Genomic Instability in Somatic Hybridization between Poncirus and Citrus Species Aiming to Create New Rootstocks
Abstract
:1. Introduction
2. Materials and Methods
2.1. Somatic Hybridization and Plant Regeneration
2.2. Ploidy Analysis by Flow Cytometry
2.3. GBS
2.4. Genetic Analysis
2.5. Interspecific Phylogenomic Mosaic Structures along the Nuclear Genome
3. Results
3.1. Plant Regeneration from Protoplast Fusion
3.2. Ploidy Estimation by Flow Cytometry
3.3. Inheritance of Mitochondrial Genome
3.4. Inheritance of Chloroplastic Genome
3.5. Inheritance of Nuclear Genomes
3.5.1. SNP Mining
3.5.2. First Approximation of the Nuclear Inheritance by Factorial Analysis of SNP Data
3.5.3. Search for Diagnostic SNPs of Ancestral Genomes
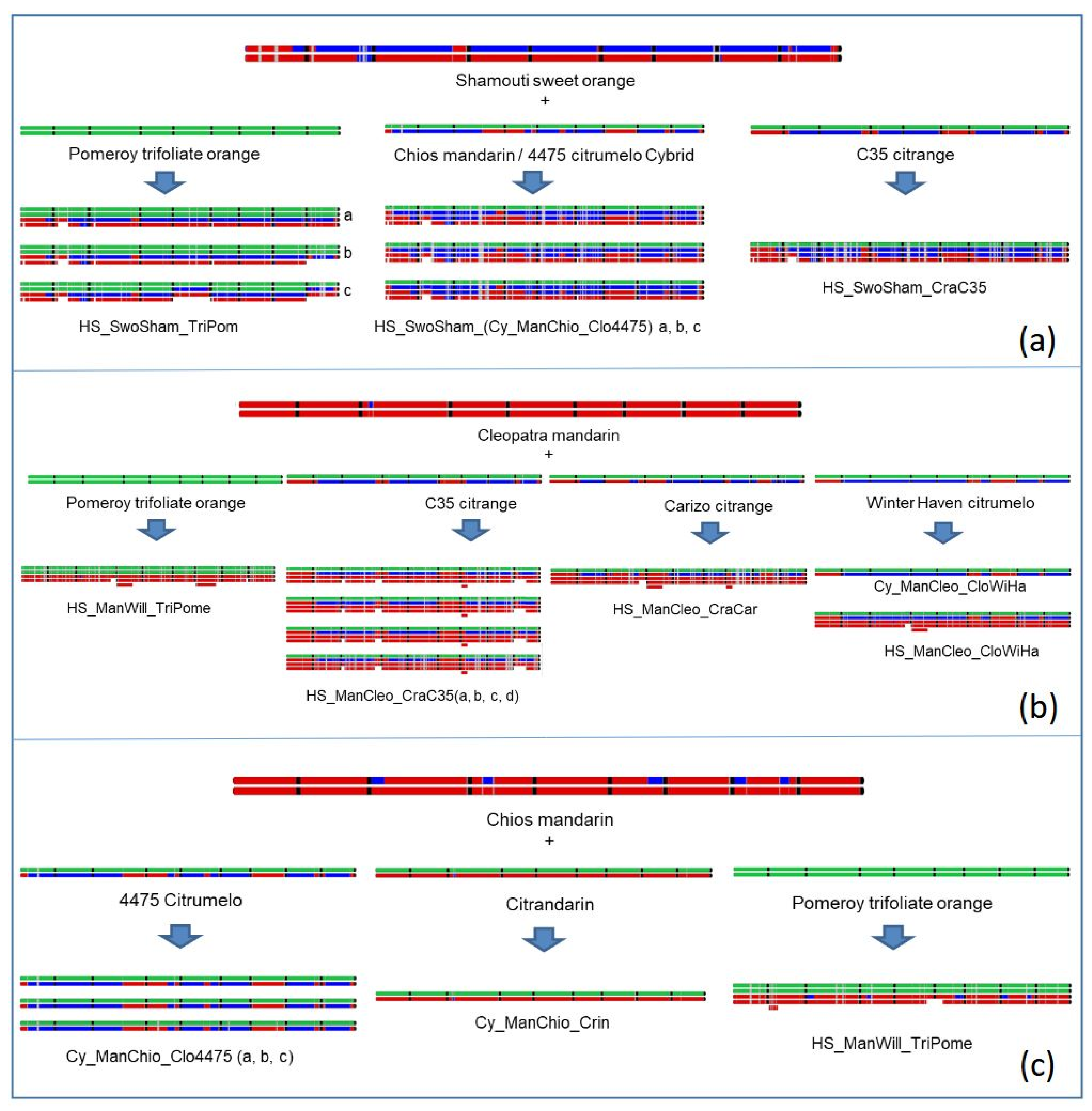
3.5.4. Interspecific Mosaic Structure of the Parental Lines, Cybrids, and Nuclear Somatic Hybrids
4. Discussion
4.1. Obtaining New Nuclear Somatic Hybrids for Three Intergeneric Parental Combinations and One New Cybrid
4.2. Efficiency of GBS with Apek1 Restriction Enzyme to Mark Two Cytoplasmic and Nuclear Genomes
4.3. Inheritance of Cytoplasmic Genomes
4.4. Inheritance and Instability of the Nuclear Genome
4.5. Implications of Genomic Instability for Rootstock Breeding
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ollitrault, P.; Gómez-Cadenas, A.; Arbona, V. Facing Climate Change: Biotechnology of Iconic Mediterranean Woody Crops. Front. Plant Sci. 2019, 10, 427. [Google Scholar] [CrossRef]
- Aritua, V.; Achor, D.; Gmitter, F.G.; Albrigo, G.; Wang, N. Transcriptional and Microscopic Analyses of Citrus Stem and Root Responses to Candidatus Liberibacter asiaticus Infection. PLoS ONE 2013, 8, e73742. [Google Scholar] [CrossRef] [Green Version]
- Johnson, E.G.; Wu, J.; Bright, D.B.; Graham, J.H. Association of “CandidatusLiberibacter asiaticus” root infection, but not phloem plugging with root loss on huanglongbing-affected trees prior to appearance of foliar symptoms. Plant Pathol. 2014, 63, 290–298. [Google Scholar] [CrossRef]
- Da Graça, J.V.; Douhan, G.W.; Halbert, S.E.; Keremane, M.L.; Lee, R.F.; Vidalakis, G.; Zhao, H. Huanglongbing: An overview of a complex pathosystem ravaging the world’s citrus. J. Integr. Plant Biol. 2016, 58, 373–387. [Google Scholar] [CrossRef] [PubMed]
- Castle, W.S.; Baldwin, J.C.; Grosser, J.W. Performance of “Washington” Navel Orange Trees in Rootstock Trials Located in Lake and St. Lucie Counties. Proc. Fla. State Hortic. Soc. 2001, 113, 106–111. [Google Scholar]
- Yang, Z.N.; Ye, X.R.; Choi, S.; Molina, J.; Moonan, F.; Wing, R.A.; Roose, M.L.; Mirkov, T.E. Construction of a 1.2-Mb Contig Including the Citrus Tristeza Virus Resistance Gene Locus Using a Bacterial Artificial Chromosome Library of Poncirus Trifoliata (L.) Raf. Genome 2001, 44, 382–393. [Google Scholar] [CrossRef] [PubMed]
- Folimonova, S.Y.; Robertson, C.J.; Garnsey, S.M.; Gowda, S.; Dawson, W.O. Examination of the Responses of Different Genotypes of Citrus to Huanglongbing (Citrus Greening) Under Different Conditions. Phytopathology 2009, 99, 1346–1354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albrecht, U.; Bowman, K.D. Tolerance of the Trifoliate Citrus Hybrid US-897 (Citrus reticulata Blanco × Poncirus trifoliata L. Raf.) to Huanglongbing. HortScience 2011, 46, 16–22. [Google Scholar] [CrossRef] [Green Version]
- Albrecht, U.; Bowman, K.D. Transcriptional response of susceptible and tolerant citrus to infection with Candidatus Liberibacter asiaticus. Plant Sci. 2012, 185–186, 118–130. [Google Scholar] [CrossRef]
- Ramadugu, C.; Keremane, M.L.; Halbert, S.E.; Duan, Y.P.; Roose, M.L.; Stover, E.; Lee, R.F. Long-Term Field Evaluation Reveals Huanglongbing Resistance in Citrus Relatives. Plant Dis. 2016, 100, 1858–1869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- George, J.; Lapointe, S.L. Host-plant resistance associated with Poncirus trifoliata influence oviposition, development and adult emergence of Diaphorina citri (Hemiptera: Liviidae). Pest Manag. Sci. 2019, 75, 279–285. [Google Scholar] [CrossRef] [Green Version]
- Huang, M.; Roose, M.L.; Yu, Q.; Du, D.; Yu, Y.; Zhang, Y.; Deng, Z.; Stover, E.; Gmitter, F.G.J. Construction of High-Density Genetic Maps and Detection of QTLs Associated with Huanglongbing Tolerance in Citrus. Front. Plant Sci. 2018, 9, 1694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, Z.; Bredeson, J.V.; Wu, G.A.; Shu, S.; Rawat, N.; Du, D.; Parajuli, S.; Yu, Q.; You, Q.; Rokhsar, D.S.; et al. A chromosome-scale reference genome of trifoliate orange (Poncirus trifoliata) provides insights into disease resistance, cold tolerance and genome evolution in Citrus. Plant J. 2020, 104, 1215–1232. [Google Scholar] [CrossRef] [PubMed]
- Alves, M.N.; Lopes, S.A.; Raiol-Junior, L.L.; Wulff, N.A.; Girardi, E.A.; Ollitrault, P.; Peña, L. Resistance to ‘Candidatus Liberibacter asiaticus,’ the Huanglongbing Associated Bacterium, in Sexually and/or Graft-Compatible Citrus Relatives. Front. Plant Sci. 2021, 11, 2166. [Google Scholar] [CrossRef] [PubMed]
- Biology of Horticultural Crops Pinchas Spiegel-Roy—Eliezer E Goldschmidt-Biology of Citrus-Cambridge University Press (1996) | PDF | Citrus | Orange (Fruit). Available online: https://www.scribd.com/document/279002770/Biology-of-Horticultural-Crops-Pinchas-Spiegel-Roy-Eliezer-E-Goldschmidt-Biology-of-Citrus-Cambridge-University-Press-1996 (accessed on 28 November 2021).
