Soil Compaction Drives an Intra-Genotype Leaf Economics Spectrum in Wine Grapes
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Site and Design
2.2. Functional Trait Measurements
2.3. Analysis of Leaf Trait Variation
3. Results
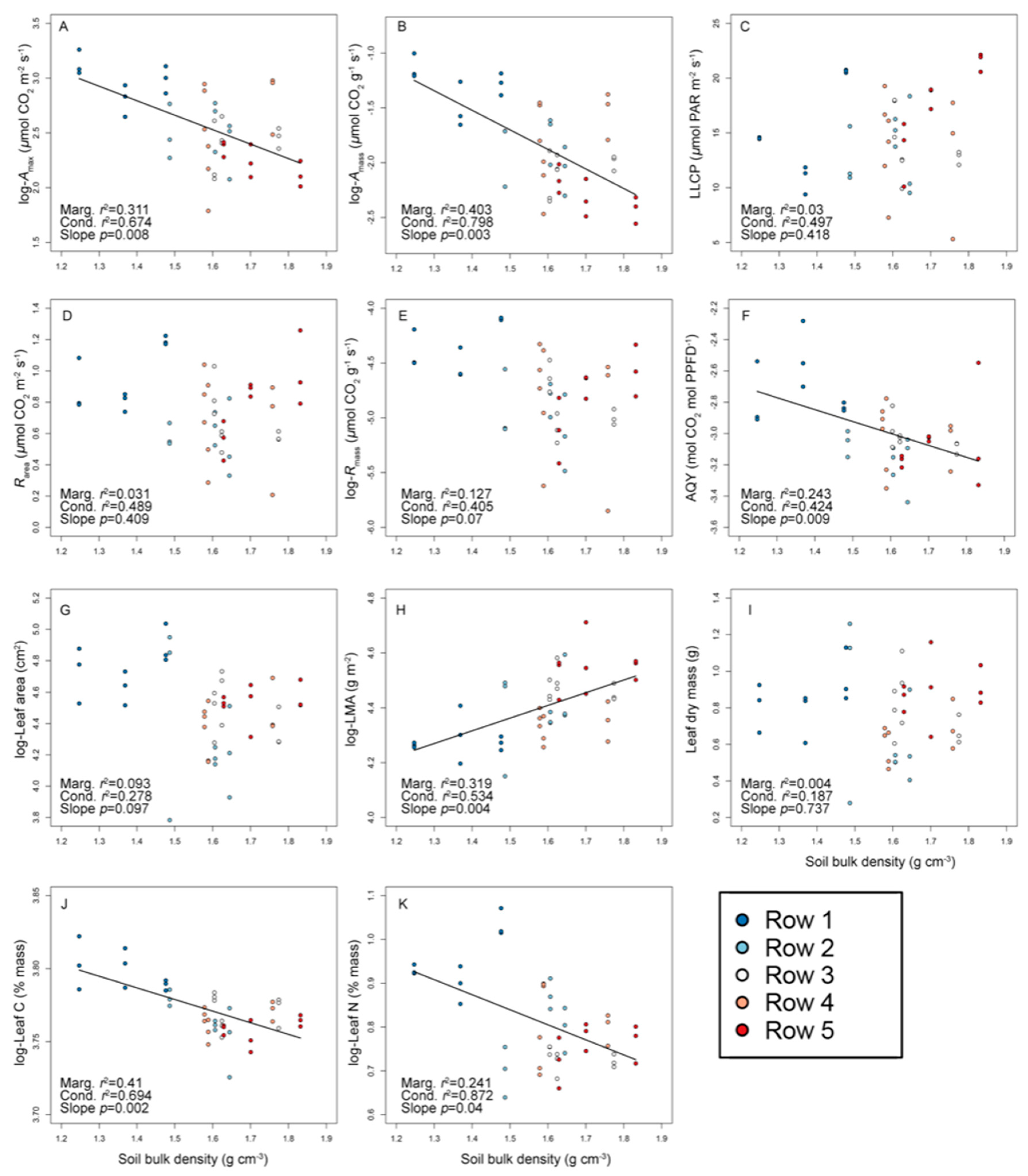
3.1. ‘Chardonnay’ Functional Trait Variation in Relation to Soil Bulk Density
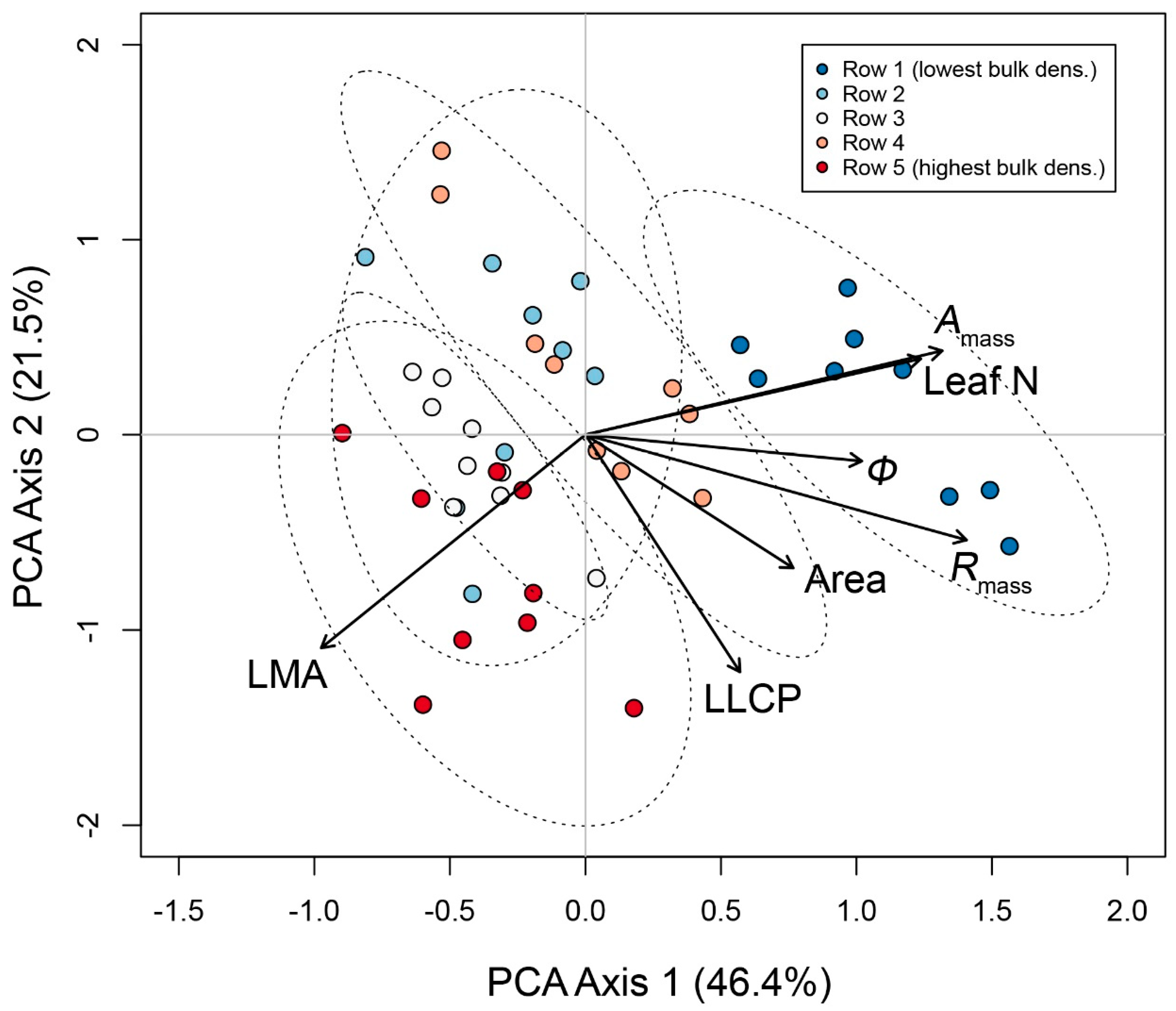
3.2. Multivariate Trait Syndromes in ‘Chardonnay’ in Relation to Soil Bulk Density
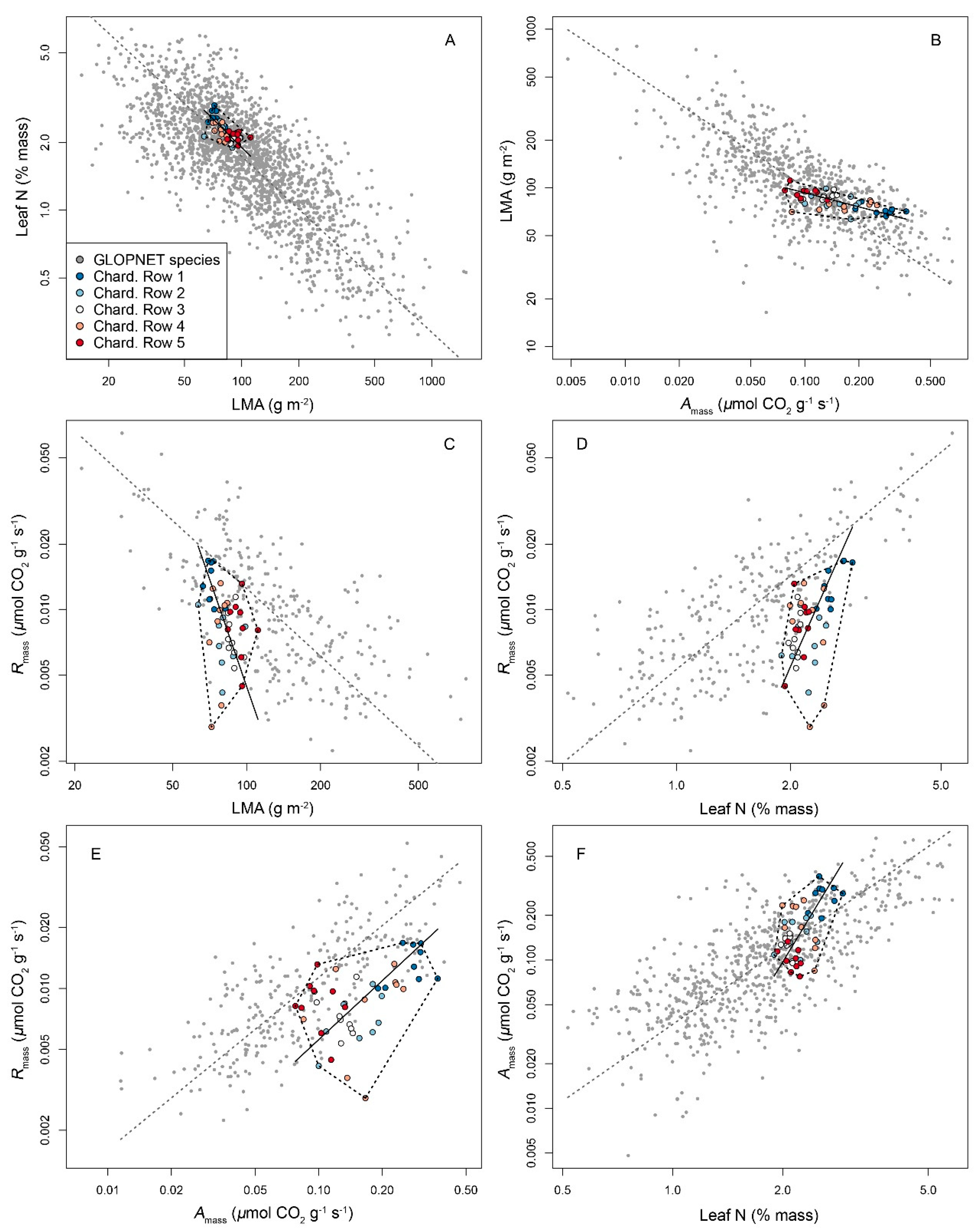
3.3. An Intragenotype LES in Chardonnay Driven by Soil Compaction
4. Discussion
4.1. Intraspecific Trait Variation and Soil Compaction
4.2. Leaf Economics Traits in Relation to Crop Domestication Syndromes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Wright, I.J.; Reich, P.B.; Westoby, M.; Ackerly, D.D.; Baruch, Z.; Bongers, F.; Cavender-Bares, J.; Chapin, T.; Cornelissen, J.H.; Diemer, M.; et al. The worldwide leaf economics spectrum. Nature 2004, 428, 821–827. [Google Scholar] [CrossRef]
- Wright, I.J.; Reich, P.B.; Cornelissen, J.H.; Falster, D.S.; Groom, P.K.; Hikosaka, K.; Lee, W.; Lusk, C.H.; Niinemets, Ü.; Oleksyn, J.; et al. Modulation of leaf economic traits and trait relationships by climate. Global Ecol. Biogeogr. 2005, 14, 411–421. [Google Scholar] [CrossRef]
- Reich, P.B.; Rich, R.L.; Lu, X.; Wang, Y.P.; Oleksyn, J. Biogeographic variation in evergreen conifer needle longevity and impacts on boreal forest carbon cycle projections. Proc. Natl. Acad. Sci. USA 2014, 111, 13703–13708. [Google Scholar] [CrossRef] [PubMed]
- Reich, P.B.; Walters, M.B.; Ellsworth, D.S. From tropics to tundra: Global convergence in plant functioning. Proc. Natl. Acad. Sci. USA 1997, 94, 13730–13734. [Google Scholar] [CrossRef] [PubMed]
- Thomas, H.J.; Bjorkman, A.D.; Myers-Smith, I.H.; Elmendorf, S.C.; Kattge, J.; Diaz, S.; Vellend, M.; Blok, D.; Cornelissen, J.H.C.; Forbes, B.C.; et al. Global plant trait relationships extend to the climatic extremes of the tundra biome. Nat. Commun. 2020, 11, 1351. [Google Scholar] [CrossRef] [PubMed]
