified an LEA protein producing full-length gene in rice; in rice, OsLEA3-2 is considered an abiotic stress induced gene with major importance in salt and drought tolerance. During drought and high salinity, sucrose has a role in osmotic adjustment [64] and consequently in stress adaptation. Lipid transfer proteins (LTPs), which are still an enigmatic family, were considered to have a role in adaptation to biotic and abiotic stresses. According to data related to gene expression, LTPs have some functions in adaptation to salt stress [59,60] and osmotic stress [65].
In citrus, the relatively deleterious effects of Na+ and Cl− are still controversial. Some studies showed that photosynthesis and transpiration were more affected by Na+ ions than Cl− [66,67]. Cleopatra mandarin (Citrus reshni hort. ex Tanaka) rootstock, grafted by ‘Valencia’ sweet orange (C. sinensis (L.) Osbeck cultivar ‘Valencia’), ‘Taylor Eureka’ lemon (C. limon (L.) Burm f.), and ‘Ellendale tangor’ hybrid mandarin (C. reticulata Blanco × C. sinensis) (C. reticulata Blanco ‘Tangor’ Swingle variety ‘Ellendale’) scions exhibited decreased photosynthesis under saline conditions (75 mM NaCl), caused by Na+-dependent stomatal closure and chlorophyll degradation [68]. However, many studies have shown that citrus tolerance to salinity is basically related to the physiological Cl− exclusion capability, or to the Cl− uptake and transport limitation from root to shoot [69,70,71].
Rootstocks have an important impact on the tolerance of the rootstock–scion combination. Rootstocks, through their specific root structure, xylem anatomy, ion uptake, and hormonal and biochemical profiles, have differential effects on scion growth, development, and response to drought and salinity [34]. Thus, the selection of suitable citrus rootstocks under stress conditions is important and has a role in enhancing stress avoidance and/or tolerance mechanisms [13,72]. Studies have shown that the exclusion mechanisms of Na+ and Cl− are independent [13,23,72]. Sour orange rootstock (C. aurantium L.) is deemed a good Cl− and Na+ excluder. Cleopatra mandarin and Rangpur lime (C. limonia Osbeck) are considered Cl− excluders [73], while Carrizo citrange (C. sinensis × Poncirus trifoliata (L.) Raf) and Swingle citrumelo (P. trifoliata × C. paradisi Macfad.) are Na+ excluders. Citrus macrophylla (C. macrophylla Wester) and Rough lemon (C. jambhiri Lush.) rootstocks are Cl− and Na+ accumulators [74,75,76]. P. trifoliata and its citrange hybrids can exclude more Na+ than Cl− ions [23].
Citrus trees have a great undiscovered potential to reveal important plant growth processes and responses that may not be possible to investigate in other plants. Several previous studies have suggested that citrus plants may have a pool of genes with distinctive and exceptional roles, such as those involved in salt stress tolerance. While many studies have tackled the impact of citrus rootstock on the morphological, nutritional, and hormonal regulation of the scion related to physiological characteristics, only a few have focused on the effect of salt stress on citrus rootstock–scion combinations at the molecular level. Therefore, this study was aimed at analyzing the rootstock-specific activation of major salt tolerance pathways through transcriptome profiling of mRNA expression in different organs, for 12 key candidate genes in 5 rootstocks grafted with ‘Hernandina’ clementine (C. clementina hort. ex Tanaka) (willowleaf mandarin (C. deliciosa) × sweet orange (C. sinensis)).
2. Material and Methods
2.1. Plant Material and Salt Treatment
The National Institute for Agriculture, Food and Environment (INRAE; Corsica, France) provided the seeds for the CTV resistant rootstock cultivars used in the study: Rangpur lime (LR), Volkamer lemon (CV) (C. volkameriana V. Ten. & Pasq.), Carrizo citrange (CC), and Cleopatra mandarin (MC). Sour orange (Big)/scion combinations are CTV sensitive; however, this rootstock is still widely used in many countries. Following seed germination, the most developed seedlings, being of nucellar origin, were chosen and then grafted with virus-free budwood of ‘Hernandina’ clementine. Then, 18-month-old citrus combinations (rootstock–scion) were transferred to an insect-proof greenhouse. The experiment was carried out under semi-controlled settings, with minimum and maximum temperatures of 11.5 and 23.5 °C and average humidity of 62.5%.
During the entire experimental assay, plants were irrigated twice weekly with 250 to 350 mL of water with half-strength Hoagland’s solution [77] for 1 month before starting treatments, depending on water loss as measured by pot weight during the experiment. Salinity treatments consisted of adding NaCl to the nutrient solution to a final concentration of 2.0 g L−1 (35 mM NaCl) and 4.0 g L−1 (70 mM NaCl) to achieve final electrical conductivity of 4.0 and 7.0 dS/m, respectively. The basic nutrient solution, used as a control, had an electrical conductivity of 1 dS/m and a pH of 6.2. To avoid osmotic shock, plants were acclimated to their final NaCl concentrations over 2 weeks by a progression of 15 to 20 mM of NaCl in 2-day intervals. After 14 weeks of treatment (at final concentrations), samples of leaves, shoots, and roots from 3 independent biological replicates were collected, instantly frozen in liquid nitrogen, and preserved at −80 °C until use.
The experimental layout was a randomized block design where each rootstock/scion combination was represented by 9 plants, 3 biological and 3 technical replicates, which were subjected to each of the 3 treatments: T0: only nutrient solution (control, 1 dS/m); T1: moderate salt (4.0 dS/m); and T2: severe salt (7.0 dS/m).
2.2. Identification and Sequence Analysis of Citrus Candidate Genes
C. clementina and C. sinensis sequences with high similarity to the original A. thaliana sequence were identified using the Basic Local Alignment Search Tool (BLAST) in the International Citrus Genome Consortium (ICGC) database (http://www.phytozome.jgi.doe.gov (accessed on 28 December 2021); NCBI reference sequence database, http://www.ncbi.nlm.nih.gov) (accessed on 28 December 2021) for candidate genes SOS1, NHX1, HKT1, CCC1, APX, CAT, P5CS, CMO, Lea2, SPS, LTP, and V-PPiase. For each gene, sequences found in both C. clementina and C. sinensis were aligned using multialin software (multialin.toulouse.inra.fr) [78]. The common part of the two sequences showing maximum homology far from mismatches and intron zones served for primer designation. Using the Primer 3.0 program [79,80], forward and reverse gene-specific primers were constructed to amplify chosen candidate genes (Table 1). Selected gene expression to be analyzed included NHX1, HKT1, CCC1, CMO, P5CS, CAT, APX, Lea2, LTP, and SPS. To standardize the qPCR results, the citrus housekeeping gene β-actin was used.

Table 1.
List of primers used for quantitative real-time PCR.
2.3. RNA Extraction and Quantitative Real-Time RT-PCR Analysis
Using Geno/Grinder 2010 (SPEX SamplePrep, Metuchen, NJ, USA), frozen plant tissue was ground to a fine powder in liquid nitrogen. Total RNA was isolated from 100 mg of frozen ground tissues (leaf and shoot samples) of ‘Hernandina’ clementine grafted onto 5 different rootstocks and of root samples of the 5 citrus rootstocks (Rangpur lime, Volkamer lemon, Carrizo citrange, Cleopatra mandarin, and sour orange), using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. The quality and quantity of RNA extracts were examined by UV absorption with a NanoDrop 2000c spectrophotometer (NanoDrop Technologies, Thermo Fisher Scientific, Wilmington, DE, USA). Reverse transcription quantitative PCR (RT-qPCR) was conducted in two steps:
First, cDNA synthesis was performed with an iScriptTM cDNA synthesis kit (Bio-Rad, Hercules, CA, USA). Reactions of 20 μL total volume contained 4 μL 5× iScript reaction mix, 1 μL of iScript reverse transcriptase, 11 μL of water, and 4 μL of template total RNA (20–600 ng/μL). Incubation was carried out in a C1000 Touch thermal cycler (Bio-Rad) using the following protocol: priming for 5 min at 25 °C, reverse transcription for 30 min at 42 °C, RT inactivation for 5 min at 85 °C. All synthetized cDNA was diluted to a final concentration equal to 200 ng/μL.
Second, quantitative RT-PCR was performed using a CFX 96 TouchTM Real-Time PCR Detection System for accurate and repeatable real-time PCR. The C1000 Touch Thermal Cycler (Bio-Rad) coupled with the CFX 96 optical reaction module detects SYBR® Green dye based real-time PCR reactions. With a total volume of 10 μL, the PCR reaction comprised 5 μL of SSO Advanced Universal SYBR® Green Supermix (2X) (Bio-Rad), 2.5 μL of diluted cDNA (200 ng/μL), and 1 μL each of the upstream and downstream primers of each gene (10 mM). The following PCR procedure was used: initial DNA denaturation and polymerase activation for 3 min at 95 °C, amplification through 39 cycles of denaturation for 10 s at 95 °C, followed by an optimal annealing temperature gradient for each primer (55–65 °C) for 30 s; annealing/extension and plate read were performed in each cycle of this step at 60 °C.
To test the specificity of the amplification reactions, melt-curve analysis was conducted immediately after amplification 61 times for 5 s at 65–95 °C with 0.5 °C increments, in 96-well iCycler iQTM PCR plates (Bio-Rad) sealed with optical sealing tape (Bio-Rad).
Complete gene layout, cycling protocol information for all assays, and results analysis were performed by the instrument onboard CFX Maestro software (CFX 96 TouchTM Real-Time PCR Detection System, Bio-Rad).
2.4. Data and Statistical Analysis
The Ct value of the β-actin gene was used to normalize all qPCR data. The relative gene expression was computed using the 2−ΔΔCT method (fold change) [81]. Data are given as the means of 3 biological replicates. RT-qPCR analysis was conducted with 3 biological and 3 independent technical replicates for each RNA sample, to ensure the reproducibility of the results, and the data are presented as means ± SE (n = 3).
ΔCt for each gene by treatment, rootstock, and tissue was computed by subtracting the Ct number of the target sample from the Ct number of the control sample. A statistical comparison of ΔCt values under different salinity treatments was performed using the one-way ANOVA parametric test followed by Student–Newman–Keuls (SNK) post hoc test. The assumptions for ANOVA were that the ΔCt groups had Gaussian distribution and equal variance. However, when these assumptions were not valid, as in many real-time PCR experiments, as a non-parametric alternative to ANOVA, the Kruskal–Wallis test was used, followed by Dunn’s test. Differences between treatments were considered statistically significant at p-value < 0.05 using RStudio statistical software. Figure illustration was performed using SigmaPlot 14.0 (Systat Software, San Jose, CA, USA).
The potential genes implicated in salt tolerance were compared in terms of their relative expression, for the same tissue, at several levels between unstressed and stressed plants: depending on the 5 rootstock/scion combinations, depending on treatment (T1, T2), and depending on gene, by real-time quantitative reverse transcription PCR (qRT-PCR).
3. Results and Discussion
The transcript abundance of genes associated with functional processes was quantified by qRT-PCR in leaf, shoot, and root tissues of five citrus ‘Hernandina’ rootstock/scion combinations challenged with salt stress. Considerable increases or decreases in the transcript levels of particular genes were detected in specific plant tissues subjected to salt treatment (T1 and T2) in comparison to the control (T0).
3.1. Ion Homeostasis Pathways
3.1.1. SOS1 Gene Expression
The Salt Overly Sensitive (SOS) pathway is activated by salt treatment, in which the Na+/H+ antiporter SOS1 is a major regulator of Na+ transport from the cytoplasm to the apoplast [40]. SOS1 encodes a plasma membrane Na+/H+ antiporter that plays a critical role in sodium extrusion and in controlling long-distance Na+ transport from the root to the shoot, while protecting individual cells from sodium toxicity [82,83]. Studies have shown that overexpression of SOS1 can increase the salt tolerance of plants [84,85].
To further understand the behavior of the SOS pathway in salt-stressed citrus, the expression of putative SOS1 in aerial and root tissues was examined in Big (Cl− and Na+ excluder), CC (Cl− includer), CV, LR (Cl− excluder), and MC (Cl− excluder) under salt stress with 35–70 mM NaCl.