- Syvertsen, J.P.; Garcia-Sanchez, F. Multiple abiotic stresses occurring with salinity stress in citrus. Environ. Exp. Bot. 2014, 103, 128–137. [Google Scholar] [CrossRef]
- Grosser, J.W. Citrus Scion and Rootstock Improvement via Somatic Hybridization. Acta Hortic. 1993, 297–305. [Google Scholar] [CrossRef]
- Dambier, D.; Benyahia, H.; Pensabene-Bellavia, G.; Kaçar, Y.A.; Froelicher, Y.; Belfalah, Z.; Lhou, B.; Handaji, N.; Printz, B.; Morillon, R.; et al. Somatic hybridization for citrus rootstock breeding: An effective tool to solve some important issues of the Mediterranean citrus industry. Plant Cell Rep. 2011, 30, 883–900. [Google Scholar] [CrossRef]
- Grosser, J.W.; Louzada, E.S.; Gmitter, F.G., Jr.; Chandler, J.; Louzada, E.S. Somatic Hybridization of Complementary Citrus Rootstock:Five New Hybrids. HortScience 1994, 29, 812–813. [Google Scholar] [CrossRef] [Green Version]
- Grosser, J.W.; Ollitrault, P.; Olivares-Fuster, O. Somatic hybridization in citrus: An effective tool to facilitate variety improvement. In Vitro Cell. Dev. Biol. Anim. 2000, 36, 434–449. [Google Scholar] [CrossRef]
- Ollitrault, P.; Dambier, D.; Froelicher, Y.; Carreel, F.; D’Hont, A.; Luro, F.; Bruyere, S.; Cabasson, C.; Lotfy, S.; Joumaa, A.; et al. Somatic hybridization potential for citrus germplasm utilization. Cah. D’etudes Rech. Francoph. Agric. 2000, 9, 223–236. [Google Scholar]
- Grosser, J.W.; Gmitter, F.G. Protoplast fusion for production of tetraploids and triploids: Applications for scion and rootstock breeding in citrus. Plant Cell Tissue Organ Cult. 2011, 104, 343–357. [Google Scholar] [CrossRef]
- Mendes, B.M.J.; de Assis, A. Mourão Filho, F.; De M. Farias, P.C.; Benedito, V.A. Citrus somatic hybridization with potential for improved blight and CTV resistance. In Vitro Cell. Dev. Biol. Anim. 2001, 37, 490–495. [Google Scholar] [CrossRef]
- Ruiz, M.; Pensabene-Bellavia, G.; Quiñones, A.; García-Lor, A.; Morillon, R.; Ollitrault, P.; Primo-Millo, E.; Navarro, L.; Aleza, P. Molecular Characterization and Stress Tolerance Evaluation of New Allotetraploid Somatic Hybrids Between Carrizo Citrange and Citrus macrophylla W. rootstocks. Front. Plant Sci. 2018, 9, 901. [Google Scholar] [CrossRef] [PubMed]
- MedinaUrrutia, V.; Lopez Madera, K.F.; Serrano, P.; Ananthakrishnan, G.; Grosser, J.W.; Guo, W. New Intergeneric Somatic Hybrids Combining Amblycarpa Mandarin with Six Trifoliate/Trifoliate Hybrid Selections for Lime Rootstock Improvement. HortScience 2004, 39, 355–360. [Google Scholar] [CrossRef]
- Guo, W.; Cheng, Y.; Deng, X. Regeneration and molecular characterization of intergeneric somatic hybrids between Citrus reticulata and Poncirus trifoliata. Plant Cell Rep. 2002, 20, 829–834. [Google Scholar] [CrossRef]
- Ohgawara, T.; Kobayashi, S.; Ishii, S.; Yoshinaga, K.; Oiyama, I. Fertile fruit trees obtained by somatic hybridization: Navel orange (Citrus sinensis) and Troyer citrange (C. sinensis × Poncirus trifoliata). Theor. Appl. Genet. 1991, 81, 141–143. [Google Scholar] [CrossRef] [PubMed]
- Ollitrault, P.; Germanà, M.A.; Froelicher, Y.; Cuenca, J.; Aleza, P.; Morillon, R.; Grosser, J.W.; Guo, W. Ploidy Manipulation for Citrus Breeding, Genetics, and Genomics. In The Citrus Genome; Compendium of Plant Genomes; Gentile, A., La Malfa, S., Deng, Z., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 75–105. ISBN 978-3-030-15308-3. [Google Scholar]
- Mouhaya, W.; Allario, T.; Costantino, G.; Andres, F.; Talon, M.; Luro, F.; Ollitrault, P.; Morillon, R. Citrus Tetraploid Rootstocks Are More Tolerant to Salt Stress than Diploid. Modern variety breeding for present and future needs 2008. In Proceedings of the 18th EUCARPIA General Congress, Alencia, Spain, 9–12 September 2008; p. 439. [Google Scholar]
- Podda, A.; Checcucci, G.; Mouhaya, W.; Centeno, D.; Rofidal, V.; del Carratore, R.; Luro, F.; Morillon, R.; Ollitrault, P.; Maserti, B.E. Salt-stress induced changes in the leaf proteome of diploid and tetraploid mandarins with contrasting Na+ and Cl− accumulation behaviour. J. Plant Physiol. 2013, 170, 1101–1112. [Google Scholar] [CrossRef]
- Ruiz, M.; Quiñones, A.; Martínez-Alcántara, B.; Aleza, P.; Morillon, R.; Navarro, L.; Primo-Millo, E.; Martínez-Cuenca, M.-R. Tetraploidy Enhances Boron-Excess Tolerance in Carrizo Citrange (Citrus sinensis L. Osb. × Poncirus trifoliata L. Raf.). Front. Plant Sci. 2016, 7, 701. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, T.M.; Yahmed, J.B.; Dutra, J.; Maserti, B.E.; Talon, M.; Navarro, L.; Ollitrault, P.; Gesteira, A.S.; Morillon, R. Better Tolerance to Water Deficit in Doubled Diploid “Carrizo Citrange” Compared to Diploid Seedlings Is Associated with More Limited Water Consumption and Better H2O2 Scavenging. Acta Physiol. Plant. 2017, 39, e204. [Google Scholar] [CrossRef]
- Allario, T.; Brumos, J.; Colmenero-Flores, J.M.; Iglesias, D.J.; Pina, J.A.; Navarro, L.; Talon, M.; Ollitrault, P.; Morillon, R. Tetraploid Rangpur lime rootstock increases drought tolerance via enhanced constitutive root abscisic acid production. Plant Cell Environ. 2013, 36, 856–868. [Google Scholar] [CrossRef] [Green Version]