- Reich, P.B.; Ellsworth, D.S.; Walters, M.B.; Vose, J.M.; Gresham, C.; Volin, J.C.; Bowman, W.D. Generality of leaf trait relationships: A test across six biomes. Ecology 1999, 80, 1955–1969. [Google Scholar] [CrossRef]
- Shipley, B.; Lechowicz, M.J.; Wright, I.; Reich, P.B. Fundamental trade-offs generating the worldwide leaf economics spectrum. Ecology 2006, 87, 535–541. [Google Scholar] [CrossRef]
- Donovan, L.A.; Maherali, H.; Caruso, C.M.; Huber, H.; de Kroon, H. The evolution of the worldwide leaf economics spectrum. Trends Ecol. Evol. 2011, 26, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Reich, P.B.; Wright, I.J.; Cavender-Bares, J.; Craine, J.M.; Oleksyn, J.; Westoby, M.; Walters, M.B. The evolution of plant functional variation: Traits, spectra, and strategies. Int. J. Plant Sci. 2003, 164, S143–S164. [Google Scholar] [CrossRef]
- Albert, C.H.; Thuiller, W.; Yoccoz, N.G.; Soudant, A.; Boucher, F.; Saccone, P.; Lavorel, S. Intraspecific functional variability: Extent, structure and sources of variation. J. Ecol. 2010, 98, 604–613. [Google Scholar] [CrossRef]
- Siefert, A.; Violle, C.; Chalmandrier, L.; Albert, C.H.; Taudiere, A.; Fajardo, A.; Aarssen, L.W.; Baraloto, C.; Carlucci, M.B.; Cianciaruso, M.V.; et al. A global meta-analysis of the relative extent of intraspecific trait variation in plant communities. Ecol. Lett. 2015, 18, 1406–1419. [Google Scholar] [CrossRef]
- Fajardo, A.; Siefert, A. Intraspecific trait variation and the leaf economics spectrum across resource gradients and levels of organization. Ecology 2018, 99, 1024–1030. [Google Scholar] [CrossRef] [PubMed]
- Hayes, F.J.; Buchanan, S.W.; Coleman, B.; Gordon, A.M.; Reich, P.B.; Thevathasan, N.V.; Wright, I.J.; Martin, A.R. Intraspecific variation in soy across the leaf economics spectrum. Ann. Bot. 2019, 123, 107–120. [Google Scholar] [CrossRef]
- Martin, A.R.; Rapidel, B.; Roupsard, O.; Van den Meersche, K.; de Melo Virginio Filho, E.; Barrios, M.; Isaac, M.E. Intraspecific trait variation across multiple scales: The leaf economics spectrum in coffee. Funct. Ecol. 2017, 31, 604–612. [Google Scholar] [CrossRef]