In leaves, SOS1 expression was not significantly modified by salt treatment (Figure 1) in all rootstock–scion combinations except for Big. Indeed, Brumós et al. [31] showed that in citrus leaves subjected to salt stress for several weeks, Cl− toxicity, rather than Na+ toxicity and/or simultaneous osmotic disturbance, was the predominant component implicated in the salinity-induced molecular reactions. Citrus and other fruit tree crops (e.g., peaches) are rather uncommon in this regard, since Cl− buildup in citrus leaves is typically a larger problem than Na+ accumulation, though this varies depending on the rootstock [3]. Among the most widely used commercial citrus rootstocks, sour orange and Rangpur lime are considered good Cl− excluders, and thus tolerant to salinity [76,86]. Grosser et al. [87] showed that under well-watered saline circumstances in a greenhouse, leaf Cl− concentration levels in young ‘Valencia’ sweet orange trees grafted on Carrizo citrange could reach 3.5% after 5 months without apparent toxicity symptoms.
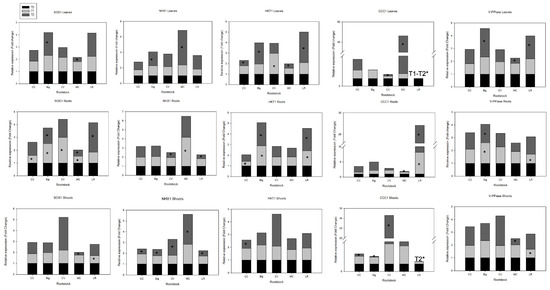
Figure 1.
Expression profiles of SOS1, NHX1, HKT1, CCC1, and V-PPiase of ‘Hernandina’ clementine (Citrus clementina hort. ex Tanaka) (willowleaf mandarin (C. deliciosa) × sweet orange (C. sinensis (L.) Osbeck)) grafted on 5 citrus rootstocks in response to salt. Big: sour orange (C. aurantium L.); CC: Carrizo citrange (C. sinensis × Poncirus trifoliata (L.) Raf); CV: Volkamer lemon (C. volkameriana V. Ten. & Pasq.); LR: Rangpur lime (C. limonia Osbeck); and MC: Cleopatra mandarin (Citrus reshni hort. ex Tanaka). For each combination (rootstock/‘Hernandina’), stacked panels showed gene expression in plant tissue (leaves, shoots, and roots) treated with NaCl (T1: 35 mM NaCl and T2: 70 mM NaCl) relative to untreated samples (T0 = only nutrient solution without salt treatment). Relative gene expression was calculated using 2−ΔΔCT method (fold change). * Significant differences for p-values < 0.05.
There were no significant variations in SOS1 expression in shoots across all combinations following NaCl treatment (Figure 1). There was no abundance of SOS1 mRNA transcripts observed in root tissues in CC and MC genotypes under salt stress. CV, Big, and LR showed significantly increased SOS1 expression following salt treatment. SOS1 increased by 2- and 1.4-fold in CV and Big, respectively, with T1 (35 mM NaCl), while it increased by 2.3-fold in LR at high salinity (70 mM). This is consistent with a study showing that compared to Cleopatra mandarin leaves, sour orange leaves absorbed less Na+, indicating that sour orange may partially exclude Na+ from the leaves by collecting it in the roots [16]. Under mild NaCl conditions, the primary component appears to be osmotic, with no significant Cl− or Na+ tissue accumulation [34]. Under more severe salinity stress (T1-T2), the ion toxicity contribution was more relevant. Shoot Cl− exclusion is primarily regulated by the rootstock, most likely because it is dependent on anion transporters found in the root. However, both scion and rootstock are implicated in Na+ exclusion, most likely because important genes for Na+ exclusion in plants work at both the root and shoot level [88]. From our results, severe salt stress increased the transcript level of SOS1 in roots of Big, CV, and LR, which can be considered Na-tolerant rootstocks (Na+ excluders). Similar findings were reported for trifoliate orange (P. trifoliata) rootstock [89], Populus euphratica Oliv. [90], and A. thaliana [82]. SOS1 enhances Na+ exclusion by extruding the cation from root tip epidermal cells [91]. SOS1 loads Na+ into the xylem in roots under mild stress, allowing for regulated Na+ delivery to shoots and storage in leaf mesophyll vacuoles. SOS1 appears to work under excessive Na+ stress by reducing net absorption at the root tip and limiting Na+ loading of xylem sap in roots to reduce damage to leaves that may occur when the capacity of Na+ sequestration in leaf cell vacuoles is exceeded [89]. Our results also suggest increased Na+ root-to-shoot transport in Big, CV, and LR genotypes.
The net Na+/Cl− uptake rates with the concentrations of Na+/Cl− in root/shoot tissues of tolerant rootstocks could provide more information to support these hypotheses. More research on citrus SOS genes is needed to determine the functional implications of the SOS pathway in citrus.
3.1.2. NHX1 Gene Expression
The other adaptive mechanism for regulating ion homeostasis under high salinity is vacuolar Na+ partitioning, which is mediated by the vacuolar Na+/H+ antiporter (NHX) [92,93], which is expressed in the roots and leaves and selectively carries Na+ into the vacuoles [42]. Transgenic kiwifruit, tomato (Solanum esculentum L.), and rice (Oryza sativa L.) overexpressing NHX1 were shown to be more tolerant than wild types [94,95,96].
We have shown that in leaves, salt stress increases mRNA transcripts of NHX1 in Big (twofold increase) and more sharply in MC (4.5-fold increase) rootstock (Figure 1). Similar findings were observed in cotton [97], wheat [98], and A. thaliana [99]. NHX1 gene expression was also shown to be very responsive to salt stress in shoots and roots as well as in leaves.
It was shown that tissue and vacuole compartmentalization is important in salt stress. Studies have shown a preferential sequestration of recovered Na+ from xylem, primarily in roots, into the vacuole, thus avoiding the harmful effects of this cation and preserving cell osmotic equilibrium [89]. Moreover, dye fluorescence investigations on Na+-sensitive rough lemon and Na+-tolerant Swingle citrumelo revealed higher Na+ sequestration in epidermal and pericycle root cell vacuoles in Na+-tolerant rootstock, as compared to Na+-sensitive rootstock, as a strategy to maintain lower Na+ levels in the leaves [76]. However, it has also been reported that compartmentalization of Cl−/Na+ ions in leaf cell vacuoles can be considered as a tolerance mechanism that allows the plant to withstand osmotic and ionic stresses, and that citrus resistance to salinity mostly relies on avoidance processes (Cl−/Na+ exclusion) rather than efficient vacuolar partitioning of these ions in leaf cells [34]. In our study, MC showed an increase in NHX1 expression in the three tissues, suggesting that the main tolerance strategy of MC is through Na+ vacuolar compartmentalization.
3.1.3. Vacuolar H+-Pump
NHX activity is tightly controlled by the electrochemical vacuolar proton gradient established across the tonoplast by two vacuolar H+-pumps, H+-adenosine triphosphate (V-H+-ATPase) and H+-inorganic pyrophosphatase (V-H+-PPiase) [100,101]. A recent study showed that in response to salt stress, a vacuolar H+-pump pyrophosphatase gene (HVP10) was upregulated in barley (Hordeum vulgare L.) [102]. In our study, as a result of high salt stress, V-PPiase gene was upregulated in leaves of both Big and LR rootstocks (2.3- and 1.8-fold increase, respectively), while expression of this gene did not change in response to NaCl exposure for all of the other genotypes (Figure 1). In roots, Big maintained a constant 1.5-fold increase in gene expression under salt stress (T1 and T2), while LR mRNA transcripts increased only under severe salt stress (1.4-fold increase) (Figure 1).
Several studies have shown that sustained overexpression of the H+-PPase gene accelerates growth and enhances tolerance to biotic and abiotic challenges in A. thaliana [103] as well as many other plants: e.g., cotton [104], barley [105], poplar [106], and tobacco [107]. Salinity tolerance attributed to the tissue tolerance mechanism was positively correlated with expression of A. thaliana vacuolar H+-pyrophosphatase gene (AtAVP1) [108].
Therefore, the increased V-PPiase expression in Big and LR, especially in leaves, may partly explain their salt tolerance pattern. This was confirmed, as Big and LR also showed a concomitant increase in the expression of tandem vacuolar NHX1 antiporter gene (in leaves), arguing for a preferential sequestration of recovered Na+ from xylem into vacuole, mostly in leaves.
3.1.4. HKT1 Gene Expression
Among the class of sodium channels, membrane transporter-class gene HKT1 (high-affinity K+), which regulates the uptake and translocation of Na+, is important in plant salt tolerance and root-to-shoot Na+ partitioning [109,110]. HKT-type transporters are key ion channels that have been shown to selectively enable Na+ and/or K+ transit, hence are potential salt tolerance candidates [111]. In our study, real-time PCR analysis revealed increased transcript levels of the putative HKT1 gene in T2-stressed Big (threefold increase) and LR (threefold increase) leaf tissues (Figure 1). Expression in CV increased under moderate salinity. These findings imply that this gene is overexpressed in Na+ excluder genotypes. Gene expression analysis of seedlings stressed with 70 mM NaCl revealed low gene expression with no significant variations for genotypes CC and MC from their respective control values. These findings complement previous salinity studies demonstrating that the citrus rootstock Cleopatra mandarin is a Cl− excluder, accumulating more Na+ than other genotypes [13]. Big, CV, and LR, on the contrary, might be considered as Na+-tolerant.
Real-time PCR analysis revealed increased transcript levels of the putative HKT1 gene in stressed Big and LR root tissues (2.2- and 2.1-fold increase, respectively) (T2). No significant differences were recorded for the roots of other genotypes. In shoots, none of the rootstocks showed activation or downregulation of HKT1 expression (Figure 1). Martinez-Alcántara et al. [89] discovered increased transcript levels of AtHKT1-like transporter HKT1 in root and shoot tissues of salt-stressed plants with the Na+ excluder trifoliate orange genotype compared to Cleopatra mandarin, which would be responsible for the reduced root-to-shoot Na+ transfer and consequently Na+ sequestration in the roots. Moreover, studies of A. thaliana have shown that AtHKT1;1 gene contributes to the control of both root accumulation of Na+ and retrieval of Na+ from the xylem, which control and decrease shoot Na+ concentration and increase plant salinity tolerance [112,113].
Interestingly, in Na+-tolerant rootstocks Big, CV, and LR, coordinated action of Na+ transporters HKT1 and SOS1 under salt stress was observed. The coordination between HKT1 and SOS1 Na+ transporters enables appropriate partitioning of this harmful cation between leaves, roots, and shoots in Big, CV, and LR rootstock genotypes. Based on our results, we hypothesized that overexpression of putative SOS1 (in roots) and HKT1 (in roots and leaves) might indicate improved xylem Na+ retrieval and impaired translocation to shoot tissues in these rootstocks, which implies that the Na+ transport pathway varies depending on the rootstock. Indeed, the capacity of particular genotypes, such as sour orange, to exclude Na+ has been connected to the expression of SOS1 and HKT1 genes [89].
For all tissues, results for SOS1, NHX1, and HKT1 showed that all rootstocks except CC overexpressed one or several genes involved in ion homeostasis, which advocates for their salt tolerance [114]. This is consistent with the well-documented salt susceptibility of CC [115,116].
3.1.5. Chloride Homeostasis
A. thaliana cation–Cl− cotransporter (AtCCC) is thought to catalyze the coordinated symport of K+, Na+, and Cl−, and has been proposed as a gene that participates in xylem delivery and/or re-absorption of Cl− [35]. Transcript abundance of this gene following T2 salt treatment increased in leaf tissues of only MC rootstock (42-fold increase) (Figure 1). Our results do not agree with those revealed by Brumós et al. [36], showing that the citrus CCC1 gene does not respond to salt stress (90 mM NaCl) in CC and CM. Only the CcSLAH1 gene showed a substantial response to salt stress, and was particularly activated by salt stress in CC, whereas it was somewhat repressed in CM. The fact that the citrus CcCCC1 gene was not sensitive to Cl− treatment or salt stress in the rootstocks tested (MC and CC) does not necessarily mean that it is not engaged in Cl− homeostasis, according to the authors, because plant CCCs can be controlled at the level of protein activity [116]. However, such an increase in CCC1 gene expression (42-fold increase) in our case gives strong evidence that in MC, salt stress particularly activates this gene, whereas it is inhibited in the other genotypes.
In shoot tissues and under severe conditions (T2), a very high, strong increase in mRNA transcripts of CCC1 was recorded in the CV genotype (13-fold increase) in comparison to its basal levels in unstressed plants (Figure 1).