- Oustric, J.; Morillon, R.; Luro, F.; Herbette, S.; Lourkisti, R.; Giannettini, J.; Berti, L.; Santini, J. Tetraploid Carrizo Citrange Rootstock (Citrus Sinensis Osb. × Poncirus Trifoliata, L. Raf.) Enhances Natural Chilling Stress Tolerance of Common Clementine (Citrus Clementina Hort. Ex Tan). J. Plant Physiol. 2017, 214, 108–115. [Google Scholar] [CrossRef]
- Oustric, J.; Morillon, R.; Ollitrault, P.; Herbette, S.; Luro, F.; Froelicher, Y.; Tur, I.; Dambier, D.; Giannettini, J.; Berti, L.; et al. Somatic hybridization between diploid Poncirus and Citrus improves natural chilling and light stress tolerances compared with equivalent doubled-diploid genotypes. Trees 2018, 32, 883–895. [Google Scholar] [CrossRef]
- Khalid, M.F.; Hussain, S.; Anjum, M.A.; Ahmad, S.; Ali, M.A.; Ejaz, S.; Morillon, R. Better salinity tolerance in tetraploid vs diploid volkamer lemon seedlings is associated with robust antioxidant and osmotic adjustment mechanisms. J. Plant Physiol. 2020, 244, 153071. [Google Scholar] [CrossRef]
- Ollitrault, P.; Guo, W.; Grosser, J.W. Somatic Hybridization. In Citrus Genetics, Breeding and Biotechnology; CABI Publishing: Wallingford/Oxfordshire, UK, 2007; pp. 235–260. [Google Scholar]
- Moreira, C.D.; Chase, C.D.; Gmitter, F.G.; Grosser, J.W. Inheritance of organelle genomes in citrus somatic cybrids. Mol. Breed. 2000, 6, 401–405. [Google Scholar] [CrossRef]
- Moreira, C.D.; Chase, C.D.; Gmitter, F.G.; Grosser, J.W. Transmission of organelle genomes in citrus somatic hybrids. Plant Cell Tissue Organ Cult. 2000, 61, 165–168. [Google Scholar] [CrossRef]
- Cheng, Y.J.; Guo, W.W.; Deng, X.X. Inter-generic mitochondrial genome recombination in somatic hybrids between Citrus and Poncirus. J. Hortic. Sci. Biotechnol. 2007, 82, 849–854. [Google Scholar] [CrossRef]
- Zhang, S.; Yin, Z.-P.; Wu, X.-M.; Li, C.-C.; Xie, K.-D.; Deng, X.-X.; Grosser, J.W.; Guo, W.-W. Assembly of Satsuma mandarin mitochondrial genome and identification of cytoplasmic male sterility–specific ORFs in a somatic cybrid of pummelo. Tree Genet. Genomes 2020, 16, 84. [Google Scholar] [CrossRef]
- Guo, W.W.; Deng, X.X. Wide somatic hybrids of Citrus with its related genera and their potential in genetic improvement. Euphytica 2001, 118, 175–183. [Google Scholar] [CrossRef]
- Pensabene-Bellavia, G.; Ruiz, M.; Aleza, P.; Olivares-Fuster, O.; Ollitrault, P.; Navarro, L. Chromosome Instability in “Carrizo” Citrange + Citrus Macrophylla Somatic Hybrids. Acta Hortic. 2015, 1065, 677–685. [Google Scholar] [CrossRef]
- Sundberg, E.; Glimelius, K. Effects of parental ploidy level and genetic divergence on chromosome elimination and chloroplast segregation in somatic hybrids within Brassicaceae. Theor. Appl. Genet. 1991, 83, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Walters, T.W.; Earle, E.D. Organellar segregation, rearrangement and recombination in protoplast fusion-derived Brassica oleracea calli. Theor. Appl. Genet. 1993, 85, 761–769. [Google Scholar] [CrossRef] [PubMed]
- Buiteveld, J.; Kassies, W.; Geels, R.; van Lookeren Campagne, M.; Jacobsen, E.; Creemers-Molenaar, J. Biased chloroplast and mitochondrial transmission in somatic hybrids of Allium ampeloprasum L. and Allium cepa L. Plant Sci. 1998, 131, 219–228. [Google Scholar] [CrossRef]
- Babiychuk, E.; Kushnir, S.; Gleba, Y.Y. Spontaneous extensive chromosome elimination in somatic hybrids between somatically congruent species Nicotiana tabacum L. and Atropa belladonna L. Theor. Appl. Genet. 1992, 84, 87–91. [Google Scholar] [CrossRef] [PubMed]
- Buiteveld, J.; Suo, Y.; Campagne, M.M.L.; Creemers-Molenaar, J. Production and characterization of somatic hybrid plants between leek (Allium ampeloprasum L.) and onion (Allium cepa L.). Theor. Appl. Genet. 1998, 96, 765–775. [Google Scholar] [CrossRef]
- Minqin, W.; Junsheng, Z.; Zhenying, P.; Wei, G.; Yun, W.; Le, W.; Guangmin, X. Chromosomes are eliminated in the symmetric fusion between Arabidopsis thaliana L. and Bupleurum scorzonerifolium Willd. Plant Cell Tissue Organ Cult. 2008, 92, 121–130. [Google Scholar] [CrossRef]
- Cui, H.; Sun, Y.; Deng, J.; Wang, M.; Xia, G. Chromosome elimination and introgression following somatic hybridization between bread wheat and other grass species. Plant Cell Tissue Organ Cult. 2015, 120, 203–210. [Google Scholar] [CrossRef]
- Trabelsi, S.; Gargouri-Bouzid, R.; Vedel, F.; Nato, A.; Lakhoua, L.; Drira, N. Somatic hybrids between potato Solanum tuberosum and wild species Solanum verneï exhibit a recombination in the plastome. Plant Cell Tissue Organ Cult. 2005, 83, 1–11. [Google Scholar] [CrossRef]