- Niinemets, Ü. Is there a species spectrum within the world-wide leaf economics spectrum? Major variations in leaf functional traits in the Mediterranean sclerophyll Quercus ilex. New Phytol. 2015, 205, 79–96. [Google Scholar] [CrossRef] [PubMed]
- Siefert, A.; Ritchie, M.E. Intraspecific trait variation drives functional responses of old-field plant communities to nutrient enrichment. Oecologia 2016, 181, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Laforest-Lapointe, I.; Martínez-Vilalta, J.; Retana, J. Intraspecific variability in functional traits matters: Case study of Scots pine. Oecologia 2014, 175, 1337–1348. [Google Scholar] [CrossRef]
- Mitchell, R.M.; Ames, G.M.; Wright, J.P. Intraspecific trait variability shapes leaf trait response to altered fire regimes. Ann. Bot. 2021, 127, 543–552. [Google Scholar] [CrossRef]
- Westerband, A.C.; Funk, J.L.; Barton, K.E. Intraspecific trait variation in plants: A renewed focus on its role in ecological processes. Ann. Bot. 2021, 127, 397–410. [Google Scholar] [CrossRef]
- Xiong, D.; Flexas, J. Leaf economics spectrum in rice: Leaf anatomical, biochemical, and physiological trait trade-offs. J. Exp. Bot. 2018, 69, 5599–5609. [Google Scholar] [CrossRef]
- Gagliardi, S.; Martin, A.R.; Virginio Filho, E.D.M.; Rapidel, B.; Isaac, M.E. Intraspecific leaf economic trait variation partially explains coffee performance across agroforestry management regimes. Agric. Ecosyst. Environ. 2015, 200, 151–160. [Google Scholar] [CrossRef]
- Roucou, A.; Violle, C.; Fort, F.; Roumet, P.; Ecarnot, M.; Vile, D. Shifts in plant functional strategies over the course of wheat domestication. J. Appl. Ecol. 2018, 55, 25–37. [Google Scholar] [CrossRef]
- Martin, A.R.; Hale, C.E.; Cerabolini, B.E.; Cornelissen, J.H.; Craine, J.; Gough, W.A.; Kattge, J.; Tirona, C.K. Inter-and intraspecific variation in leaf economic traits in wheat and maize. AoB Plants 2018, 10, ply006. [Google Scholar] [CrossRef]