In roots, other rootstock genotypes were found to have highly expressed CCC1 gene. Under severe salt stress (T2), the data showed high expression levels of CCC1 in Big and CC (threefold and twofold increases, respectively). Noteworthy expression of this gene under salt stress was revealed in LR rootstock (7-fold increase with T1 and 19-fold increase with T2) compared to unstressed plants.
Effective Cl− exclusion from the shoot is widely accepted as the most important strategy for preventing salt stress in citrus plants [34]. In recent years, an important breakthrough was achieved in understanding the uptake and functions of Cl−, which progressed from being seen as a detrimental ion that is inadvertently absorbed by plants via passive processes or nitrate transporters to being regarded as a preferred macronutrient whose transport is carefully regulated by plants [31,117].
Brumós et al. [36] demonstrated a model for symplastic regulation of Cl− homeostasis in the roots of includer Carrizo citrange and excluder Cleopatra mandarin rootstocks. Cl− xylem transfer is clearly limited in the salt-tolerant Cleopatra mandarin but not the sensitive Carrizo citrange rootstock. Indeed, membrane transporter genes are differently regulated in tolerant (Cl− excluder) and sensitive (Cl− accumulator) rootstocks. This suggests that in salt-tolerant rootstocks, decreased Cl− translocation may result in greater Cl− buildup in distal roots and, most likely, more Cl− release to the rhizosphere via Cl− channels in epidermal cells.
In our study, at the root level, LR and (to a lesser degree) Big displayed salt-tolerant behavior. This was highly expressed in MC at the leaf level and in CV and to a lesser degree at the shoot level. In addition, for salt-tolerant rootstocks such as CV, the expression of Cl− transporter gene CCC1 was highly induced in shoots but repressed in leaves and to a lesser degree in roots, and for MC, the expression of CCC1 gene was highly induced in leaves, slightly induced in shoots, but repressed in roots. This suggests preferential tissue for CCC1 gene expression: leaves for MC, roots for LR and Big, and shoots for CV. Such patterns have been reported; for example, Martins et al. [118] investigated the expression of 34 aquaporins and other major intrinsic proteins (MIPs) involved in salinity tolerance in sweet orange (C. sinensis) and discovered that the majority of these genes were upregulated in the roots, but had both increased and decreased transcript levels in the leaves.
Our results show no significant modification of Carrizo citrange CCC1 gene expression in any tissue under moderate or severe stress. This is consistent with the study of Brumós et al. [36] showing that CC is a Cl− includer because of its limited ability to exclude the anion. In fact, after Cl− supply, the Cl− includer genotype CC showed extremely active translocation of Cl− to the shoot (at a rate six-fold greater than CM), but root-to-shoot Cl- transport was substantially reduced in the excluder rootstock CM. Compared to CC, Cleopatra mandarin had a less effective root system for water absorption and a larger shoot-to-root ratio. Otherwise, the comparatively strong salt tolerance of citrus rootstocks such as Rangpur lime, Cleopatra mandarin, and CV is linked to their ability to minimize Cl− buildup in scion leaves [119]. Salinity stress from 50 mM of NaCl for 8 to 9 weeks has been shown to diminish growth and water usage in well-irrigated citrus seedlings and grafted plants. Salinity-induced growth inhibition was more important in ‘Valencia’ sweet orange trees grafted on CC rootstock than in plants grafted on MC, and this was related to leaf Cl− concentrations [114].
The key element influencing the degree of chloride exclusion appears to be the regulation of root-to-shoot Cl− transport. The Cl− exclusion mechanism of salt-tolerant rootstocks is primarily based on decreased Cl− unloading into the xylem, which correlates with a lower absorption capacity and increased Cl− retention in the distal regions of the root following Cl− administration [36].
3.2. Tolerance Pathways to Oxidative Stress
A recent study, aimed at identifying and analyzing salinity-induced genes in Rangpur lime, used functional categorization and revealed 13 different genes associated with antioxidant activity that were induced in response to high salinity [39]. In this study, in leaf tissue, two genes encoding CAT and APX were analyzed; APX was upregulated in CC (3.5-fold increase, T1), LR (2-fold increase, T1; 3-fold increase, T2), and significantly in CV (2.5-fold increase, T1), while it was downregulated in Big and MC. Salt treatment showed no significant impact on CAT gene expression, except a slight upregulation in Big (Figure 2).
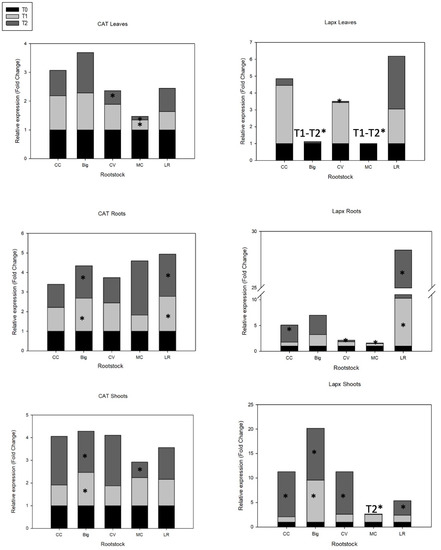
Figure 2.
Expression profiles of APX and CAT genes of ‘Hernandina’ clementine (Citrus clementina hort. ex Tanaka) (willowleaf mandarin (C. deliciosa) × sweet orange (C. sinensis (L.) Osbeck)) grafted on 5 citrus rootstocks in response to salt. Big: sour orange (C. aurantium L.); CC: Carrizo citrange (C. sinensis × Poncirus trifoliata (L.) Raf); CV: Volkamer lemon (C. volkameriana V. Ten. & Pasq.); LR: Rangpur lime (C. limonia Osbeck); MC: Cleopatra mandarin (Citrus reshni hort. ex Tanaka). For each combination (rootstock/‘Hernandina’), stacked panels showed gene expression in plant tissue (leaves, shoots, and roots) treated with NaCl (T1: 35 mM NaCl; T2: 70 mM NaCl) relative to untreated samples (T0, only nutrient solution without salt treatment). Relative gene expression was calculated using 2−ΔΔCT method (fold change). * Significant differences for p values < 0.05.
Seday et al. [120] investigated the antioxidative enzyme responses of six ungrafted citrus rootstocks under various NaCl concentrations (from 45 to 135 mM) and showed that leaf APX activity in the rootstocks (Big, CC, CV, MC) increased with elevated salt levels. CAT activity increased in salt-tolerant Plantago maritima L. and Lycopersicon pennellii Correll in response to salt stress [121,122]. However, CAT activity was found to decrease in response to various stresses [123,124]. Abiotic oxidative stress tolerance was modulated from ROS detoxification and scavenging [25].
Our findings revealed that increasing the salt dosage reduced CAT activity in all rootstocks except Big. Similar trends were also reported for SOD activity in citrus leaves under salinity [120]. Stressed Big, LR, and MC plants showed higher transcript levels of candidate CAT gene (1.8-, 2-, and 2.8-fold increase, respectively) in roots and shoots in Big, CC, and LR (1.8-, 2.2-, and 1.5-fold increase, respectively). Salt-tolerant rootstocks Big, LR, and MC and, to a lesser extent, CV upregulate CAT gene in roots, while only Big showed a significant increase in shoots. Even being sensitive to salt, under severe salt treatment (T2), CC rootstock showed an increase in CAT gene expression in shoots. On the other hand, APX expression was notably higher in shoots and roots for almost all citrus rootstocks grown under salt stress compared to controls. Sour orange upregulated APX gene in roots (3.8-fold increase) and shoots (8.5-fold increase in T1; 10.6-fold increase in T2), although this gene was repressed in leaves. This result shows the strong and highly significant involvement of roots and mainly shoots in expressing antioxidant enzyme APX in stressed Big.
For CC, although it is considered to be sensitive to salinity, APX gene expression was increased 3.5-fold in leaves (T1), and 3.4-fold in roots and 9.2-fold in shoots (T2), compared with their respective basal values. These data suggest the involvement of the three tissues in the gene expression of ROS scavenging enzymes in CC (Figure 2).
Under salt treatment, CV rootstock showed a significant response to salt (T1) by overexpressing APX in leaves, and this was accentuated in shoots with increased salinity (T2). This gene was downregulated in roots. These results might also suggest a preferential tissue response of CV in expressing APX gene with gradual salinity. Similar results were shown with LR rootstock, in which APX was overexpressed in leaves and shoots (3-fold increase with T2), but much more in roots (9.3-fold increase with T1, 18-fold increase with T2). This emphasizes the quick response of LR to salt stress and the strong involvement of the antioxidant defense mechanism, mainly in roots. This confirms a recent study showing that rootstocks of Volkamer lemon grafted with Kinnow hybrid mandarin (‘King’ (C. nobilis) × ‘willowleaf’ (C. deliciosa)) exhibited high and significant increases in antioxidant enzymes and osmoprotectants following abiotic stress [46].
In MC rootstock, APX is either downregulated or repressed in all tissues. MC is considered to be a very tolerant rootstock, suggesting that APX plays a very limited role in this tolerance. Contrarily, according to Sekmen et al. [122], salt-tolerant plant species exhibit a considerable increase in APX activity in response to salt stress, whereas salt-sensitive genotypes exhibit a reduction or stay stable.
The differences observed between studies regarding the analysis of genes involved in ROS detoxifying pathways have been discussed. Singh and Sharma [1] reported that antioxidants are linked not only to salt tolerance but also to other factors, including crop growth stage and experimental management practices. It was shown that the simultaneous occurrence of other abiotic stresses (e.g., salinity, drought, and high light intensity), as compared to a single stimulus, changes the expression of genes controlling antioxidant activity. Moreover, while it was shown that ROS and reactive nitrogen species (RNS) together govern plant stress responses to unfavorable environmental conditions, the mechanisms underlying this role have not yet been completely uncovered. This is also the case for the involvement of antioxidant enzymes [120]. Antioxidant enzyme responses are influenced by two- or three-way interactions between rootstock, salt concentration, and sampling time, revealing the intricacy of the tolerance mechanisms of citrus rootstocks. Plant species most likely evolved distinct defensive mechanisms against salty environments. The stress-related gene families found in cytoplasmic organelles of cells (mitochondria, chloroplasts) were shown to evolve at different rates and diversified in different ways than those found in the nucleus, resulting in diverse defensive strategies among citrus species [125]. All of these ROS-scavenging enzyme results suggest the involvement of this defense system under salt stress in all genotypes, whether they are salt-tolerant or -sensitive, and a preferential tissue response in expressing CAT and APX genes under gradual salinity (T1, T2).
Salt stress enhances antioxidant enzyme activity, particularly in salt-tolerant citrus rootstocks [18]. In this context, salinity causes oxidative stress at the subcellular level in plants, and chloroplasts and mitochondria are significant producers of ROS in salt-stressed plants. Some authors have identified coordinated upregulation of the anti-oxidative machinery as one of the processes involved in the salt tolerance response, while others have linked salt tolerance to greater constitutive levels of certain antioxidant enzymes. Salt-sensitive species, on the other hand, exhibit no change or even a decline in antioxidant defenses, and they have lower constitutive antioxidant enzyme levels than salt-tolerant species [126].
Balfagón et al. [127] demonstrated that the rootstock altered APX and CAT activity in citrus scions under stress conditions. Their findings suggest that activation of the antioxidant system under stress conditions is a transmissible feature that can be passed down from rootstock to scion, emphasizing the importance of rootstock selection in improving crop performance and maintaining citrus yield under the current climate change scenario.
3.3. Compatible Osmolyte Biosynthesis
Under salt stress, compatible osmolytes such as proline and glycine betaine are produced and accumulated, a process that is species- and tissue-specific [40]. Compatible osmolytes can minimize water loss in response to short-term osmotic stress and improve cell turgor and expansion in response to long-term osmotic stress (continued growth under salt stress) [128]. Higher transcript levels of CMO in CC and CV rootstocks were recorded for T1 in leaves compared to the other genotypes (Figure 3). Tolerance to salt and temperature stress in rice was improved by the production of a CMO gene from spinach (Spinacia oleracea L.) [129]. Transgenic tobacco plants that overexpress the beet BvCMO gene accumulate GlyBet and have increased resistance to salt (150 mM NaCl) and drought stress [130]. Our results show the early involvement of this gene in leaves for both CC and CV in response to salt stress.