- Tomiczak, K.; Sliwinska, E.; Rybczyński, J.J. Protoplast fusion in the genus Gentiana: Genomic composition and genetic stability of somatic hybrids between Gentiana kurroo Royle and G. cruciata L. Plant Cell Tissue Organ Cult. 2017, 131, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Tomiczak, K. Molecular and cytogenetic description of somatic hybrids between Gentiana cruciata L. and G. tibetica King. J. Appl. Genet. 2020, 61, 13–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Xu, C.H.; Wang, M.Q.; Zhi, D.Y.; Xia, G.M. Genomic changes at the early stage of somatic hybridization. Genet. Mol. Res. 2014, 13, 1938–1948. [Google Scholar] [CrossRef]
- Liu, S.; Li, F.; Kong, L.; Sun, Y.; Qin, L.; Chen, S.; Cui, H.; Huang, Y.; Xia, G. Genetic and Epigenetic Changes in Somatic Hybrid Introgression Lines Between Wheat and Tall Wheatgrass. Genetics 2015, 199, 1035–1045. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.-C.; Zhai, H.; He, S.-Z.; Yang, Y.-F.; Liu, Q.-C. Analysis of drought tolerance and genetic and epigenetic variations in a somatic hybrid between Ipomoea batatas (L.) Lam. and I. triloba L. J. Integr. Agric. 2017, 16, 36–46. [Google Scholar] [CrossRef]
- Grosser, J.W.; Gmitter, F.G., Jr. Protoplast Fusion and Citrus Improvement. In Plant Breeding Reviews; John Wiley & Sons Ltd.: Hoboken, NJ, USA, 1990; pp. 339–374. ISBN 978-1-118-06105-3. [Google Scholar]
- Kao, K.N.; Michayluk, M.R. A method for high-frequency intergeneric fusion of plant protoplasts. Planta 1974, 115, 355–367. [Google Scholar] [CrossRef]
- Murashige, T.; Tucker, D. Growth Factor Requirements of Citrus Tissue Culture. In Proceedings of the 1st International Citrus Symposium, Riverside, CA, USA, 16–26 March 1968; Volume 3, pp. 1155–1160. [Google Scholar]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A Robust Simple Genotyping-by-Sequencing (GBS) Approach for High Diversity Species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sonah, H.; Bastien, M.; Iquira, E.; Tardivel, A.; Légaré, G.; Boyle, B.; Normandeau, É.; Laroche, J.; LaRose, S.; Jean, M.; et al. An Improved Genotyping by Sequencing (GBS) Approach Offering Increased Versatility and Efficiency of SNP Discovery and Genotyping. PLoS ONE 2013, 8, e54603. [Google Scholar] [CrossRef] [Green Version]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Herten, K.; Hestand, M.S.; Vermeesch, J.R.; Van Houdt, J.K. GBSX: A toolkit for experimental design and demultiplexing genotyping by sequencing experiments. BMC Bioinform. 2015, 16, 73. [Google Scholar] [CrossRef] [Green Version]
- Baurens, F.-C.; Martin, G.; Hervouet, C.; Salmon, F.; Yohomé, D.; Ricci, S.; Rouard, M.; Habas, R.; Lemainque, A.; Yahiaoui, N.; et al. Recombination and Large Structural Variations Shape Interspecific Edible Bananas Genomes. Mol. Biol. Evol. 2019, 36, 97–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, F.; Bi, C.; Wang, X.; Qian, X.; Ye, N. The complete mitochondrial genome of Citrus sinensis. Mitochondrial DNA Part B 2018, 3, 592–593. [Google Scholar] [CrossRef] [Green Version]
- Bausher, M.G.; Singh, N.D.; Lee, S.-B.; Jansen, R.K.; Daniell, H. The complete chloroplast genome sequence of Citrus sinensis (L.) Osbeck var “Ridge Pineapple”: Organization and phylogenetic relationships to other angiosperms. BMC Plant Biol. 2006, 6, 21. [Google Scholar] [CrossRef] [Green Version]
- Calvez, L.; Dereeper, A.; Mournet, P.; Froelicher, Y.; Bruyère, S.; Morillon, R.; Ollitrault, P. Intermediate Inheritance with Disomic Tendency in Tetraploid Intergeneric Citrus × Poncirus Hybrids Enhances the Efficiency of Citrus Rootstock Breeding. Agronomy 2020, 10, 1961. [Google Scholar] [CrossRef]
- Wu, G.A.; Terol, J.; Ibanez, V.; López-García, A.; Pérez-Román, E.; Borredá, C.; Domingo, C.; Tadeo, F.; Carbonell-Caballero, J.; Alonso, R.; et al. Genomics of the origin and evolution of Citrus. Nature 2018, 554, 311–316. [Google Scholar] [CrossRef] [Green Version]
- Oueslati, A.; Salhi-Hannachi, A.; Luro, F.; Vignes, H.; Mournet, P.; Ollitrault, P. Genotyping by sequencing reveals the interspecific C. maxima/C. reticulata admixture along the genomes of modern citrus varieties of mandarins, tangors, tangelos, orangelos and grapefruits. PLoS ONE 2017, 12, e0185618. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, D.; Comte, A.; Curk, F.; Costantino, G.; Luro, F.; Dereeper, A.; Mournet, P.; Froelicher, Y.; Ollitrault, P. Genotyping by sequencing can reveal the complex mosaic genomes in gene pools resulting from reticulate evolution: A case study in diploid and polyploid citrus. Ann. Bot. 2019, 123, 1231–1251. [Google Scholar] [CrossRef]
- Ohgawara, T.; Kobayashi, S.; Ishii, S.; Yoshinaga, K.; Oiyama, I. Somatic hybridization in Citrus: Navel orange (C. sinensis Osb.) and grapefruit (C. paradisi Macf.). Theor. Appl. Genet. 1989, 78, 609–612. [Google Scholar] [CrossRef]
- Tusa, N.; Grosser, J.W.; Gmitter, F.G., Jr. Plant Regeneration of “Valencia” Sweet Orange, “Femminello” Lemon, and the Interspecific Somatic Hybrid Following Protoplast Fusion. J. Am. Soc. Hort. Sci. 1990, 115, 1043–1046. [Google Scholar] [CrossRef] [Green Version]