- Coleman, B.R.; Martin, A.R.; Thevathasan, N.V.; Gordon, A.M.; Isaac, M.E. Leaf trait variation and decomposition in short-rotation woody biomass crops under agroforestry management. Agric. Ecosyst. Environ. 2020, 298, 106971. [Google Scholar] [CrossRef]
- Martin, A.R.; Hayes, F.J.; Borden, K.A.; Buchanan, S.W.; Gordon, A.M.; Isaac, M.E.; Thevathasan, N.V. Integrating nitrogen fixing structures into above-and belowground functional trait spectra in soy (Glycine max). Plant Soil 2019, 440, 53–69. [Google Scholar] [CrossRef]
- Fulthorpe, R.R.; Martin, A.R.; Isaac, M.E. Root endophytes of coffee (Coffea arabica): Variation across climatic gradients and relationships with functional traits. Phytobiomes J. 2019, 4, 27–39. [Google Scholar] [CrossRef]
- Buchanan, S.; Isaac, M.E.; Van den Meersche, K.; Martin, A.R. Functional traits of coffee along a shade and fertility gradient in coffee agroforestry systems. Agrofor. Syst. 2019, 93, 1261–1273. [Google Scholar] [CrossRef]
- Martin, A.R.; Isaac, M.E. The leaf economics spectrum’s morning coffee: Plant size-dependent changes in leaf traits and reproductive onset in a perennial tree crop. Ann. Bot. 2021, 127, 483–493. [Google Scholar] [CrossRef]
- Hamza, M.A.; Anderson, W.K. Soil compaction in cropping systems: A review of the nature, causes and possible solutions. Soil Tillage Res. 2005, 82, 121–145. [Google Scholar] [CrossRef]
- Nawaz, M.F.; Bourrie, G.; Trolard, F. Soil compaction impact and modelling. A review. Agron. Sustain. Dev. 2013, 33, 291–309. [Google Scholar] [CrossRef]
- Colombi, T.; Keller, T. Developing strategies to recover crop productivity after soil compaction—A plant eco-physiological perspective. Soil Tillage Res. 2019, 191, 156–161. [Google Scholar] [CrossRef]
- Morales, F.; Pavlovič, A.; Abadía, A.; Abadía, J. Photosynthesis in poor nutrient soils, in compacted soils, and under drought. In The Leaf: A Platform for Performing Photosynthesis; Adams, W., III, Terashima, I., Eds.; Springer: Cham, Switzerland, 2018; pp. 371–399. [Google Scholar]
- Sadras, V.O.; O’Leary, G.J.; Roget, D.K. Crop responses to compacted soil: Capture and efficiency in the use of water and radiation. Field Crops Res. 2005, 91, 131–148. [Google Scholar] [CrossRef]