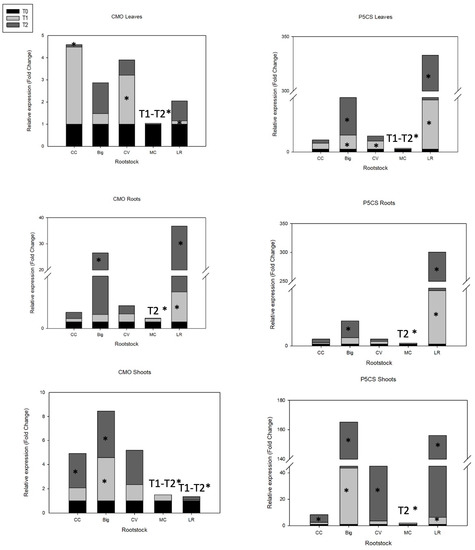
Figure 3.
Expression profiles of CMO and P5CS genes of ‘Hernandina’ clementine (Citrus clementina hort. ex Tanaka) (willowleaf mandarin (C. deliciosa) × sweet orange (C. sinensis (L.) Osbeck)) grafted on 5 citrus rootstocks in response to salt stress. Big: sour orange (C. aurantium L.); CC: Carrizo citrange (C. sinensis × Poncirus trifoliata (L.) Raf); CV: Volkamer lemon (C. volkameriana V. Ten. & Pasq.); LR: Rangpur lime (C. limonia Osbeck); MC: Cleopatra mandarin (Citrus reshni hort. ex Tanaka). For each combination (rootstock/’Hernandina’), stacked panels show gene expression in plant tissues (leaves, shoots, and roots) treated with NaCl (T1: 35 mM NaCl; T2: 70 mM NaCl) relative to untreated samples (T0, only nutrient solution without salt treatment). Relative gene expression was calculated using 2−ΔΔCT method (fold change). * Significant differences for p values < 0.05.
The accumulation of another compatible osmolyte, proline, catalyzed by P5CS, plays a key role in reducing the cell osmotic potential and stabilizing proteins and cellular structures under salt stress [40]. A positive correlation has been demonstrated between salt tolerance and proline concentration [131]. The increment of sodium chloride levels (up to 5000 ppm) in culture medium led to a significant accumulation of proline in leaves of citrus rootstocks [132]. Our results reveal that the P5CS gene was differently expressed in citrus rootstock leaves during salt treatment, but transcript abundance was generally similar between genotypes under control conditions. Interestingly, three genotypes showed moderate to very high levels in mRNA transcripts: Big (5-fold increase with T1, 13-fold increase with T2), CV (3-fold increase with T1, 2-fold increase with T2), and LR (18-fold increase with T1, 314-fold increase with T2). P5CS gene was clearly downregulated in MC. Our results suggest the involvement of this gene in leaves of Big, CV, and mainly LR in response to salt stress. It was previously demonstrated that sour orange highly accumulates proline under severe salinity stress (5 g/L NaCl) [132].
P5CS gene expression increased in roots of Big (3-fold increase with T1, 9-fold increase with T2), and LR (27.8-fold increase with T1, 271-fold increase with T2). In shoots, all genotypes except MC showed high levels of P5CS mRNA transcripts under gradual salinity: Big: 42.5-fold increase with T1, 121.7-fold increase with T2; CC: 1.5-fold increase with T1, 5.8-fold increase with T2; CV: 2.5-fold increase with T1, 20-fold increase with T2; and LR: 5.3-fold increase with T1, 150-fold increase with T2 (Figure 3). These results confirm the strong and major impact of salt stress on upregulating P5CS gene expression.
Many studies have used proline content to evaluate the performance of rootstocks under saline stress conditions [71,86,132]. Mostly, it was shown that proline concentration is higher in salt-tolerant rootstocks than other rootstocks [133,134]. This finding is consistent with our results for Big, CV, and LR rootstocks, but MC rootstock was an exception. Other investigations showed that the highest salt tolerance (to a salinity level of 75 mM NaCl), as determined by maximal proline accumulation, was recorded in Rangpur lime rootstock, while Carrizo citrange and sour orange showed minimum salt tolerance by accumulating the lowest amounts of proline [17]. The degree of alteration varied according to rootstock and salt level. A recent study attempted to investigate the physiological responses of three recently produced citrus rootstocks, Cleopatra mandarin, Volkamer lemon, and Carrizo citrange, after salinity treatment (0–100 mM NaCl). There were no statistically significant differences in proline content between Cleopatra mandarin and Carrizo leaves. Compared to MC, Volkamer lemon and Carrizo showed a considerable increase in proline concentration [71]. This agrees with our results, suggesting that even though CC is a salt-sensitive rootstock, it may show relative proline accumulation.
CMO gene expression was less significant than P5CS expression for all genotypes. However, in roots, Big showed a 24.3-fold increase in CMO mRNA transcript levels under 70 mM NaCl treatment, and LR showed a 4.5-fold increase in CMO mRNA transcripts with T1 and a 31.2-fold increase with T2 (Figure 3). In shoots, only Big and CC exhibited significant increases in CMO mRNA transcripts under salt stress (4-fold and 3-fold increase, respectively). These results further confirm the differential expression of CMO gene according to the rootstock, tissue, and salt stress level. Interestingly, the induction of expression was always more marked in tolerant rootstocks, reflecting the involvement of CMO gene in the process of regulating salt tolerance in citrus rootstocks. Our results show that in CC rootstock, despite its salt sensitivity, CMO gene is significantly involved in the early response to salt stress in leaves; a second response to more severe salinity emanates from shoots tissue.
Lv et al. [135] discovered various salt tolerance determinants and signaling pathways in sugar beet, and demonstrated that the CMO gene was one of the important candidate genes to test for enhanced sugar beet salt tolerance.
3.4. Stress-Induced Proteins
3.4.1. Late Embryogenesis Abundant Protein Gene
Despite the correlation shown between late embryogenesis abundant (LEA) protein expression and salt tolerance, no significant increase in LEA2 protein coding gene expression in leaves was recorded for any citrus rootstock genotypes under salt treatment. However, LEA2 gene was highly expressed in roots and shoots of all genotypes. Real-time PCR analysis revealed greater transcript levels of the LEA2 candidate gene under T1 salt stress conditions in roots of stressed rootstocks in Big (2.7-fold increase) and CV (1.9-fold increase) compared to their basal values under control conditions. Almost all rootstocks showed high to extremely high activation of gene expression of LEA2 in shoots in response to salt stress: Big: 72-fold increase; CC: 9.6-fold increase; CV: 21.5-fold increase; and MC: 31-fold increase (Figure 4). This result suggests that under salt stress, LEA2 gene is initially induced and upregulated in roots and shoots of Big and CV rootstocks, with no expression change in leaves compared to control basal values, under severe salt stress (T2). In CV, LEA2 was upregulated in shoots (21.4-fold increase) in response to severe salt treatment (T2). Interestingly, in MC rootstock, this gene was highly upregulated in shoots under severe salt stress (1050-fold increase), while it was downregulated in leaves and roots under both levels of salinity. In LR rootstock, there was significantly increased expression of LEA2 under severe salinity (T2) only in roots (2.4-fold increase) (Figure 4).
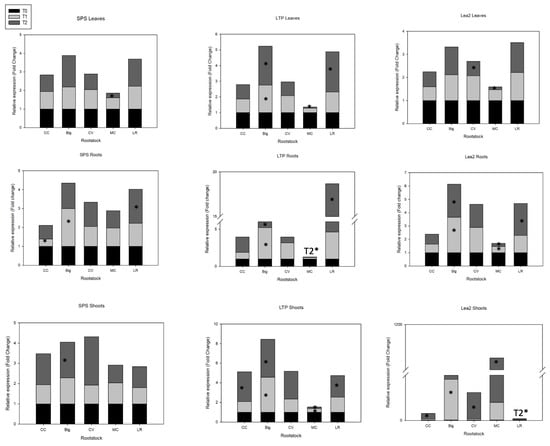
Figure 4.
Expression profiles of SPS, LTP, and LEA2 genes in ‘Hernandina’ clementine (Citrus clementina hort. ex Tanaka) (willowleaf mandarin (C. deliciosa) × sweet orange (C. sinensis (L.) Osbeck)) grafted on 5 citrus rootstocks in response to salt. Big: sour orange (C. aurantium L.); CC: Carrizo citrange (C. sinensis × Poncirus trifoliata (L.) Raf); CV: Volkamer lemon (C. volkameriana V. Ten. & Pasq.); LR: Rangpur lime (C. limonia Osbeck); MC: Cleopatra mandarin (Citrus reshni hort. ex Tanaka). For each combination (rootstock/’Hernandina’), stacked panels showed gene expression in plant tissues (leaves, shoots, and roots) treated with NaCl (T1: 35 mM NaCl; T2: 70 mM NaCl) relative to untreated samples (T0, only nutrient solution without salt treatment). Relative gene expression was calculated using 2−ΔΔCT method (fold change). * Significant differences for p values < 0.05.
Late embryogenesis abundant (LEA) proteins are a large and varied class of proteins that are thought to perform in normal plant growth and development and in cell protection against abiotic stress [136,137]. LEA proteins play a key role in plant drought tolerance and have been positively correlated with several abiotic stresses in many plants, such as brassicas [138], cotton [139], and rice [140]. Several investigations on particular group 2 LEA proteins have demonstrated that they are present in practically all vegetative tissues [141]. Their expression and accumulation is triggered by seed desiccation and various abiotic stressors, including water deficiency and cold [142,143]. We showed that increased expression of LEA2 proteins was observed mainly in T2 salt treatment. This argues that for the studied rootstock/‘Hernandina’ combinations, activation of the LEA2 pathway is involved in the response to severe saline stress.
In this context, the potential role of Solanum tuberosum (L.) LEA (StLEA) genes in abiotic stress was recently investigated. For the investigation, 17 candidate genes with high expression levels were selected, and their expression in potato roots and leaves after drought, salt, heavy metal, and high and low temperature treatment was analyzed by qRT-PCR [136]. The response of LEA genes to NaCl stress was similar to the response to drought stress. As in our study, StLEA genes had differential expression in different tissues and in response to stress.
3.4.2. Sucrose Phosphate Synthase Gene
Salinity had no significant impact on sucrose phosphate synthase (SPS, isoform 1) gene expression in leaves, except a slight repression in MC (T2). SPS gene expression in roots increased in Big (2-fold increase) in response to moderate salinity (T1), and in LR (1.8-fold increase) under severe salinity (T2). Only Big exhibited significantly higher SPS gene expression in shoots (1.8-fold increase) under severe salinity (T2) compared to control.
In plants, SPS genes are encoded by a gene family [144], and although several SPS genes have been studied for their functions in growth and development, their specific reactions to environmental stressors are mostly unknown, and information on their activity under stress circumstances is limited. Indeed, the transcriptional control of SPS genes is complex, and it has been demonstrated that the promoter regions of distinct SPS genes confer expression that varies completely or partially across plant tissues and organs [58]. Studies in maize [145], A. thaliana [58], litchi fruits [146], apple [147], and tomato [148] have shown that different isoforms are expressed differentially according to developmental stage in various organs and tissues, and that they are either induced or repressed or insensitive to abiotic stresses. Similarly, the relation between tolerance to salinity and the activation of expression of different SPS isoforms was not clearly demonstrated; Solís-Guzmán et al. [58] reported an upregulation of AtSPS2F and AtSPS4F expression in A. thaliana under osmotic stress, but a downregulation of AtSPS1F and AtSPS3F expression.
The absence of any clear pattern that would coincide with the tolerance level of the rootstock can be explained, as we used primers to detect citrus homologs for SPS isoform 1. Besides that, for the three tissue types, the increased SPS gene expression was not higher than twofold compared to its expression under control conditions, and it was lower compared with the expression intensity of other genes (CCC1, CMO, APX, P5CS). This argues that SPS isoform 1 does not have a very important role in salt tolerance for all rootstock/‘Hernandina’ combinations.
3.4.3. Lipid Transfer Protein Gene
In comparison to the other rootstocks, sour orange showed a 13.5-fold increase in lipid transfer protein (LTP) overexpression in response to T1 salt treatment (35 mM NaCl), but only 3-fold in T2. LR displayed a less pronounced 2.5-fold increase in LTP mRNA transcripts in response to T2 salt treatment. The gene expression analysis of LTP gene in roots revealed significant increases in most rootstocks. Indeed, higher transcript levels of LTP gene were recorded for Big (4.2- and 5.2-fold increase with T1 and T2, respectively), CV (2-fold increase with T1), and LR (3.6- and 14-fold increase with T1 and T2, respectively). On the other hand, CC induced upregulation of this gene in roots (2-fold increase) and shoots (3-fold increase) when under severe salinity stress (T2), while MC showed downregulated LTP gene in all tissues and under salt stress.