- Moriguchi, T.; Hidaka, T.; Omura, M.; Motomura, T.; Akihama, T. Genotype and Parental Combination Influence Efficiency of Cybrid Induction in Citrus by Electrofusion: A publication of the American Society for Horticultural Science (USA). HortScience 1996, 31, 275–278. [Google Scholar] [CrossRef] [Green Version]
- Saito, W.; Ohgawara, T.; Shimizu, J.; Ishii, S.; Kobayashi, S. Citrus cybrid regeneration following cell fusion between nucellar cells and mesophyll cells. Plant Sci. 1993, 88, 195–201. [Google Scholar] [CrossRef]
- Yamamoto, M.; Kobayashi, S. A Cybrid Plant Produced by Electrofusion between Citrus unshiu (satsuma mandarin) and C. sinensis (sweet orange). Plant Tissue Cult. Lett. 1995, 12, 131–137. [Google Scholar] [CrossRef] [Green Version]
- Ollitrault, P.; Dambier, D.; Froelicher, Y.; Carreel, F.; d’Hont, A.; Francois, L.; Bruyère, S.; Cabasson, C.; Lotfy, S.; Joumaa, A.; et al. Apport de l’hybridation Somatique Pour l’exploitation Des Ressources Génétiques Des Agrumes. Cah. Agric. 2000, 9, 223–236. [Google Scholar]
- Grosser, J.W.; Chandler, J.L. Somatic hybridization of high yield, cold-hardy and disease resistant parents for citrus rootstock improvement. J. Hortic. Sci. Biotechnol. 2000, 75, 641–644. [Google Scholar] [CrossRef]
- Ohgawara, T.; Kobayashi, S.; Ohgawara, E.; Uchimiya, H.; Ishii, S. Somatic hybrid plants obtained by protoplast fusion between Citrus sinensis and Poncirus trifoliata. Theor. Appl. Genet. 1985, 71, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Wen-Wu, G.U.O.; Chang-He, Z.; Hua-Lin, Y.I.; Xiu-Xin, D. Intergeneric Somatic Hybrid Plants Between Citrus and Poncirus Trifoliata and Evaluation of Their Root Rot Resistance. J. Integr. Plant Biol. 2000, 42, 668. [Google Scholar]
- Fu, C.H.; Chen, C.L.; Guo, W.W.; Deng, X.X. GISH, AFLP and PCR-RFLP analysis of an intergeneric somatic hybrid combining Goutou sour orange and Poncirus trifoliata. Plant Cell Rep. 2004, 23, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.W.; Wu, R.C.; Cheng, Y.J.; Deng, X.X. Production and molecular characterization of Citrus intergeneric somatic hybrids between red tangerine and citrange. Plant Breed. 2007, 126, 72–76. [Google Scholar] [CrossRef]
- Froelicher, Y.; Mouhaya, W.; Bassene, J.-B.; Costantino, G.; Kamiri, M.; Luro, F.; Morillon, R.; Ollitrault, P. New universal mitochondrial PCR markers reveal new information on maternal citrus phylogeny. Tree Genet. Genomes 2011, 7, 49–61. [Google Scholar] [CrossRef]
- Kobayashi, S.; Ohgawara, T.; Fujiwara, K.; Oiyama, I. Analysis of cytoplasmic genomes in somatic hybrids between navel orange (Citrus sinensis Osb.) and “Murcott” tangor. Theor. Appl. Genet. 1991, 82, 6–10. [Google Scholar] [CrossRef]
- Moriguchi, T.; Motomura, T.; Hidaka, T.; Akihama, T.; Omura, M. Analysis of mitochondrial genomes amongCitrus plants produced by the interspecific somatic fusion of ‘Seminole’ tangelo with rough lemon. Plant Cell Rep. 1997, 16, 397–400. [Google Scholar] [CrossRef] [PubMed]
- Cabasson, C.M.; Luro, F.; Ollitrault, P.; Grosser, J.W. Non-random inheritance of mitochondrial genomes in Citrus hybrids produced by protoplast fusion. Plant Cell Rep. 2001, 20, 604–609. [Google Scholar] [CrossRef]
- Cai, X.; Fu, J.; Guo, W. Mitochondrial Genome of Callus Protoplast Has a Role in Mesophyll Protoplast Regeneration in Citrus: Evidence From Transgenic GFP Somatic Homo-Fusion. Hortic. Plant J. 2017, 3, 177–182. [Google Scholar] [CrossRef]
- Omar, A.A.; Murata, M.; Yu, Q.; Gmitter, F.G.; Chase, C.D.; Graham, J.H.; Grosser, J.W. Production of three new grapefruit cybrids with potential for improved citrus canker resistance. In Vitro Cell. Dev. Biol. Anim. 2017, 53, 256–269. [Google Scholar] [CrossRef]
- Motomura, T.; Hidaka, T.; Monguchi, T.; Akihama, T.; Omura, M. Intergeneric Somatic Hybrids between Citrus and Atalantia or Severinia by Electrofusion, and Recombination of Mitochondrial Genomes. Jpn. J. Breed. 1995, 45, 309–314. [Google Scholar] [CrossRef] [Green Version]
- Belliard, G.; Vedel, F.; Pelletier, G. Mitochondrial recombination in cytoplasmic hybrids of Nicotiana tabacum by protoplast fusion. Nature 1979, 281, 401–403. [Google Scholar] [CrossRef]
- Vedel, F.; Chétrit, P.; Mathieu, C.; Pelletier, G.; Primard, C. Several different mitochondrial DNA regions are involved in intergenomic recombination in Brassica napus cybrid plants. Curr. Genet. 1986, 11, 17–24. [Google Scholar] [CrossRef]
- Zubko, M.K.; Zubko, E.I.; Patskovsky, Y.V.; Khvedynich, O.A.; Fisahn, J.; Gleba, Y.Y.; Schieder, O. Novel “homeotic” CMS patterns generated in Nicotianavia cybridization with HyoscyamusandScopolia. J. Exp. Bot. 1996, 47, 1101–1110. [Google Scholar] [CrossRef] [Green Version]
- Sanchez-Puerta, M.V.; Cho, Y.; Mower, J.P.; Alverson, A.J.; Palmer, J.D. Frequent, Phylogenetically Local Horizontal Transfer of the cox1 Group I Intron in Flowering Plant Mitochondria. Mol. Biol. Evol. 2008, 25, 1762–1777. [Google Scholar] [CrossRef] [Green Version]