- Lipiec, J.; Stępniewski, W. Effects of soil compaction and tillage systems on uptake and losses of nutrients. Soil Tillage Res. 1995, 35, 37–52. [Google Scholar] [CrossRef]
- Kozlowski, T.T. Soil compaction and growth of woody plants. Scand. J. For. Res. 1999, 14, 596–619. [Google Scholar] [CrossRef]
- Milla, R.; Osborne, C.P.; Turcotte, M.M.; Violle, C. Plant domestication through an ecological lens. Trends Ecol. Evol. 2015, 30, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Wright, I.J.; Reich, P.B.; Atkin, O.K.; Lusk, C.H.; Tjoelker, M.G.; Westoby, M. Irradiance, temperature and rainfall influence leaf dark respiration in woody plants: Evidence from comparisons across 20 sites. New Phytol. 2006, 169, 309–319. [Google Scholar] [CrossRef]
- Reich, P.B.; Walters, M.B.; Ellsworth, D.S.; Vose, J.M.; Volin, J.C.; Gresham, C.; Bowman, W.D. Relationships of leaf dark respiration to leaf nitrogen, specific leaf area and leaf life-span: A test across biomes and functional groups. Oecologia 1998, 114, 471–482. [Google Scholar] [CrossRef]
- Nunes-Nesi, A.; Carrari, F.; Lytovchenko, A.; Smith, A.M.; Loureiro, M.E.; Ratcliffe, R.G.; Sweetlove, L.J.; Fernie, A.R. Enhanced photosynthetic performance and growth as a consequence of decreasing mitochondrial malate dehydrogenase activity in transgenic tomato plants. Plant Physiol. 2005, 137, 611–622. [Google Scholar] [CrossRef]
- Hauben, M.; Haesendonckx, B.; Standaert, E.; Van Der Kelen, K.; Azmi, A.; Akpo, H.; Van Breusegem, F.; Guisez, Y.; Bots, M.; Lambert, B.; et al. Energy use efficiency is characterized by an epigenetic component that can be directed through artificial selection to increase yield. Proc. Natl. Acad. Sci. USA 2009, 106, 20109–20114. [Google Scholar] [CrossRef]