LTPs are implicated in adaptation to salt and osmotic stress, according to gene expression data [60,65,149]. In A. thaliana, some LTP genes were activated by salt and osmotic stress but not by drought, indicating that changed LTP gene expression should result in variations in stress sensitivity. It was demonstrated that A. thaliana plants that overexpress LTP genes (LTP1, LTP3) had better growth on NaCl-containing media, implying a function in abiotic stress adaption [150]. Only a few papers indicate increased sensitivity to salt and osmotic stress as a result of altered LTP gene expression. So far, LTP mutants have shown some modifications in their susceptibility to salt stress, but there is no explanation for how LTPs mediate this impact [151,152].
4. Conclusions
In this study, we analyzed the expression of potential genes involved in salt tolerance in citrus and provided insights on the regulation of these genes according to the rootstock used, the salinity level, and the tissue.
We observed that, independent of the tissue, the highest values of increased upregulated genes were obtained with LR, MC, and sour orange, followed by CV and CC. This coincides with the salt tolerance ranking of these rootstocks. LR rootstock was remarkable, as almost all studied genes showed increased expression upon salt stress. This rootstock is known to be very tolerant to salinity [7,39,153], and our findings support that this resistance is the result of the activation of various pathways tackling salt-induced ionic, osmotic, and oxidative stresses.
Big rootstock showed significant gene expression activation in most of the genes. This was particularly the case for sodium-related pathways (SOS1, KHT1, NHX1, V-PPiase). Indeed, it was reported that sour orange is a good Na+ excluder [76] and, thus, a salt-tolerant rootstock [7]. Big is also a Cl− excluder. However, we report only a slight increase in CCC1 gene expression. This might indicate that this gene, with other chloride regulatory mechanisms, could be involved in the regulation of Cl− homeostasis of the whole plant in Big.
MC is considered to be one of the best Cl− excluders and a very salt-tolerant rootstock [39,153]. In our assay, when grafted with ‘Hernandina’ clementine, MC showed very high induction of CCC1 gene in leaves and remarkable expression of NHX1 gene in all tissues. Almost all other genes were either not induced or repressed, suggesting that in MC, the Na+ sequestration pathway and Cl− homeostasis are the major mechanisms responsible for salt tolerance.
CV showed high activation of CCC1 expression in shoots. Sodium homeostasis genes (SOS1, HKT1, NHX1, V-PPiase) were also activated in shoots. This might indicate that this tissue is important for the salt tolerance of CV, potentially through recirculation and compartmentalization. In CC, the studied genes involved in ion homeostasis did not show a response to salt treatment, globally. For all other genes, only increased expression of APX and CMO in all tissues was observed. It is well documented that the ionic component is the most important in salt tolerance, and indeed, trifoliate orange and its hybrids, such as Carrizo citrange, are considered salt-sensitive [18,74].
This study provides insights on the differential expression of genes in citrus under moderate and high salt stress. It reveals that the major genes of functional groups responsible for encoding salt-stress proteins showing important induction of expression were those involved in ion transporter proteins (mainly NHX1 and HKT1 genes), Cl− homeostasis (CCC1 gene), biosynthesis and accumulation of compatible osmolytes, proline (P5CS gene) and glycine betaine (CMO gene), accumulation of proteins (LEA2 gene), and ROS scavenging antioxidant activity (mainly APX). The expression patterns could explain the relative tolerance of the used rootstocks. However, further investigations should be carried out on morphological measurements and ion content, uptake, and transport rate in different plant tissues in order to more deeply understand and confirm the research findings and the extent to which these genes participate in the whole process of salt tolerance.
Author Contributions
Conceptualization, H.S., A.N. and W.E.K.; Data curation, H.S., D.N. and W.E.K.; Formal analysis, H.S., D.N. and W.E.K.; Funding acquisition, H.S. and A.A.; Investigation, H.S., H.A., A.A., A.N. and W.E.K.; Methodology, H.S., H.A., I.O., A.A. and A.N.; Project administration, A.A. and A.N.; Resources, A.A. and A.N.; Software, D.N. and W.E.K.; Supervision, H.S., A.A. and W.E.K.; Validation, H.S., H.A., A.N. and W.E.K.; Writing—original draft, H.S.; Writing—review and editing, H.S., H.A., A.A., A.N. and W.E.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the 2017 Norman E. Borlaug International Agricultural Science and Technology Fellowship Program (Borlaug Fellowship) funded by the U.S. Department of Agriculture (USDA). The APC was supported by Florida A&M University.
Institutional Review Board Statement
Not applicable.
Acknowledgments
This work was supported by the Ministry of Research and Higher Education of Tunisia and by the Borlaug Program of the US Department of Agriculture. The authors would also like to thank Mabrouka Company, Khelidia, Tunisia, for assistance with the development of plant material.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Singh, A.; Sharma, P.C. Recent insights into physiological and molecular regulation of salt stress in fruit crops. Adv. Plants Agric. Res. 2018, 8, 171–183. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.; Zhu, J.-K. Salt stress signaling and mechanisms of plant salt tolerance. In Genetic Engineering, Principles and Methods; Setlow, J.K., Hollaender, A., Eds.; Springer Science+Business Media, Inc.: New York, NY, USA, 2006; Volume 27, pp. 141–177. [Google Scholar]
- Syvertsen, J.P.; Garcia-Sanchez, F. Multiple abiotic stresses occurring with salinity stress in citrus. Environ. Exp. Bot. 2014, 103, 128–137. [Google Scholar] [CrossRef]
- Machado, R.M.A.; Serralheiro, R.P. Soil Salinity: Effect on Vegetable Crop Growth. Management Practices to Prevent and Mitigate Soil Salinization. Horticulturae 2017, 3, 30. [Google Scholar] [CrossRef]
- Ruiz, M.; Pensabene-Bellavia, G.; Quiñones, A.; García-Lor, A.; Morillon, R.; Ollitrault, P.; Primo-Millo, E.; Navarro, L.; Aleza, P. Molecular Characterization and Stress Tolerance Evaluation of New Allotetraploid Somatic Hybrids Between Carrizo Citrange and Citrus macrophylla W. rootstocks. Front. Plant Sci. 2018, 9, 901. [Google Scholar] [CrossRef]
- Najar, A.; Hamdi, I.; Mahmoud, S.; Medhioub, L.; Jaouadi, I.; Varsani, A.; Jemmali, A. First report of citrus tristeza virus in commercial citrus orchards in Tunisia. J. Plant Pathol. 2021, 103, 1051–1052. [Google Scholar] [CrossRef]
- Cimen, B.; Yesiloglu, T. Rootstock Breeding for Abiotic Stress Tolerance in Citrus. In Abiotic and Biotic Stress in Plants-Recent Advances and Future Perspectives; Shanker, A., Ed.; IntechOpen: London, UK, 2016; pp. 527–563. [Google Scholar]
- Liu, X.-Y.; Li, J.; Liu, M.-M.; Yao, Q.; Chen, J.-Z. Transcriptome Profiling to Understand the Effect of Citrus Rootstocks on the Growth of ‘Shatangju’ Mandarin. PLoS ONE 2017, 12, e0169897. [Google Scholar] [CrossRef] [Green Version]
- Domingues, A.R.; Marcolini, C.D.M.; da Silva Gonçalves, C.H.; de Resende, J.T.V.; Roberto, S.R.; Carlos, E.F. Rootstocks Genotypes Impact on Tree Development and Industrial Properties of ‘Valencia’ Sweet Orange Juice. Horticulturae 2021, 7, 141. [Google Scholar] [CrossRef]
- Liu, X.; Li, J.; Huang, M.; Chen, J. Mechanisms for the influence of citrus rootstocks on fruit size. J. Agric. Food Chem. 2015, 63, 2618–2627. [Google Scholar] [CrossRef]
- García-Muñoz, M.C.; Henao-Rojas, J.C.; Moreno-Rodríguez, J.M.; Botina-Azain, B.L.; Romero-Barrera, Y. Effect of rootstock and environmental factors on fruit quality of Persian lime (Citrus latifolia Tanaka) grown in tropical regions. J. Food Compos. Anal. 2021, 103, 104081. [Google Scholar] [CrossRef]
- Alfaro, J.M.; Bermejo, A.; Navarro, P.; Quiñones, A.; Salvador, A. Effect of Rootstock on Citrus Fruit Quality: A Review. Food Rev. Int. 2021, 1–9. [Google Scholar] [CrossRef]
- Storey, R.; Walker, R.R. Citrus and salinity. Sci. Hortic. 1999, 78, 39–81. [Google Scholar] [CrossRef]
- Ziogas, V.; Tanou, G.; Morianou, G.; Kourgialas, N. Drought and Salinity in Citriculture: Optimal Practices to Alleviate Salinity and Water Stress. Agronomy 2021, 11, 1283. [Google Scholar] [CrossRef]
- Levy, Y.; Syvertsen, J.P. Irrigation water quality and salinity effects in citrus trees. Hort. Rev. 2004, 30, 37–82. [Google Scholar]
- Camara-Zapata, J.M.; Garcίa-Sánchez, F.; Martinez, V.; Nieves, M.; Cerdá, A. Effect of NaCl on citrus cultivars. Agronomie 2004, 24, 155–160. [Google Scholar] [CrossRef]
- Balal, R.M.; Ashraf, M.Y.; Khan, M.M.; Jaskani, M.J.; Ashfaq, M. Influence of salt stress on growth and biochemical parameters of citrus rootstocks. Pak. J. Bot. 2011, 43, 2135–2141. [Google Scholar]
- Balal, R.M.; Khan, M.M.; Shahid, M.A.; Mattson, N.S.; Abbas, T.; Ashfaq, M.; Garcia-Sanchez, F.; Ghazanfer, U.; Gimeno, V.; Iqbal, Z. Comparative Studies on the Physiobiochemical, Enzymatic, and Ionic modifications in Salt-tolerant and Salt-sensitive Citrus Rootstocks under NaCl Stress. J. Am. Soc. Hort. Sci. 2012, 137, 86–95. [Google Scholar] [CrossRef] [Green Version]
- Yildiz, M.; Poyraz, I.; Çavdar, A.; Özgen, Y.; Beyaz, R. Plant Responses to Salt Stress. In Plant Breeding—Current and Future Views; Abdurakhmonov, I.Y., Ed.; IntechOpen: London, UK, 2020; pp. 1–18. [Google Scholar] [CrossRef]
- Munns, R. Comparative physiology of salt and water stress. Plant Cell Environ. 2002, 25, 239–250. [Google Scholar] [CrossRef]
- Zhu, J.K. Plant Salt Stress. In Encyclopedia of Life Sciences; O’Daly, A., Ed.; Wiley: Chichester, UK, 2007; pp. 1–3. [Google Scholar]
- Carillo, P.; Annunziata, M.G.; Pontecorvo, G.; Fuggi, A.; Woodrow, P. Salinity Stress and Salt Tolerance. In Abiotic Stress in Plants—Mechanisms and Adaptations; Shanker, A., Venkateswarlu, B., Eds.; IntechOpen: London, UK, 2011; Chapter 2; pp. 1–21. [Google Scholar]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Ann. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [Green Version]