- Arimura, S.; Yanase, S.; Tsutsumi, N.; Koizuka, N. The mitochondrial genome of an asymmetrically cell-fused rapeseed, Brassica napus, containing a radish-derived cytoplasmic male sterility-associated gene. Genes Genet. Syst. 2018, 93, 143–148. [Google Scholar] [CrossRef] [Green Version]
- Garcia, L.E.; Zubko, M.K.; Zubko, E.I.; Sanchez-Puerta, M.V. Elucidating genomic patterns and recombination events in plant cybrid mitochondria. Plant Mol. Biol. 2019, 100, 433–450. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Puerta, M.V.; Zubko, M.K.; Palmer, J.D. Homologous recombination and retention of a single form of most genes shape the highly chimeric mitochondrial genome of a cybrid plant. New Phytol. 2015, 206, 381–396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grosser, J.W.; Mourao-Fo, F.A.A.; Gmitter, F.G.; Louzada, E.S.; Jiang, J.; Baergen, K.; Quiros, A.; Cabasson, C.; Schell, J.L.; Chandler, J.L. Allotetraploid hybrids between citrus and seven related genera produced by somatic hybridization. Theor. Appl. Genet. 1996, 92, 577–582. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Grosser, J.W.; Ćalović, M.; Serrano, P.; Pasquali, G.; Gmitter, J.; Gmitter, F.G. Verification of Mandarin and Pummelo Somatic Hybrids by Expressed Sequence Tag–Simple Sequence Repeat Marker Analysis. J. Am. Soc. Hortic. Sci. 2008, 133, 794–800. [Google Scholar] [CrossRef] [Green Version]
- Scarano, M.-T.; Abbate, L.; Ferrante, S.; Lucretti, S.; Tusa, N. ISSR-PCR technique: A useful method for characterizing new allotetraploid somatic hybrids of mandarin. Plant Cell Rep. 2002, 20, 1162–1166. [Google Scholar] [CrossRef]
- Shi, Y.Z.; Deng, X.X.; Yi, H.L. Variation of Intergeneric Somatic Hybrids Between Citrus Sinensis Cv. Valencia and Fortunella Crassifolia Cv. Meiwa. J. Integr. Plant Biol. 1998, 40. Available online: https://www.jipb.net/EN/abstract/abstract26260.shtml (accessed on 28 November 2021).
- Grosser, J.W.; Gmitter, F.G., Jr.; Sesto, F.; Deng, X.X.; Chandler, J.L. Six New Somatic Citrus Hybrids and Their Potential for Cultivar Improvement. J. Am. Soc. Hortic. Sci. 1992, 117, 169–173. [Google Scholar] [CrossRef] [Green Version]
- Miranda, M.; Motomura, T.; Ikeda, F.; Ohgawara, T.; Saito, W.; Endo, T.; Omura, M.; Moriguchi, T. Somatic hybrids obtained by fusion between Poncirus trifoliata (2×) and Fortunella hindsii (4×) protoplasts. Plant Cell Rep. 1997, 16, 401–405. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.W.; Deng, X.-X. Unexpected Ploidy Variation of the Somatic Hybrid Plants Between Citrus Sinensis and Clausena Lansium. J. Integr. Plant Biol. 1999, 41. Available online: https://www.semanticscholar.org/paper/Unexpected-Ploidy-Variation-of-the-Somatic-Hybrid-Wen-wu-Xiu-xin/cf1bd40da12763978e75ee20dc983e0714dea38c (accessed on 28 November 2021).
- Guo, W.W.; Deng, X.X. Intertribal hexaploid somatic hybrid plants regeneration from electrofusion between diploids of Citrus sinensis and its sexually incompatible relative, Clausena lansium. Theor. Appl. Genet. 1999, 98, 581–585. [Google Scholar] [CrossRef]
- Fu, C.H.; Guo, W.W.; Liu, J.H.; Deng, X.X. Regeneration of citrus sinensis (+) clausena lansium intergeneric triploid and tetraploid somatic hybrids and their identification by molecular markers. In Vitro Cell. Dev. Biol. Anim. 2003, 39, 360–364. [Google Scholar] [CrossRef]
- Starrantino, A.; Russo, F. Reproduction of Seedless Orange Cultivars from Undeveloped Ovules Raised “In Vitro”. Acta Hortic. 1983, 131, 253–258. [Google Scholar] [CrossRef]
- Kobayashi, S. Uniformity of plants regenerated from orange (Citrus sinensis Osb.) protoplasts. Theor. Appl. Genet. 1987, 74, 10–14. [Google Scholar] [CrossRef]
- Grosser, J.W.; Deng, X.X.; Goodrich, R.M. Somaclonal variation in sweet orange: Practical applications for variety improvement and possible causes. In Citrus Genetics, Breeding and Biotechnology; Khan, I.A., Ed.; CABI Publishing: Wallingford, UK, 2007; pp. 219–233. ISBN 978-0-85199-019-4. [Google Scholar]
- McClintock, B. The significance of responses of the genome to challenge. Science 1984, 226, 792–801. [Google Scholar] [CrossRef] [Green Version]
- Cai, Q.; Guy, C.L.; Moore, G. Extension of the Genetic Linkage Map in Citrus Using Random Amplified Polymorphic DNA (RAPD) Markers and RFLP Mapping of Cold-Acclimation Responsive Loci. Theor. Appl. Genet. 1994, 89, 606–614. [Google Scholar] [CrossRef] [PubMed]
- Weber, C.A.; Moore, G.A.; Deng, Z.N.; Gmitter, F.G., Jr. Mapping Freeze Tolerance Quantitative Trait Loci in a Citrus Grandis × Poncirus Trifoliata F1 Pseudo-Testcross Using Molecular Markers. J. Am. Soc. Hortic. Sci. 2003, 128, 508–514. [Google Scholar] [CrossRef] [Green Version]
- Hong, Q.-B.; Ma, X.-J.; Gong, G.-Z.; Peng, Z.-C.; He, Y.-R. Qtl Mapping of Citrus Freeze Tolerance. Acta Hortic. 2015, 467–474. [Google Scholar] [CrossRef]
- Raga, V.; Intrigliolo, D.S.; Bernet, G.P.; Carbonell, E.A.; & Asins, M.J. Genetic Analysis of Salt Tolerance in a Progeny Derived from the Citrus Rootstocks Cleopatra Mandarin and Trifoliate Orange. Tree Genet. Genomes 2016, 12, 34. [Google Scholar] [CrossRef]