- Juszczuk, I.M.; Flexas, J.; Szal, B.; Dąbrowska, Z.; Ribas-Carbo, M.; Rychter, A.M. Effect of mitochondrial genome rearrangement on respiratory activity, photosynthesis, photorespiration and energy status of MSC16 cucumber (Cucumis sativus) mutant. Physiol. Plant. 2007, 131, 527–541. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.; Jones, J.G. Effect of selection for dark respiration rate of mature leaves on crop yields of Lolium perenne cv. S23. Ann. Bot. 1982, 49, 313–320. [Google Scholar] [CrossRef]
- Amthor, J.S. Respiration and Crop Productivity; Springer Science & Business Media: New York, NY, USA, 2012. [Google Scholar]
- Ramankutty, N.; Evan, A.T.; Monfreda, C.; Foley, J.A. Farming the planet: 1. Geographic distribution of global agricultural lands in the year 2000. Glob. Biogeochem. Cycles 2008, 22, GB1003. [Google Scholar] [CrossRef]
- Atkin, O.K.; Bloomfield, K.J.; Reich, P.B.; Tjoelker, M.G.; Asner, G.P.; Bonal, D.; Bonisch, G.; Bradford, M.G.; Cernusak, L.A.; Cosio, E.G.; et al. Global variability in leaf respiration in relation to climate, plant functional types and leaf traits. New Phytol. 2015, 206, 614–636. [Google Scholar] [CrossRef]
- Aryal, N.R.; Anderson, K. Which Winegrape Varieties Are Grown Where? A Global Empirical Picture; University of Adelaide Press: Adelaide, Australia, 2013; p. 700. [Google Scholar]
- Coombe, B.G. Growth stages of the grapevine: Adoption of a system for identifying grapevine growth stages. Aust. J. Grape Wine Res. 1995, 1, 104–110. [Google Scholar] [CrossRef]
- Berry, Z.C.; Goldsmith, G.R. Diffuse light and wetting differentially affect tropical tree leaf photosynthesis. New Phytol. 2020, 225, 143–153. [Google Scholar] [CrossRef]
- Salter, W.T.; Merchant, A.M.; Richards, R.A.; Trethowan, R.; Buckley, T.N. Rate of photosynthetic induction in fluctuating light varies widely among genotypes of wheat. J. Exp. Bot. 2019, 70, 2787–2796. [Google Scholar] [CrossRef] [PubMed]
- Delignette-Muller, M.L.; Dutang, C. fitdistrplus: An R package for fitting distributions. J. Stat. Softw. 2015, 64, 1–34. [Google Scholar] [CrossRef]
- Pinheiro, J.; Bates, D.; DebRoy, S.; Sarkar, D.; Team, R.C. nlme: Linear and Nonlinear Mixed Effects Models. R Package Version 3.1-131. 2017. Available online: https://CRAN.R-project.org/package=nlme (accessed on 10 January 2021).