- Tanou, G.; Job, C.; Rajjou, L.; Arc, E.; Belghazi, M.; Diamantidis, G.; Molassiotis, A.; Job, D. Proteomics reveals the overlapping roles of hydrogen peroxide and nitric oxide in the acclimation of citrus plants to salinity. Plant J. 2009, 60, 795–804. [Google Scholar] [CrossRef]
- Hossain, M.A.; Bhattacharjee, S.; Armin, S.-M.; Qian, P.; Xin, W.; Li, H.-Y.; Burritt, D.J.; Fujita, M.; Tran, L.-S.P. Hydrogen peroxide priming modulates abiotic oxidative stress tolerance: Insights from ROS detoxification and scavenging. Front. Plant Sci. 2015, 6, 420. [Google Scholar] [CrossRef] [Green Version]
- de la Garma, J.G.; Fernandez-Garcia, N.; Bardisi, E.; Pallol, B.; Asensio-Rubio, J.S.; Bru, R.; Olmos, E. New insights into plant salt acclimation: The roles of vesicle trafficking and reactive oxygen species signalling in mitochondria and the endomembrane system. New Phytol. 2015, 205, 216–239. [Google Scholar] [CrossRef]
- Molassiotis, A.; Job, D.; Ziogas, V.; Tanou, G. Citrus Plants: A Model System for Unlocking the Secrets of NO and ROS-Inspired Priming Against Salinity and Drought. Front. Plant Sci. 2016, 7, 229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, P.; Jha, A.B.; Dubey, R.S.; Pessarakli, M. Reactive Oxygen Species, Oxidative Damage, and Antioxidative Defense Mechanism in Plants under Stressful Conditions. J. Bot. 2012, 2012, 217037. [Google Scholar] [CrossRef] [Green Version]
- Krasensky, J.; Jonak, C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 2012, 63, 1593–1608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, R.; Zhang, J.; Ma, Y.; Pan, X.; Dong, C.; Pang, S.; He, S.; Deng, L.; Yi, S.; Zheng, Y.; et al. Combined analysis of mRNA and miRNA identifies dehydration and salinity responsive key molecular players in citrus roots. Sci. Rep. 2017, 7, 42094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brumós, J.; Colmenero-Flores, J.M.; Conesa, A.; Izquierdo, P.; Sánchez, G.; Iglesias, D.J.; López-Climent, M.F.; Gómez-Cadenas, A.; Talón, M. Membrane transporters and carbon metabolism implicated in chloride homeostasis differentiate salt stress responses in tolerant and sensitive Citrus rootstocks. Funct. Integr. Genom. 2009, 9, 293–309. [Google Scholar] [CrossRef]
- López-Climent, M.F.; Arbona, V.; Pérez-Clemente, R.M.; Gómez-Cadenas, A. Relationship between salt tolerance and photosynthetic machinery performance in Citrus. Environ. Exp. Bot. 2008, 62, 176–184. [Google Scholar] [CrossRef]
- Apse, M.P.; Blumwald, E. Na+ transport in plants. FEBS Lett. 2007, 581, 2247–2254. [Google Scholar] [CrossRef] [Green Version]
- Colmenero-Flores, J.M.; Arbona, V.; Morillon, R.; Gomez-Cadenas, A. Salinity and water deficit. In The Genus Citrus, 1st ed.; Talon, M., Caruso, M., Gmitter, F.G., Jr., Eds.; Elsevier: Oxford, UK, 2020; Chapter 14; pp. 291–309. [Google Scholar]
- Colmenero-Flores, J.M.; Martínez, G.; Gamba, G.; Vázquez, N.; Iglesias, D.J.; Brumós, J.; Talón, M. Identification and functional characterization of cation–chloride cotransporters in plants. Plant J. 2007, 50, 278–292. [Google Scholar] [CrossRef]
- Brumós, J.; Talón, M.; Bouhlal, R.; Colmenero-Flores, J.M. Cl- homeostasis in includer and excluder citrus rootstocks: Transport mechanisms and identification of candidate genes. Plant Cell Environ. 2010, 33, 2012–2027. [Google Scholar] [CrossRef]
- Mahmoud, L.M.; Huyck, P.J.; Vincent, C.I.; Gmitter, F.G., Jr.; Grosser, J.W.; Dutt, M. Physiological Responses and Gene Expression Patterns in Open-Pollinated Seedlings of a Pummelo-Mandarin Hybrid Rootstock Exposed to Salt Stress and Huanglongbing. Plants 2021, 10, 1439. [Google Scholar] [CrossRef] [PubMed]
- Volkov, V.; Beilby, M.J. Editorial: Salinity Tolerance in Plants: Mechanisms and Regulation of Ion Transport. Front. Plant Sci. 2017, 8, 1795. [Google Scholar] [CrossRef] [PubMed]
- Şahin-Çevik, M.; Çevik, B.; Coşkan, A. Identification and Expression Analysis of Salinity-induced Genes in Rangpur lime (Citrus limonia). Hortic. Plant J. 2020, 6, 267–276. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018, 217, 523–539. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Li, Q. Effect of Environmental Salt Stress on Plants and the Molecular Mechanism of Salt Stress Tolerance. Int. J. Environ. Sci. Nat. Res. 2017, 7, 555714. [Google Scholar] [CrossRef]
- Munns, R. Genes and salt tolerance: Bringing them together. New Phytol. 2005, 167, 645–663. [Google Scholar] [CrossRef]
- Xie, R.; Pan, X.; Zhang, J.; Ma, Y.; He, S.; Zheng, Y.; Ma, Y. Effect of salt-stress on gene expression in citrus roots revealed by RNA-seq. Funct. Integr. Genom. 2018, 18, 155–173. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.K. Plant salt tolerance. In Plant Responses to Abiotic Stress; Topics in Current Genetics; Hirt, H., Shinozaki, K., Eds.; Springer: Berlin/Heidelberg, Germany, 2003; Volume 4, pp. 241–270. [Google Scholar]
- Noreen, Z.; Ashraf, M. Assessment of variation in antioxidative defense system in salt-treated pea (Pisum sativum) cultivars and its putative use as salinity tolerance markers. J. Plant Physiol. 2009, 166, 1764–1774. [Google Scholar] [CrossRef]
- Khalid, M.F.; Morillon, R.; Anjum, M.A.; Ejaz, S.; Rao, M.J.; Ahmad, S.; Hussain, S. Volkamer Lemon Tetraploid Rootstock Transmits the Salt Tolerance When Grafted with Diploid Kinnow Mandarin by Strong Antioxidant Defense Mechanism and Efficient Osmotic Adjustment. J. Plant Growth Regul. 2021, 1–13. [Google Scholar] [CrossRef]
- Ashfaque, F.; Iqbal, M.; Khan, R.; Khan, N.A. Exogenously applied H2O2 promotes proline accumulation, water relations, photosynthetic efficiency and growth of wheat (Triticum aestivum L.) under salt stress. Annu. Res. Rev. Biol. 2014, 4, 105–120. [Google Scholar] [CrossRef]
- Nazar, R.; Iqbal, N.; Syeed, S.; Khan, N.A. Salicylic acid alleviates decreases in photosynthesis under salt stress by enhancing nitrogen and sulfur assimilation and antioxidant metabolism differentially in two mungbean cultivars. J. Plant Physiol. 2011, 168, 807–815. [Google Scholar] [CrossRef] [PubMed]
- AbdElgawad, H.; Zinta, G.; Hegab, M.M.; Pandey, R.; Asard, H.; Abuelsoud, W. High Salinity Induces Different Oxidative Stress and Antioxidant Responses in Maize Seedlings Organs. Front. Plant Sci. 2016, 7, 276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blumwald, E. Engineering salt tolerance in plants. Biotechnol. Genetic Eng. Rev. 2013, 20, 261–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verslues, P.E.; Agarwal, M.; Katiyar-Agarwal, S.; Zhu, J.; Zhu, J.K. Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 2006, 45, 523–539. [Google Scholar] [CrossRef]
- Hong, Z.; Lakkineni, K.; Zhang, Z.; Verma, D.P. Removal of feedback inhibition of D1-pyrroline-5-carboxylate synthetase results in increased proline accumulation and protection of plants from osmotic stress. Plant Physiol. 2000, 122, 1129–1136. [Google Scholar] [CrossRef] [Green Version]
- Székely, G.; Abraham, E.; Cséplo, A.; Rigó, G.; Zsigmond, L.; Csiszár, J.; Ayaydin, F.; Strizhov, N.; Jásik, J.; Schmelzer, E.; et al. Duplicated p5cs genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J. 2008, 53, 11–28. [Google Scholar] [CrossRef] [Green Version]
- Annunziata, M.G.; Ciarmiello, L.F.; Woodrow, P.; Dell’Aversana, E.; Carillo, P. Spatial and Temporal Profile of Glycine Betaine Accumulation in Plants under Abiotic Stresses. Front. Plant Sci. 2019, 10, 230. [Google Scholar] [CrossRef] [Green Version]
- Mäkelä, P.S.A.; Munns, R.; Colmer, T.D.; Condon, A.G.; Peltonen-Sainio, P. Effect of foliar applications of glycinebetaine on stomatal conductance, abscisic acid and solute concentrations in leaves of salt- or drought-stressed tomato. Funct. Plant Biol. 1998, 25, 655–663. [Google Scholar] [CrossRef]
- Guo, L.; Yang, H.; Xiaoyan, Z.; Yang, S. Lipid transfer protein 3 as a target of MYB96 mediates freezing and drought stress in Arabidopsis. J. Exp. Bot. 2013, 64, 1755–1767. [Google Scholar] [CrossRef]
- Liu, F.; Zhang, X.; Lu, C.; Zeng, X.; Li, Y.; Fu, D.; Wu, G. Non-specific lipid transfer proteins in plants: Presenting new advances and an integrated functional analysis. J. Exp. Bot. 2015, 66, 5663–5681. [Google Scholar] [CrossRef] [Green Version]
- Solís-Guzmán, M.G.; Argüello-Astorga, G.; López-Bucio, J.; Ruiz-Herrera, L.F.; López-Meza, J.E.; Sánchez-Calderón, L.; Carreón-Abud, Y.; Martínez-Trujillo, M. Arabidopsis thaliana sucrose phosphate synthase (sps) genes are expressed differentially in organs and tissues, and their transcription is regulated by osmotic stress. Gene Expr. Patterns 2017, 25–26, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Jang, C.S.; Lee, H.J.; Chang, S.J.; Seo, Y.W. Expression and promoter analysis of the TaLTP1 gene induced by drought and salt stress in wheat (Triticum aestivum L.). Plant Sci. 2004, 167, 995–1001. [Google Scholar] [CrossRef]
- Wang, H.; Kwon, H.; Yim, W.; Lim, S.; Moon, J.; Lee, B.; Seo, Y.; Kim, W.; Jang, C. Expressional diversity of wheat nsLTP genes: Evidence of subfunctionalization via cis-regulatory divergence. Genetica 2010, 138, 843–852. [Google Scholar] [CrossRef] [PubMed]
- Shih, M.D.; Hoekstra, F.A.; Hsing, Y.I.C. Late Embryogenesis Abundant Proteins. Adv. Bot. Res. 2008, 48, 211–255. [Google Scholar] [CrossRef]
- Battaglia, M.; Olvera-Carrillo, Y.; Garciarrubio, A.; Campos, F.; Covarrubias, A.A. The Enigmatic LEA Proteins and Other Hydrophilins. Plant Physiol. 2008, 148, 6–24. [Google Scholar] [CrossRef] [Green Version]
- Duan, J.; Cai, W. OsLEA3-2, an abiotic stress induced gene of rice plays a key role in salt and drought tolerance. PLoS ONE 2012, 7, e45117. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Dias, M.C.; Freitas, H. Drought and Salinity Stress Responses and Microbe-Induced Tolerance in Plants. Front. Plant Sci. 2020, 11, 591911. [Google Scholar] [CrossRef]
- Choi, A.M.; Lee, S.B.; Cho, S.H.; Hwang, I.; Hur, C.G.; Suh, M.C. Isolation and characterization of multiple abundant lipid transfer protein isoforms in developing sesame (Sesamum indicum L.) seeds. Plant Physiol. Biochem. 2008, 46, 127–139. [Google Scholar] [CrossRef]
- Singh, A.; Prakash, J.; Srivastav, M.; Singh, S.K.; Awasthi, O.P.; Singh, A.K.; Chaudhari, S.K.; Sharma, D.K. Physiological and biochemical responses of citrus rootstocks under salinity stress. Indian J. Hort. 2014, 71, 162–167. [Google Scholar]
- Cimen, B.; Yesiloglu, T.; Yilmaz, B.; Incesu, M. Effects of different salinity levels on photosynthetic performances of some citrus rootstocks. Res. J. Agric. Sci. 2013, 6, 13–18. [Google Scholar]