- Gulsen, O.; Uzun, A.; Canan, I.; Seday, U.; Canihos, E. A new citrus linkage map based on SRAP, SSR, ISSR, POGP, RGA and RAPD markers. Euphytica 2010, 173, 265–277. [Google Scholar] [CrossRef]
- Cuenca, J.; Aleza, P.; Garcia-Lor, A.; Ollitrault, P.; Navarro, L. Fine Mapping for Identification of Citrus Alternaria Brown Spot Candidate Resistance Genes and Development of New SNP Markers for Marker-Assisted Selection. Front. Plant Sci. 2016, 7, 1948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cristofani, M.; Machado, M.A.; Grattapaglia, D. Genetic linkage maps of Citrus sunki Hort. ex. Tan. and Poncirus trifoliata (L.) Raf. and mapping of citrus tristeza virus resistance gene. Euphytica 1999, 109, 25–32. [Google Scholar] [CrossRef]
- Bernet, G.P.; Bretó, M.P.; Asins, M.J. Expressed sequence enrichment for candidate gene analysis of citrus tristeza virus resistance. Theor. Appl. Genet. 2004, 108, 592–602. [Google Scholar] [CrossRef] [PubMed]
- Ling, P.; Duncan, L.W.; Deng, Z.; Dunn, D.; Hu, X.; Huang, S.; Gmitter, F.G., Jr. Inheritance of Citrus Nematode Resistance and Its Linkage with Molecular Markers. Theor. Appl. Genet. 2000, 100, 1010–1017. [Google Scholar] [CrossRef]
- Gmitter, F.G.; Xiao, S.Y.; Huang, S.; Hu, X.L.; Garnsey, S.M.; Deng, Z. A localized linkage map of the citrus tristeza virus resistance gene region. Theor. Appl. Genet. 1996, 92, 688–695. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Huang, S.; Ling, P.; Yu, C.; Tao, Q.; Chen, C.; Wendell, M.K.; Zhang, H.B.; Gmitter, F.G., Jr. Fine Genetic Mapping and BAC Contig Development for the Citrus Tristeza Virus Resistance Gene Locus in Poncirus Trifoliata (Raf.). Mol. Genet. Genom. 2001, 265, 739–747. [Google Scholar] [CrossRef]
- Yang, Z.-N.; Ye, X.-R.; Molina, J.; Roose, M.L.; Mirkov, T.E. Sequence Analysis of a 282-Kilobase Region Surrounding the Citrus Tristeza Virus Resistance Gene (Ctv) Locus in Poncirus trifoliata L. Raf. Plant Physiol. 2003, 131, 482–492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiang, X.; Deng, Z.; Chen, C.; Gmitter, F.G.; Bowman, K. Marker Assisted Selection in Citrus Rootstock Breeding Based on a Major Gene Locus “Tyr1” Controlling Citrus Nematode Resistance. Agric. Sci. China 2010, 9, 557–567. [Google Scholar] [CrossRef]
- Xiang, X.; Deng, Z.; Zheng, Q.; Chen, C.; Gmitter, F. Developing Specific Markers and Improving Genetic Mapping for a Major Locus Tyr1 of Citrus Nematode Resistance. Biosci. Methods 2010, 1, 1–8. [Google Scholar] [CrossRef]
- Comai, L.; Tyagi, A.P.; Winter, K.; Holmes-Davis, R.; Reynolds, S.H.; Stevens, Y.; Byers, B. Phenotypic Instability and Rapid Gene Silencing in Newly Formed Arabidopsis Allotetraploids. Plant Cell 2000, 12, 1551–1567. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Tian, L.; Lee, H.-S.; Wei, N.E.; Jiang, H.; Watson, B.; Madlung, A.; Osborn, T.C.; Doerge, R.W.; Comai, L.; et al. Genomewide Nonadditive Gene Regulation in Arabidopsis Allotetraploids. Genetics 2006, 172, 507–517. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.J. Genetic and Epigenetic Mechanisms for Gene Expression and Phenotypic Variation in Plant Polyploids. Annu. Rev. Plant Biol. 2007, 58, 377–406. [Google Scholar] [CrossRef] [Green Version]
- Jiang, X.; Song, Q.; Ye, W.; Chen, Z.J. Concerted genomic and epigenomic changes accompany stabilization of Arabidopsis allopolyploids. Nat. Ecol. Evol. 2021, 5, 1382–1393. [Google Scholar] [CrossRef]
- Dutt, M.; Mahmoud, L.M.; Chamusco, K.; Stanton, D.; Chase, C.D.; Nielsen, E.; Quirico, M.; Yu, Q.; Gmitter, F.G., Jr.; Grosser, J.W. Utilization of somatic fusion techniques for the development of HLB tolerant breeding resources employing the Australian finger lime (Citrus australasica). PLoS ONE 2021, 16, e0255842. [Google Scholar] [CrossRef]
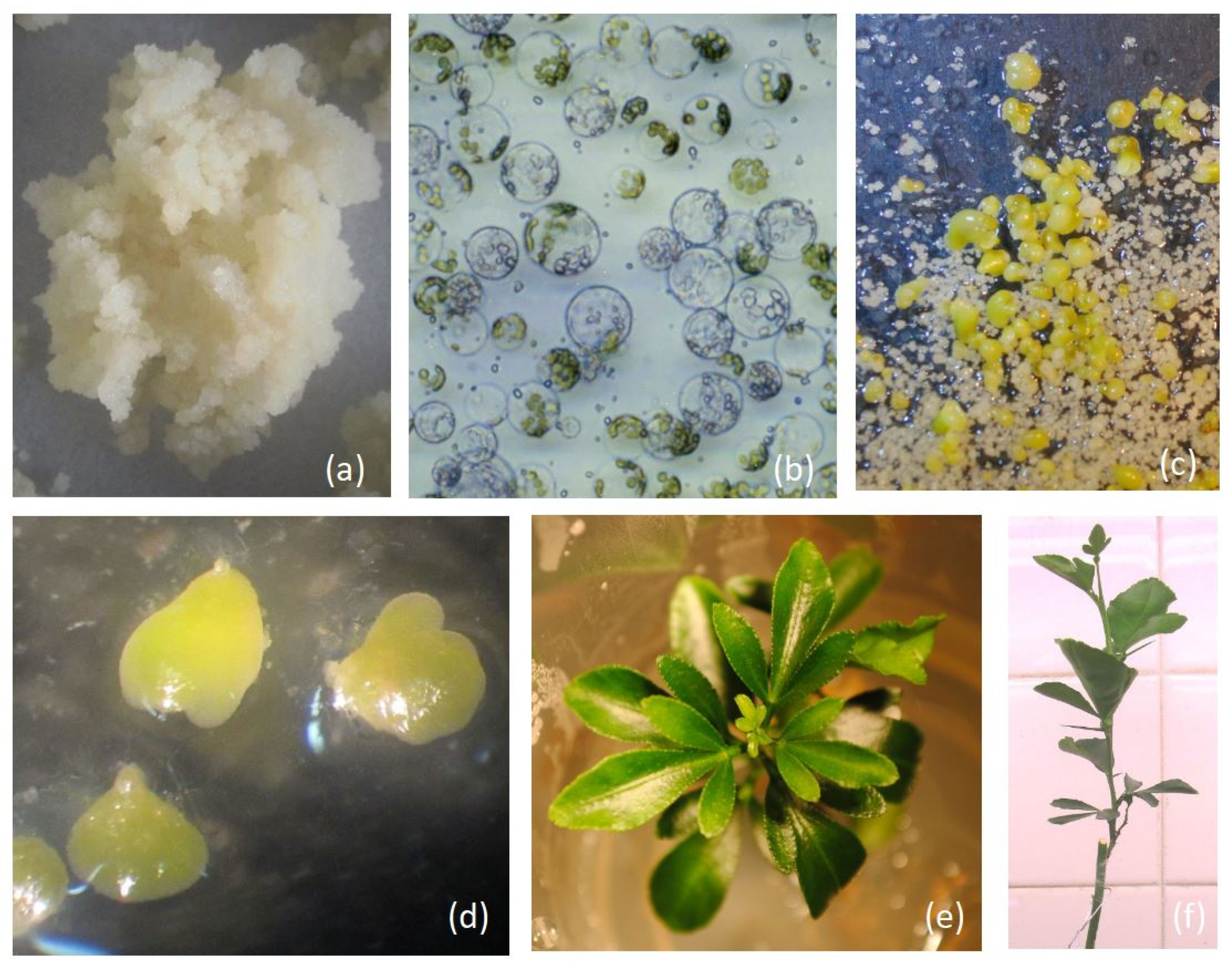
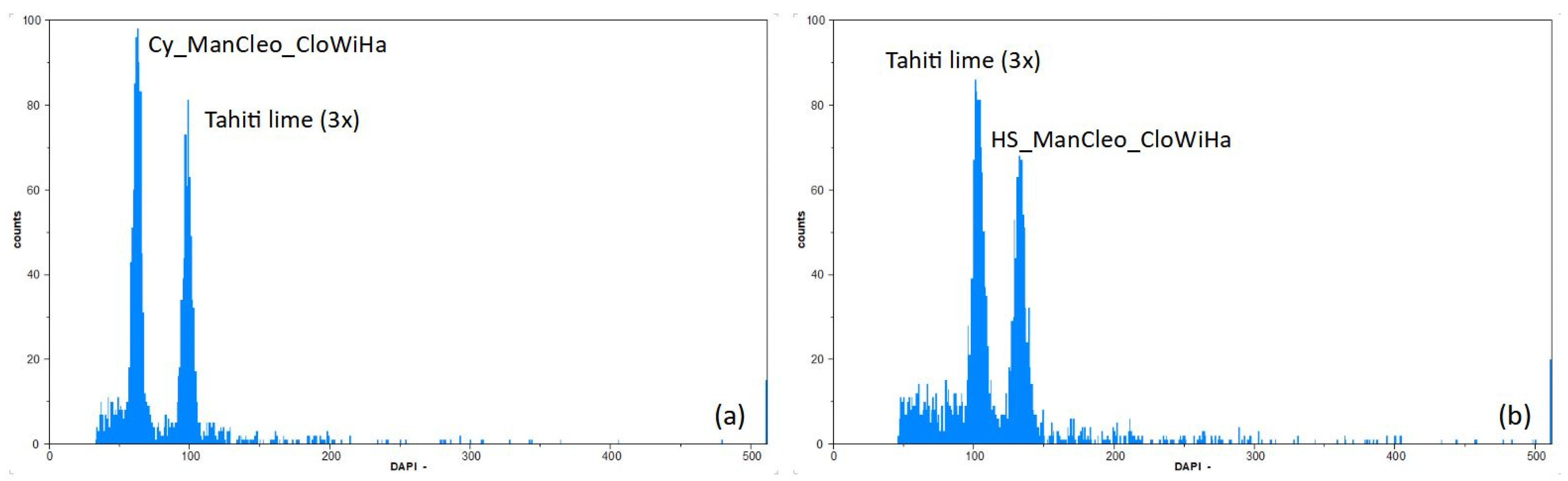

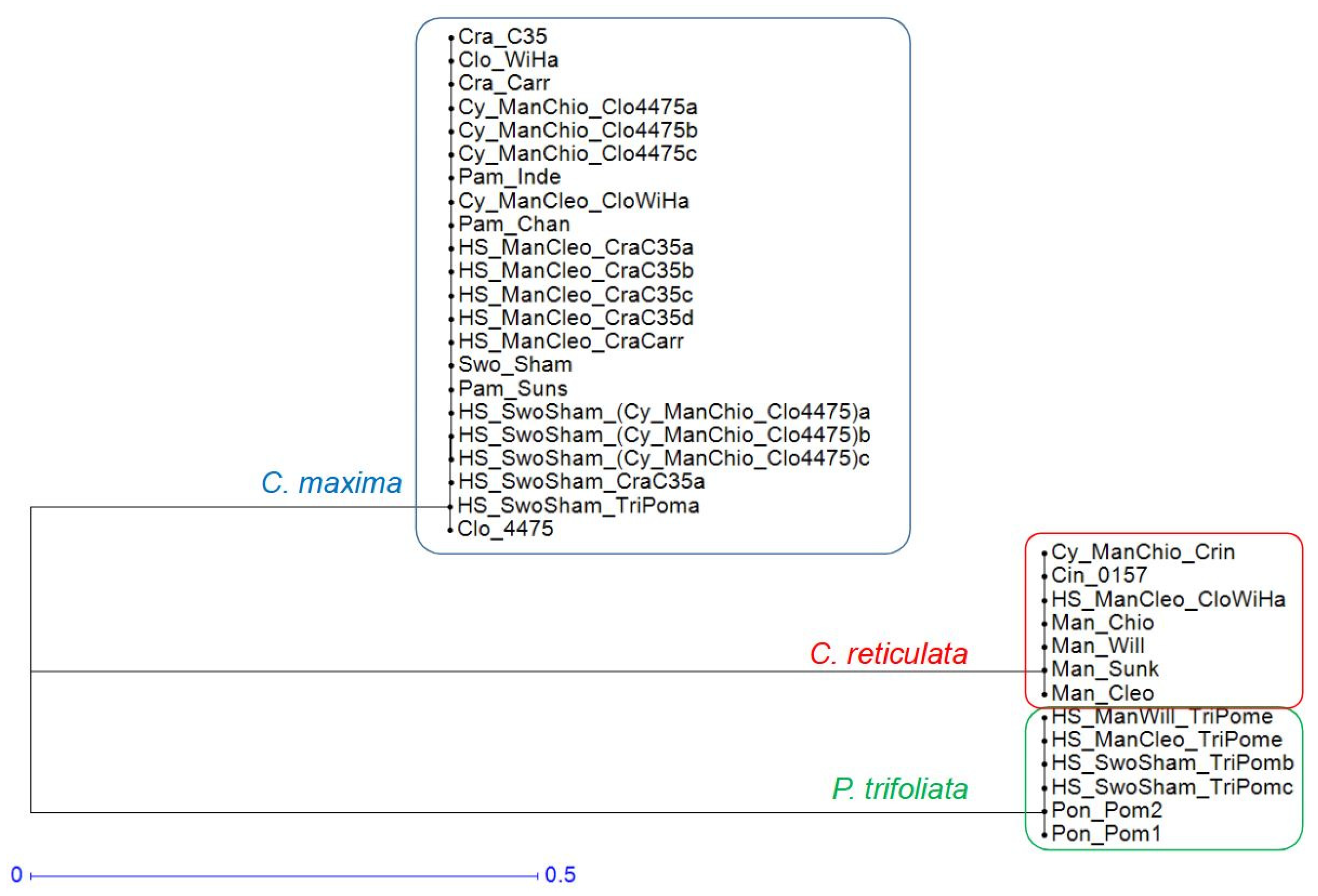


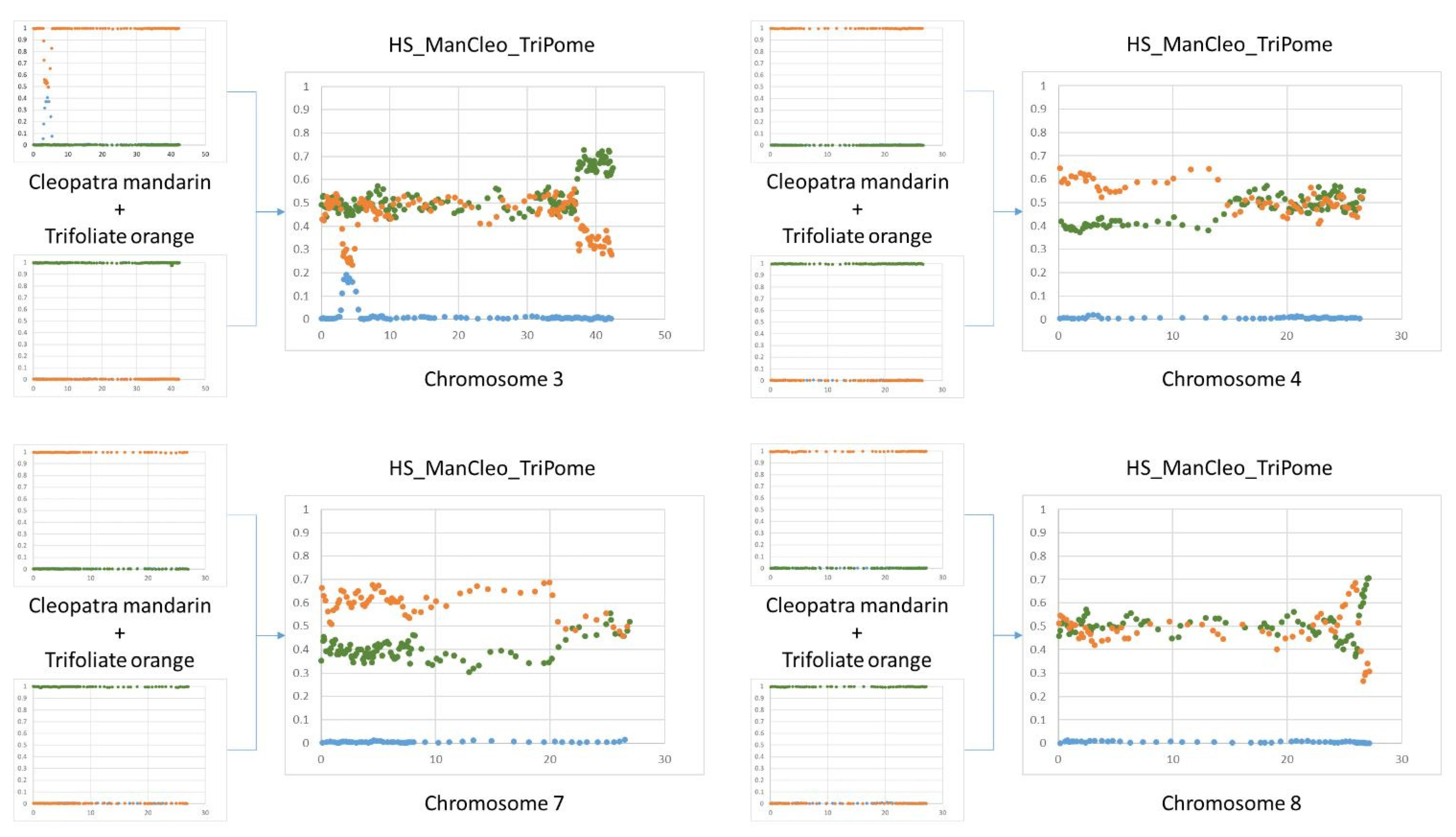
| Callus Parent | Leaf Parent | Code | Ref. | |
|---|---|---|---|---|
| Parents | Citrandarin | Cin_0157 | ICVN1100157 | |
| Winter Haven Citrumelo | Clo_WiHa | ICVN0110147 | ||
| 4475 Citrumelo | Clo_4475 | SRA1112 | ||
| C35 Citrange | Cra_C35 | SRA731 | ||
| Carrizo Citrange | Cra_Carr | SRA989 | ||
| Shamouti Sweet orange | Swo_Sham | SRA299 | ||
| Cleopatra Mandarin | Man_Cleo | SRA991 | ||
| Willow leaf Mandarin | Man_Will | SRA133 | ||
| Chios Mandarin | Man_Chio | SRA598 | ||
| Pomeroy Trifoliate orange | Pon_Pom1 | SRA1040 | ||
| Cybrids | Chios mandarin | 4475 Citrumelo | Cy_ManChio_Clo4475a | [18] |
| Chios mandarin | 4475 Citrumelo | Cy_ManChio_Clo4475b | [18] | |
| Chios mandarin | 0157 Citrandarin | Cy_ManChio_Crin | [18] | |
| Chios mandarin | 4475 Citrumelo | Cy_ManChio_Clo4475c | [18] | |
| Cleopatra mandarin | Winter Haven Citrumelo | Cy_ManCleo_CloWiHa | New | |