- Paradis, E.; Claude, J.; Strimmer, K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 2004, 20, 289–290. [Google Scholar] [CrossRef]
- Le, S.; Josse, J.; Husson, F. FactoMineR: An R Package for Multivariate Analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Oksanen, J.; Guillaume Blanchet, F.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R Package Version 2.4-5. 2017. Available online: CRAN.R-project.org/package=vegan (accessed on 10 January 2021).
- Warton, D.I.; Duursma, R.A.; Falster, D.S.; Taskinen, S. smatr 3—An R package for estimation and inference about allometric lines. Methods Ecol. Evol. 2012, 3, 257–259. [Google Scholar] [CrossRef]
- Keller, M. The Science of Grapevines; Academic Press: Cambridge, MA, USA, 2020. [Google Scholar]
- Gambetta, G.A.; Herrera, J.C.; Dayer, S.; Feng, Q.; Hochberg, U.; Castellarin, S.D. The physiology of drought stress in grapevine: Towards an integrative definition of drought tolerance. J. Exp. Bot. 2020, 71, 4658–4676. [Google Scholar] [CrossRef] [PubMed]
- Isaac, M.E.; Martin, A.R.; de Melo Virginio Filho, E.; Rapidel, B.; Roupsard, O.; van den Meersche, K. Intraspecific trait variation and coordination: Root and leaf economics spectra in coffee across environmental gradients. Front. Plant Sci. 2017, 8, 1196. [Google Scholar] [CrossRef] [PubMed]
- Milla, R.; Morente-López, J.; Alonso-Rodrigo, J.M.; Martín-Robles, N.; Stuart Chapin, F., III. Shifts and disruptions in resource-use trait syndromes during the evolution of herbaceous crops. Proc. R. Soc. B Biol. Sci. 2014, 281, 20141429. [Google Scholar] [CrossRef] [PubMed]
- Morales-Castilla, I.; de Cortázar-Atauri, I.G.; Cook, B.I.; Lacombe, T.; Parker, A.; van Leeuwen, C.; Nicholas, K.A.; Wolkovich, E.M. Diversity buffers winegrowing regions from climate change losses. Proc. Natl. Acad. Sci. USA 2020, 117, 2864–2869. [Google Scholar] [CrossRef] [PubMed]
- Greer, D.H. Temperature and CO2 dependency of the photosynthetic photon flux density responses of leaves of Vitis vinifera cvs. Chardonnay and Merlot grown in a hot climate. Plant Physiol. Biochem. 2017, 111, 295–303. [Google Scholar] [CrossRef] [PubMed]
| Trait Information | Distribution Fitting | Descriptives | Variance Partitioning | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Trait Group | Trait | Normal | Log-Norm. | Mean/Median ±SD | Range | CV | Row | Plant | Unexplained |
| Physiological | Amax (μmol CO2 m−2 s−1) | −133.4 | −129.8 | 12.6 ± 4.7 | 6.0–26.1 | 35.4 | 0.454 | 0.235 | 0.311 |
| Amass (μmol CO2 g−1 s−1) | 54.8 | 60.9 | 0.154 ± 0.072 | 0.078–0.367 | 43.2 | 0.648 | 0.166 | 0.186 | |
| Rarea (μmol CO2 m−2 s−1) | 0.002 | −3.1 | 0.74 ± 0.24 | 0.21–1.26 | 33.0 | 0.193 | 0.288 | 0.519 | |