- Behboudian, M.; Torokfalvy, E.; Walker, R. Effects of salinity on ionic content, water relations and gas exchange parameters in some citrus scion—Rootstock combinations. Sci. Hortic. 1986, 28, 105–116. [Google Scholar] [CrossRef]
- Iglesias, D.J.; Cercós, M.; Colmenero-Flores, J.M.; Naranjo, M.A.; Ríos, G.; Carrera, E.; Ruiz-Rivero, O.; Lliso, I.; Morillon, R.; Tadeo, F.R.; et al. Physiology of citrus fruiting. Braz. J. Plant Physiol. 2007, 19, 333–362. [Google Scholar] [CrossRef]
- Rodríguez-Gamir, J.; Ancillo, G.; Legaz, F.; Primo-Millo, E.; Forner-Giner, M.A. Influence of salinity on pip gene expression in citrus roots and its relationship with root hydraulic conductance, transpiration and chloride exclusion from leaves. Environ. Exp. Bot. 2012, 78, 163–166. [Google Scholar] [CrossRef]
- Mahmoud, L.M.; Dutt, M.; Vincent, C.I.; Grosser, J.W. Salinity-induced physiological responses of three putative salt tolerant citrus rootstocks. Horticulturae 2020, 6, 90. [Google Scholar] [CrossRef]
- Romero-Aranda, R.; Moya, J.L.; Tadeo, F.R.; Legaz, F.; Primo-Millo, E.; Talon, M. Physiological and anatomical disturbances induced by chloride salts in sensitive and tolerant citrus: Beneficial and detrimental effects of cations. Plant Cell Environ. 1998, 21, 1243–1253. [Google Scholar] [CrossRef]
- Walker, R.R.; Douglas, T.J. Effect of salinity level on uptake and distribution of chloride, sodium and potassium ions in Citrus plants. Aust. J. Agric. Res. 1983, 34, 145–153. [Google Scholar] [CrossRef]
- Hussain, S.; Morillon, R.; Muhammad, A.A.; Ollitrault, P.; Costantino, G.; Luro, F. Genetic diversity revealed by physiological behavior of citrus genotypes subjected to salt stress. Acta Physiol. Plant. 2015, 37, 1740. [Google Scholar] [CrossRef]
- Moya, J.L.; Gómez-Cadenas, A.; Primo-Millo, E.; Talon, M. Chloride absorption in salt-sensitive Carrizo citrange and salt-tolerant Cleopatra mandarin citrus rootstocks is linked to water use. J. Exp. Bot. 2003, 54, 825–833. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, P.; Syvertsen, J.P.; Etxeberria, E. Sodium distribution in salt-stressed citrus rootstock seedlings. HortScience 2012, 47, 1504–1511. [Google Scholar] [CrossRef] [Green Version]
- Hoagland, D.R.; Arnon, D.I. The water-culture method for growing plants without soil. Calif. Agric. Exp. Stn. Circ. 1950, 347, 1–32. [Google Scholar]
- Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988, 16, 10881–10890. [Google Scholar] [CrossRef] [PubMed]
- Koressaar, T.; Remm, M. Enhancements and modifications of primer design program Primer3. Bioinformatics 2007, 23, 1289–1291. [Google Scholar] [CrossRef] [Green Version]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Quintero, F.J.; Pardo, J.M.; Zhu, J.K. The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 2002, 14, 465–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quintero, F.J.; Martinez-Atienza, J.; Villalta, I.; Jiang, X.; Kim, W.-Y.; Ali, Z.; Fujii, H.; Mendoza, I.; Yun, D.-J.; Zhu, J.; et al. Activation of the plasma membrane Na/H antiporter Salt-Overly-Sensitive 1 (SOS1) by phosphorylation of an auto-inhibitory C-terminal domain. Proc. Natl. Acad. Sci. USA 2011, 108, 2611–2616. [Google Scholar] [CrossRef] [Green Version]
- Yang, Q.; Chen, Z.Z.; Zhou, X.F.; Yin, H.B.; Li, X.; Xin, X.F.; Hong, X.H.; Zhu, J.K.; Gong, Z. Overexpression of SOS (Salt Overly Sensitive) genes increases salt tolerance in transgenic Arabidopsis. Mol. Plant 2009, 2, 22–31. [Google Scholar] [CrossRef] [Green Version]
- Ma, D.M.; Xu, W.R.; Li, H.W.; Jin, F.X.; Guo, L.N.; Wang, J.; Da, H.J.; Xu, X. Co-expression of the Arabidopsis SOS genes enhances salt tolerance in transgenic tall fescue (Festuca arundinacea Schreb.). Protoplasma 2014, 251, 219–231. [Google Scholar] [CrossRef] [Green Version]
- Khoshbakht, D.; Ramin, A.A.; Baninasab, B. Citrus rootstocks response to salinity: Physio-biochemical parameters changes. Res. J. Environ. Sci. 2014, 8, 29–38. [Google Scholar] [CrossRef] [Green Version]
- Grosser, J.W.; Omar, A.A.; Gmitter, J.A.; Syvertsen, J.P. Salinity Tolerance of ‘Valencia’ Orange Trees on Allotetraploid Rootstocks. Proc. Fla. State Hort. Soc. 2012, 125, 50–55. [Google Scholar]
- Pardo, J.M. Biotechnology of water and salinity stress tolerance. Curr. Opin. Biotechnol. 2010, 21, 185–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez-Alcántara, B.; Martinez-Cuenca, M.R.; Quinones, A.; Iglesias, D.J.; Primo-Millo, E.; Forner-Giner, M.A. Comparative expression of candidate genes involved in sodium transport and compartmentation in citrus. Environ. Exp. Bot. 2015, 111, 52–62. [Google Scholar] [CrossRef]
- Ding, M.; Hou, P.; Shen, X.; Wang, M.; Deng, S.; Sun, J.; Xiao, F.; Wang, R.; Zhou, X.; Lu, C.; et al. Salt induced expression of genes related to Na+/K+ and ROS homeostasis in leaves of salt-resistant and salt sensitive poplar species. Plant Mol. Biol. 2010, 73, 251–269. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Jiang, X.; Zhan, K.; Cheng, X.; Chen, X.; Pardo, J.M.; Cui, D. Functional characterization of a wheat plasma membrane Na+/H+ antiporter in yeast. Arch. Biochem. Biophys. 2008, 473, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Brini, F.; Masmoudi, K. Ion Transporters and Abiotic Stress Tolerance in Plants. ISRN Mol. Biol. 2012, 3, 927436. [Google Scholar] [CrossRef]
- Blumwald, E. Sodium transport and salt tolerance in plants. Curr. Opin. Cell Biol. 2000, 12, 431–434. [Google Scholar] [CrossRef]
- Tian, N.; Wang, J.; Xu, Z.Q. Overexpression of Na+/H+ antiporter gene AtNHX1 from Arabidopsis thaliana improves the salt tolerance of kiwifruit (Actinidia deliciosa). S. Afr. J. Bot. 2011, 77, 160–169. [Google Scholar] [CrossRef] [Green Version]
- Zhao, F.Y.; Zhang, X.J.; Li, P.H.; Zhao, Y.X.; Zhang, H. Co-expression of the Suaeda salsa SsNHX1 and Arabidopsis AVP1 confer greater salt tolerance to transgenic rice than the single SsNHX1. Mol. Breed. 2006, 17, 341–353. [Google Scholar] [CrossRef]
- Leidi, E.O.; Barragán, V.; Rubio, L.; El-Hamdaoui, A.; Ruiz, M.T.; Cubero, B.; Fernández, J.A.; Bressan, R.A.; Hasegawa, P.M.; Quintero, F.J.; et al. The AtNHX1 exchanger mediates potassium compartmentation in vacuoles of transgenic tomato. Plant J. 2010, 61, 495–506. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.A.; Yang, G.D.; Meng, Q.W.; Zheng, C.C. The cotton GhNHX1 gene encoding a novel putative tonoplast Na+/H+ antiporter plays an important role in salt stress. Plant Cell Physiol. 2004, 45, 600–607. [Google Scholar] [CrossRef] [Green Version]
- Saqib, M.; Zorb, C.; Rengel, Z.; Schubert, S. The expression of the endogenous vacuolar Na+/H+ antiporters in roots and shoots correlates positively with the salt resistance of wheat (Triticum aestivum L.). Plant Sci. 2005, 169, 959–965. [Google Scholar] [CrossRef]
- Yokoi, S.; Quintero, F.J.; Cubero, B.; Ruiz, M.T.; Bressan, R.A.; Hasegawa, P.M.; Pardo, J.M. Differential expression and function of Arabidopsis thaliana NHX Na+/H+ antiporters in the salt stress response. Plant J. 2002, 30, 529–539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, S.; Jiang, P.; Nie, L.; Chen, X.; Tai, F.; Wang, D.; Fan, P.; Feng, J.; Bao, H.; Wang, J.; et al. H+-pyrophosphatase from Salicornia europaea confers tolerance to simultaneously occurring salt stress and nitrogen deficiency in Arabidopsis and wheat. Plant Cell Environ. 2015, 38, 2433–2449. [Google Scholar] [CrossRef]
- Hasegawa, P.M. Sodium (Na+) homeostasis and salt tolerance of plants. Environ. Exp. Bot. 2013, 92, 19–31. [Google Scholar] [CrossRef]
- Fu, L.; Wu, D.; Zhang, X.; Xu, Y.; Kuang, L.; Cai, S.; Zhang, G.; Shen, Q. Vacuolar H+-pyrophosphatase HVP10 enhances salt tolerance via promoting Na+ translocation into root vacuoles. Plant Physiol. 2021, 188, 1248–1263. [Google Scholar] [CrossRef] [PubMed]
- Gaxiola, R.A.; Li, J.; Undurraga, S.; Dang, L.M.; Allen, G.J.; Alper, S.L.; Fink, G.R. Drought- and salt-tolerant plants result from overexpression of the AVP1 H+-pump. Proc. Natl. Acad. Sci. USA 2001, 98, 11444–11449. [Google Scholar] [CrossRef] [Green Version]
- Lv, S.; Zhang, K.; Gao, Q.; Lian, L.; Song, Y.; Zhang, J. Overexpression of an H+-PPase Gene from Thellungiella halophila in Cotton enhances Salt Tolerance and Improves Growth and Photosynthetic Performance. Plant Cell Physiol. 2008, 49, 1150–1164. [Google Scholar] [CrossRef] [Green Version]
- Schilling, R.K.; Marschner, P.; Shavrukov, Y.; Berger, B.; Tester, M.; Roy, S.J.; Plett, D.C. Expression of the Arabidopsis vacuolar H⁺-pyrophosphatase gene (AVP1) improves the shoot biomass of transgenic barley and increases grain yield in a saline field. Plant Biotechnol. J. 2014, 12, 378–386. [Google Scholar] [CrossRef]
- Yang, Y.; Tang, R.J.; Li, B.; Wang, H.H.; Jin, Y.L.; Jiang, C.M.; Bao, Y.; Su, H.Y.; Zhao, N.; Ma, X.J.; et al. Overexpression of a Populus trichocarpa H+-pyrophosphatase gene PtVP1. 1 confers salt tolerance on transgenic poplar. Tree Physiol. 2015, 35, 663–677. [Google Scholar] [CrossRef] [Green Version]
- Gao, F.; Gao, Q.; Duan, X.; Yue, G.; Yang, A.; Zhang, J. Cloning of an H+-PPase gene from Thellungiella halophila and its heterologous expression to improve tobacco salt tolerance. J. Exp. Bot. 2006, 57, 3259–3270. [Google Scholar] [CrossRef] [Green Version]
- Jha, D.; Shirley, N.; Tester, M.; Roy, S.J. Variation in salinity tolerance and shoot sodium accumulation in Arabidopsis ecotypes linked to differences in the natural expression levels of transporters involved in sodium transport. Plant Cell Environ. 2010, 33, 793–804. [Google Scholar]
- Zhang, M.; Cao, Y.; Wang, Z.; Wang, Z.Q.; Shi, J.; Liang, X.; Song, W.; Chen, Q.; Lai, J.; Jiang, C. A retrotransposon in an HKT1 family sodium transporter causes variation of leaf Na+ exclusion and salt tolerance in maize. New Phytol. 2018, 217, 1161–1176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Platten, J.D.; Cotsaftis, O.; Berthomieu, P.; Bohnert, H.; Davenport, R.J.; Fairbairn, D.J.; Horie, T.; Leigh, R.A.; Lin, H.-X.; Luan, S.; et al. Nomenclature for HKT transporters, key determinants of plant salinity tolerance. Trends Plant Sci. 2006, 11, 372–374. [Google Scholar] [CrossRef]