| Nuclear Hybrids | Cleopatra mandarin | C35 Citrange | HS_ManCleo_CraC35a | [18] |
| Cleopatra mandarin | C35 Citrange | HS_ManCleo_CraC35b | [18] | |
| Cleopatra mandarin | C35 Citrange | HS_ManCleo_CraC35c | [18] | |
| Cleopatra mandarin | Winter Haven Citrumelo | HS_ManCleo_CloWiHa | New | |
| Cleopatra mandarin | Carrizo Citrange | HS_ManCleo_CraCarb | New | |
| Shamouti Sweet orange | C35 Citrange | HS_SwoSham_CraC35a | [18] | |
| Cleopatra mandarin | C35 Citrange | HS_ManCleo_CraC35d | [18] | |
| Shamouti Sweet orange | Cybrid Chios mandarin/4475 Citrumelo | HS_SwoSham_(Cy_ManChio_Clo4475) a | New | |
| Shamouti Sweet orange | Cybrid Chios mandarin/4475 Citrumelo | HS_SwoSham_(Cy_ManChio_Clo4475) b | New | |
| Shamouti Sweet orange | Cybrid Chios mandarin/4475 Citrumelo | HS_SwoSham_(Cy_ManChio_Clo4475) c | New | |
| Shamouti Sweet orange | Pomeroy Trifoliate orange | HS_SwoSham_TriPoma | [18] | |
| Shamouti Sweet orange | Pomeroy Trifoliate orange | HS_SwoSham_TriPomb | [18] | |
| Shamouti Sweet orange | Pomeroy Trifoliate orange | HS_SwoSham_TriPomc | [18] | |
| Cleopatra mandarin | Pomeroy Trifoliate orange | HS_ManCleo_TriPome | [18] | |
| Willow leaf Mandarin | Pomeroy Trifoliate orange | HS_ManWill_TriPome | [20] | |
| Additional Ancestors | Sunki Mandarin | Man_Sunk | SRA970 | |
| Chandler Pummelo | Pam_Chan | SRA608 | ||
| Sunshine Pummelo | Pam_Suns | SRA324 | ||
| Indian Pummelo | Pam_Inde | ICVN0101133 | ||
| Pomeroy trifoliate orange | Pon_Pom2 | SRA996 | ||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | Total | |
|---|---|---|---|---|---|---|---|---|---|---|
| C. maxima | 909 | 961 | 1468 | 861 | 852 | 1057 | 824 | 752 | 732 | 8416 |
| C. reticulata | 962 | 1297 | 1919 | 1040 | 1068 | 1071 | 995 | 909 | 894 | 10,155 |
| P. trifoliata | 1849 | 2051 | 2740 | 1753 | 1587 | 1496 | 1502 | 1255 | 1341 | 15,574 |
| Code | Estimated Ploidy | Mitochondria | Chloroplast | Nucleus | |
|---|---|---|---|---|---|
| Cybrids | Cy_ManChio_Clo4475a | 2 | Chios Mand. | 4475 Citrumelo | 4475 Citrumelo |
| Cy_ManChio_Clo4475b | 2 | Chios Mand. | 4476 Citrumelo | 4476 Citrumelo | |
| Cy_ManChio_Clo4475c | 2 | Chios Mand. | 4477 Citrumelo | 4477 Citrumelo | |
| Cy_ManChio_Crin | 2 | Chios Mand. | C. reticulata type | Citrandarin | |
| Cy_ManCleo_CloWiHa | 2 | Cleopatra Mand. | Winter Haven Citru. | Winter Haven Citru. | |
| Nuclear Hybrids | HS_ManCleo_CloWiHa | 4 | Cleopatra Mand. | Cleopatra Mand. | Hybrid/Instability |
| HS_ManCleo_CraC35a | 4 | Chimeric | C35 Citrange | Hybrid/Instability | |
| HS_ManCleo_CraC35b | 4 | Chimeric | C35 Citrange | Hybrid/Instability | |
| HS_ManCleo_CraC35c | 4 | Chimeric | C35 Citrange | Hybrid/Instability | |
| HS_ManCleo_CraC35d | 4 | Chimeric | C35 Citrange | Hybrid/Instability | |
| HS_ManCleo_CraCarb | 4 | Cleopatra Mand. | Carrizo Citrange | Hybrid/Instability | |
| HS_ManCleo_TriPome | 4 | Cleopatra Mand. | Pomeroy Tr. Or. | Hybrid/Instability | |
| HS_ManWill_TriPome | 4 | Willow leaf Mand. | Pomeroy Tr. Or. | Hybrid/Instability | |
| HS_SwoSham_(Cy_ManChio_Clo4475) a | 4 | Shamouti Sw. Or. | C. maxima type | Hybrid/Instability | |
| HS_SwoSham_(Cy_ManChio_Clo4475) b | 4 | Shamouti Sw. Or. | C. maxima type | Hybrid/Instability | |
| HS_SwoSham_(Cy_ManChio_Clo4475) c | 4 | Shamouti Sw. Or. | C. maxima type | Hybrid/Instability | |
| HS_SwoSham_CraC35a | 4 | Shamouti Sw. Or. | C. maxima type | Hybrid/Instability | |
| HS_SwoSham_TriPoma | 4 | Shamouti Sw. Or. | Shamouti Sw. Or. | Hybrid/Instability | |
| HS_SwoSham_TriPomb | 4 | Shamouti Sw. Or. | Pomeroy Tr. Or. | Hybrid/Instability | |
| HS_SwoSham_TriPomc | 4 | Shamouti Sw. Or. | Pomeroy Tr. Or. | Hybrid/Instability |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dambier, D.; Barantin, P.; Boulard, G.; Costantino, G.; Mournet, P.; Perdereau, A.; Morillon, R.; Ollitrault, P. Genomic Instability in Somatic Hybridization between Poncirus and Citrus Species Aiming to Create New Rootstocks. Agriculture 2022, 12, 134. https://doi.org/10.3390/agriculture12020134
Dambier D, Barantin P, Boulard G, Costantino G, Mournet P, Perdereau A, Morillon R, Ollitrault P. Genomic Instability in Somatic Hybridization between Poncirus and Citrus Species Aiming to Create New Rootstocks. Agriculture. 2022; 12(2):134. https://doi.org/10.3390/agriculture12020134
Chicago/Turabian StyleDambier, Dominique, Pascal Barantin, Gabriel Boulard, Gilles Costantino, Pierre Mournet, Aude Perdereau, Raphaël Morillon, and Patrick Ollitrault. 2022. "Genomic Instability in Somatic Hybridization between Poncirus and Citrus Species Aiming to Create New Rootstocks" Agriculture 12, no. 2: 134. https://doi.org/10.3390/agriculture12020134
APA StyleDambier, D., Barantin, P., Boulard, G., Costantino, G., Mournet, P., Perdereau, A., Morillon, R., & Ollitrault, P. (2022). Genomic Instability in Somatic Hybridization between Poncirus and Citrus Species Aiming to Create New Rootstocks. Agriculture, 12(2), 134. https://doi.org/10.3390/agriculture12020134