| Rmass (μmol CO2 g−1 s−1) | 192.7 | 193.0 | 0.009 ± 0.003 | 0.003–0.017 | 37.1 | 0.260 | 0.150 | 0.589 | |
| LLCP (μmol PAR m−2 s−1) | −125.6 | −128.6 | 14.2 ± 4.0 | 5.3–22.2 | 27.0 | <0.001 | 0.474 | 0.526 | |
| Φ (mol CO2 mol PPFD−1) | 131.6 | 138.1 | 0.05 ± 0.013 | 0.03–0.1 | 25.6 | 0.429 | 0.022 | 0.549 | |
| Morphological | Area (cm2) | −205.3 | −205.2 | 89.6 ± 23.5 | 44.0–153.9 | 25.3 | 0.303 | <0.001 | 0.697 |
| LMA (g m−2) | −167.3 | −166.5 | 82.0 ± 10.1 | 63.4–111.2 | 12.2 | 0.629 | <0.001 | 0.371 | |
| Dry mass (g) | 5.8 | 4.2 | 0.77 ± 0.24 | 0.28–1.26 | 28.1 | 0.154 | 0.024 | 0.822 | |
| Chemical | Carbon (% mass) | −53.9 | −52.9 | 43.4 ± 0.8 | 41.5–45.7 | 1.8 | 0.555 | 0.161 | 0.285 |
| Nitrogen (% mass) | 1.8 | 3.6 | 2.24 ± 0.24 | 1.9–2.92 | 10.4 | 0.543 | 0.335 | 0.123 | |
| Traits | Chardonnay | GLOPNET | Slope Test | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Independ. | Depend. | Intercept | Slope | Model r2 | Model p | Intercept | Slope | Model r2 | Model p | n | p Value | r Value |
| Amass | LMA | 105.91 | −139.06 | 0.392 | <0.001 | 235.87 | −868.13 | 0.237 | <0.001 | 746 | <0.001 | −0.969 |
| Leaf N | Amass | −0.53 | 0.31 | 0.35 | <0.001 | −0.09 | 0.12 | 0.55 | <0.001 | 712 | <0.001 | 0.828 |
| LMA | Leaf N | 4.18 | −0.03 | 0.397 | <0.001 | 3.01 | −0.01 | 0.312 | <0.001 | 1958 | <0.001 | 0.860 |
| Amass | Rmass | 0.01 | 0.05 | 0.355 | <0.001 | 0.01 | 0.11 | 0.489 | <0.001 | 259 | <0.001 | −0.727 |
| LMA | Rmass | 0.04 | −0.01 | 0.146 | 0.01 | 0.03 | −0.01 | 0.216 | <0.001 | 274 | <0.001 | 0.911 |
| Leaf N | Rmass | −0.03 | 0.02 | 0.333 | <0.001 | −0.01 | 0.02 | 0.632 | <0.001 | 265 | 0.026 | 0.332 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martin, A.R.; Mariani, R.O.; Cathline, K.A.; Duncan, M.; Paroshy, N.J.; Robertson, G. Soil Compaction Drives an Intra-Genotype Leaf Economics Spectrum in Wine Grapes. Agriculture 2022, 12, 1675. https://doi.org/10.3390/agriculture12101675
Martin AR, Mariani RO, Cathline KA, Duncan M, Paroshy NJ, Robertson G. Soil Compaction Drives an Intra-Genotype Leaf Economics Spectrum in Wine Grapes. Agriculture. 2022; 12(10):1675. https://doi.org/10.3390/agriculture12101675
Chicago/Turabian StyleMartin, Adam R., Rachel O. Mariani, Kimberley A. Cathline, Michael Duncan, Nicholas J. Paroshy, and Gavin Robertson. 2022. "Soil Compaction Drives an Intra-Genotype Leaf Economics Spectrum in Wine Grapes" Agriculture 12, no. 10: 1675. https://doi.org/10.3390/agriculture12101675
APA StyleMartin, A. R., Mariani, R. O., Cathline, K. A., Duncan, M., Paroshy, N. J., & Robertson, G. (2022). Soil Compaction Drives an Intra-Genotype Leaf Economics Spectrum in Wine Grapes. Agriculture, 12(10), 1675. https://doi.org/10.3390/agriculture12101675