- Waters, S.; Gilliham, M.; Hrmova, M. Plant high-affinity potassium (HKT) transporters involved in salinity tolerance: Structural insights to probe differences in ion selectivity. Int. J. Mol. Sci. 2013, 14, 7660–7680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Møller, I.S.; Gilliham, M.; Jha, D.; Mayo, G.M.; Roy, S.J.; Coates, J.C.; Haseloff, J.; Tester, M. Shoot Na+ exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na+ transport in Arabidopsis. Plant Cell 2009, 21, 2163–2178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davenport, R.J.; Muñoz-Mayor, A.; Jha, D.; Essah, P.A.; Rus, A.; Tester, M. The Na+ transporter AtHKT1;1 controls retrieval of Na+ from the xylem in Arabidopsis. Plant Cell Environ. 2007, 30, 497–507. [Google Scholar] [CrossRef]
- Levy, Y.; Lifshitz, J.; De Malach, Y.; David, Y. The Response of Several Citrus Genotypes to High-salinity Irrigation Water. HortScience 1999, 34, 878–881. [Google Scholar] [CrossRef]
- García-Sánchez, F.; Syvertsen, J.P.; Martínez, V.; Melgar, J.C. Salinity tolerance of ‘Valencia’ orange trees on rootstocks with contrasting salt tolerance is not improved by moderate shade. J. Expt. Bot. 2006, 57, 3697–3706. [Google Scholar] [CrossRef]
- Gamba, G. Molecular physiology and pathophysiology of electroneutral cation-chloride cotransporters. Physiol. Rev. 2005, 85, 423–493. [Google Scholar] [CrossRef]
- Franco-Navarro, J.D.; Rosales, M.A.; Cubero-Font, P.; Calvo, P.; Álvarez, R.; Diaz-Espejo, A.; Colmenero-Flores, J.M. Chloride as macronutrient increases water use efficiency by anatomically-driven reduced stomatal conductance and increased mesophyll diffusion to CO2. Plant J. 2019, 99, 815–831. [Google Scholar]
- Martins, C.P.S.; Pedrosa, A.M.; Du, D.; Gonçalves, L.P.; Yu, Q.; Gmitter, F.G., Jr.; Costa, M.G.C. Genome-wide characterization and expression analysis of major intrinsic proteins during abiotic and biotic stresses in sweet orange (Citrus sinensis L. Osb.). PLoS ONE 2015, 10, e0138786. [Google Scholar] [CrossRef]
- Syvertsen, J.P.; Melgar, J.C.; García-Sánchez, F. Salinity Tolerance and Leaf Water Use Efficiency in Citrus. J. Am. Soc. Hortic. Sci. 2010, 135, 33–39. [Google Scholar] [CrossRef] [Green Version]
- Seday, U.; Gulsen, O.; Uzun, A.; Toprak, G. Response of citrus rootstocks to different salinity levels for morphological and antioxidative enzyme activites. J. Anim. Plant Sci. 2014, 24, 512–520. [Google Scholar]
- Mittova, V.; Tal, M.; Volokita, M.; Guy, M. Up-regulation of the leaf mitochondrial and peroxisomal antioxidative systems in response to salt-induced oxidative stress in the wild salt-tolerant tomato species Lycopersicon pennellii. Plant Cell Environ. 2003, 26, 845–856. [Google Scholar] [CrossRef] [PubMed]
- Sekmen, A.H.; Türkan, I.; Takio, S. Differential responses of antioxidative enzymes and lipid peroxidation to salt stress in salt-tolerant Plantago maritima and salt-sensitive Plantago Media. Physiol. Plant 2007, 131, 399–411. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.A.; Silva, J.A.T.d.; Fujita, M. Glyoxalase system and reactive oxygen species detoxification system in plant abiotic stress response and tolerance: An intimate relationship. In Abiotic Stress in Plants: Mechanisms and Adaptations; Shanker, A., Venkateswarlu, B., Eds.; IntechOpen: London, UK, 2011; Chapter 11; pp. 235–266. [Google Scholar]
- Hasanuzzaman, M.; Hossain, M.A.; Silva, J.A.T.D.; Fujita, M. Plant Response and Tolerance to Abiotic Oxidative Stress: Antioxidant Defense Is a Key Factor. In Crop Stress and Its Management: Perspectives and Strategies; Venkateswarlu, B., Shanker, A., Shanker, C., Maheswari, M., Eds.; Springer: Dordrecht, The Netherlands, 2012; pp. 261–315. [Google Scholar]
- Clegg, M.T.; Rawson, J.R.; Thomas, K. Chloroplast DNA variation in pearl millet and related species. Genetics 1984, 106, 449–461. [Google Scholar] [CrossRef]
- Acosta-Motos, J.R.; Ortuño, M.F.; Bernal-Vicente, A.; Diaz-Vivancos, P.; Sanchez-Blanco, M.J.; Hernandez, J.A. Plant Responses to Salt Stress: Adaptive Mechanisms. Agronomy 2017, 7, 18. [Google Scholar] [CrossRef] [Green Version]
- Balfagón, D.; Terán, F.; de Oliveira, T.D.R.; Santa-Catarina, C.; Gómez-Cadenas, A. Citrus rootstocks modify scion antioxidant system under drought and heat stress combination. Plant Cell Rep. 2021, 1–10. [Google Scholar] [CrossRef]
- Apse, M.P.; Blumwald, E. Engineering salt tolerance in plants. Curr. Opin. Biotechnol. 2002, 13, 146–150. [Google Scholar] [CrossRef]
- Shirasawa, K.; Takabe, T.; Takabe, T.; Kishitani, S. Accumulation of glycinebetaine in rice plants that overexpress choline monooxygenase from spinach and evaluation of their tolerance to abiotic stress. Ann. Bot. 2006, 98, 565–571. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Tan, W.; Yang, X.H.; Zhang, H.X. Plastid-expressed choline monooxygenase gene improves salt and drought tolerance through accumulation of glycine betaine in tobacco. Plant Cell Rep. 2008, 27, 1113–1124. [Google Scholar] [CrossRef] [PubMed]
- Surekha, C.; Kumari, K.N.; Aruna, L.V.; Suneetha, G.; Arundhati, A.; Kavi Kishor, P.B. Expression of the Vigna aconitifolia P5CSF129A gene in transgenic pigeonpea enhances proline accumulation and salt tolerance. Plant Cell Tissue Organ Cult. (PCTOC) 2014, 116, 27–36. [Google Scholar] [CrossRef]
- El-Habashy, S. In vitro Evaluation and Selection for Salinity Tolerance in Some Citrus Rootstock Seedlings. J. Hortic. Sci. Ornam. Plants 2018, 10, 17–27. [Google Scholar]
- Shafieizargar, A.; Awang, Y.; Ajamgard, F.; Juraimi, A.S.; Othman, R.; Ahmadi, A.K. Assessing Five Citrus Rootstocks for NaCl Salinity Tolerance Using Mineral Concentrations, Proline and Relative Water Contents as Indicators. Asian J. Plant Sci. 2015, 14, 20–26. [Google Scholar] [CrossRef]
- Ashraf, M.; Foolad, M.R. Roles of Glycine Betaine and Proline in Improving Plant Abiotic Stress Resistance. Environ. Exp. Bot. 2007, 59, 206–216. [Google Scholar] [CrossRef]
- Lv, X.; Chen, S.; Wang, Y. Advances in Understanding the Physiological and Molecular Responses of Sugar Beet to Salt Stress. Front. Plant Sci. 2019, 10, 1431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Li, C.; Zhang, B.; Yi, J.; Yang, Y.; Kong, C.; Lei, C.; Gong, M. The Role of the Late Embryogenesis-Abundant (LEA) Protein Family in Development and the Abiotic Stress Response: A Comprehensive Expression Analysis of Potato (Solanum Tuberosum). Genes 2019, 10, 148. [Google Scholar] [CrossRef] [Green Version]
- Hundertmark, M.; Hincha, D.K. LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics. 2008, 9, 118. [Google Scholar] [CrossRef] [Green Version]
- Dalal, M.; Tayal, D.; Chinnusamy, V.; Bansal, K.C. Abiotic stress and ABA-inducible Group 4 LEA from Brassica napus plays a key role in salt and drought tolerance. J. Biotechnol. 2009, 139, 137–145. [Google Scholar] [CrossRef]
- Magwanga, R.O.; Lu, P.; Kirungu, J.N.; Dong, Q.; Hu, Y.; Zhou, Z.; Cai, X.; Wang, X.; Hou, Y.; Wang, K.; et al. Cotton Late Embryogenesis Abundant (LEA2) Genes Promote Root Growth and Confer Drought Stress Tolerance in Transgenic Arabidopsis thaliana. Genes Genomes Genet. 2018, 8, 2781–2803. [Google Scholar] [CrossRef] [Green Version]
- He, S.; Tan, L.; Hu, Z.; Chen, G.; Wang, G.; Hu, T. Molecular characterization and functional analysis by heterologous expression in E. coli under diverse abiotic stresses for OsLEA5, the atypical hydrophobic LEA protein from Oryza sativa L. Mol. Genet. Genom. 2012, 287, 39–54. [Google Scholar] [CrossRef]
- Nylander, M.; Svensson, J.; Palva, E.T.; Welin, B.V. Stress-induced accumulation and tissue-specific localization of dehydrins in Arabidopsis thaliana. Plant Mol. Biol. 2001, 45, 263–279. [Google Scholar] [CrossRef] [PubMed]
- Hara, M.; Terashima, S.; Kuboi, T. Characterization and cryoprotective activity of cold-responsive dehydrin from Citrus unshiu. J. Plant Physiol. 2001, 58, 1333–1339. [Google Scholar] [CrossRef]
- Sivamani, E.; Bahieldin, A.; Wraith, J.M.; Al-Niemi, T.; Dyer, W.E.; Ho, T.D.; Qu, R. Improved biomass productivity and water use efficiency under water deficit conditions in transgenic wheat constitutively expressing the barley HVA1 gene. Plant Sci. 2000, 155, 1–9. [Google Scholar] [CrossRef]
- Jiang, S.Y.; Chi, Y.H.; Wang, J.Z.; Zhou, J.X.; Cheng, Y.S.; Zhang, B.L.; Ma, A.; Vanitha, J.; Ramachandran, S. Sucrose metabolism gene families and their biological functions. Sci. Rep. 2015, 5, 17583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lutfylla, L.L.; Xu, N.; D’Ordine, R.L.; Morrell, J.A.; Miller, P.W.; Duff, S.M. Phylogenetic and expression analysis of sucrose phosphate synthase isozymes in plants. J. Plant Physiol. 2007, 164, 923–933. [Google Scholar]
- Wang, D.; Zhao, J.; Hu, B.; Li, J.; Qin, Y.; Chen, L.; Qin, Y.; Hu, G. Identification and expression profile analysis of the sucrose phosphate synthase gene family in Litchi chinensis Sonn. PeerJ. 2018, 6, e4379. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Zhang, J.; Li, C.; Zhang, Z.; Ma, F.; Li, M. Response of sugar metabolism in apple leaves subjected to short-term drought stress. Plant Physiol. Biochem. 2019, 141, 164–171. [Google Scholar] [CrossRef]
- Duan, Y.; Yang, L.; Zhu, H.; Zhou, J.; Sun, H.; Gong, H. Structure and Expression Analysis of Sucrose Phosphate Synthase, Sucrose Synthase and Invertase Gene Families in Solanum lycopersicum. Int. J. Mol. Sci. 2021, 22, 4698. [Google Scholar] [CrossRef]
- Pitzschke, A.; Datta, S.; Persak, H. Salt stress in Arabidopsis: Lipid transfer protein AZI1 and its control by mitogen-activated protein kinase MPK3. Mol. Plant 2014, 7, 722–738. [Google Scholar] [CrossRef] [Green Version]
- Jülke, S.; Ludwig-Müller, J. Response of Arabidopsis thaliana Roots with Altered Lipid Transfer Protein (LTP) Gene Expression to the Clubroot Disease and Salt Stress. Plants 2016, 5, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, H.W.; Kim, W.; Hwang, B.K. Three pathogen-inducible genes encoding lipid transfer protein from pepper are differentially activated by pathogens, abiotic, and environmental stresses. Plant. Cell Environ. 2003, 26, 915–928. [Google Scholar] [CrossRef] [PubMed]
- Safi, H.; Saibi, W.; Alaoui, M.M.; Hmyene, A.; Masmoudi, K.; Hanin, M.; Brini, F. A wheat lipid transfer protein (TdLTP4) promotes tolerance to abiotic and biotic stress in Arabidopsis thaliana. Plant Physiol. Biochem. 2015, 89, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Bitters, W.P. Citrus Rootstocks: Their characters and reactions. J. Citrus Pathol. 2021, 8, 239. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).